ABSTRACT
Eucheuma denticulatum, an economically and industrially important red alga, is a valuable marine resource. Although microRNAs (miRNAs) play an essential role in gene post-transcriptional regulation, no research has been conducted to identify and characterize miRNAs in E. denticulatum. In this study, we identified 134 miRNAs (133 conserved miRNAs and one novel miRNA) from 2,997,135 small-RNA reads by high-throughput sequencing combined with bioinformatics analysis. BLAST searching against miRBase uncovered 126 potential miRNA families. A conservation and diversity analysis of predicted miRNA families in different plant species was performed by comparative alignment and homology searching. A total of 4 and 13 randomly selected miRNAs were respectively validated by northern blotting and stem-loop reverse transcription PCR, thereby demonstrating the reliability of the miRNA sequencing data. Altogether, 871 potential target genes were predicted using psRobot and TargetFinder. Target genes classification and enrichment were conducted based on Gene Ontology analysis. The functions of target gene products and associated metabolic pathways were predicted by Kyoto Encyclopedia of Genes and Genomes pathway analysis. A Cytoscape network was constructed to explore the interrelationships of miRNAs, miRNA-target genes and target genes. A large number of miRNAs with diverse target genes will play important roles for further understanding some essential biological processes in E. denticulatum. The uncovered information can serve as an important reference for the protection and utilization of this unique red alga in the future.
KEYWORDS: Bioinformatics, characterization, eucheuma denticulatum, high-throughput sequencing, identification, microRNA
Abbreviations
- miRNA
microRNA
- RT-PCR
reverse transcription polymerase chain reaction
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- HiSeq
high-throughput sequencing
- MFE
minimum free energy of folding
- BP
biological process
- CC
cellular component
- MF
molecular function
Introduction
MicroRNAs (miRNAs) are endogenous, small non-coding RNAs approximately 21 nt long that play pivotal roles in post-transcriptional processing via mRNA cleavage or translational repression.1,2 Their pri-miRNA precursors, which have a characteristic stem-loop structure, are recognized and cleaved by a Dicer-containing protein complex to yield pre-miRNAs.3 In plants, a pre-miRNA is then cleaved by Dicer-like 1 protein to generate an approximately 23-nt miRNA/miRNA* duplex.4 The mature miRNA on the 5′ end of the duplex is generally incorporated into the RNA-induced silencing complex (RISC), 5 while the miRNA* on the 3′ end is usually degraded following miRNA-RISC target recognition.6 Finally, translational repression or mRNA degradation is induced.7
Since their initial discovery in Caenorhabditis elegans,8 many miRNAs have been identified in model organisms and found to play crucial roles in various biological processes.9 With the rapid development of next-generation sequencing technology10 and new bioinformatics software,11 an increasing number of miRNAs have been detected in crop plant species such as rice, tobacco, maize, potato and wheat.4-7,12 Much evidence is accruing that miRNAs play an essential role in processes such as tissue development, signal transduction and environmental stress response in model organisms and higher plants.12-17 In non-model algae, however, studies on unique small RNAs have been limited.18
Eucheuma denticulatum or eucheuma, frequently dubbed a “marine crop,” is one of the main economic marine algae typically growing on reefs in tropical areas.19 As the main raw material of carrageenan,20 eucheuma has been extensively used in the food industry.21 In addition to carrageenan, eucheuma is rich in dietary fiber, a substance that according to some reported studies can reduce blood lipids.22,23 Notably, some research has demonstrated that seleno-polysaccharides of eucheuma can inhibit tumor cells and virus growth,24-27 possibly by blocking cell growth and inducing cell apoptosis in the former case and by preventing viral adsorption normal cells in the latter.28 Despite its importance, eucheuma germplasm resources are undergoing severe degradation as a result of frequent introduction and over-reproduction, environmental pollution, disease interference, competition from epiphytic algae and herbivore animal invasion.29-32 The protection and development of eucheuma resources is an urgent priority in the main countries of production, such as South Africa, Australia, Japan, the Philippines, Indonesia and China.33,34 Identifying as many miRNAs as possible in this red alga is very important for this purpose.
Thus far, most studies on eucheuma have focused on its growth and cultivation,35,36 photosynthesis and respiration,37 processing technology,38 bioactive components,39 genetic variation,40 and cloning and expression of key enzyme genes.41 Although over 35,000 miRNAs from 223 species have been submitted to the miRNA database (miRBase 21.0, June 2014, http://www.mirbase.org/),42 no miRNAs have yet been found in E. denticulatum. Because the E. denticulatum genome has not been sequenced, the lack of extensive sequence information in the database impedes miRNA identification using bioinformatics technology. However, new high-throughput sequencing (HiSeq) combined with bioinformatics43,44 may be an effective strategy for identification of as many miRNAs as possible in this red alga. Their identification would be extremely important to comprehensively understand the roles of these non-coding RNAs in various biological regulatory processes and metabolic pathways.45 Capture of useful information from miRNA data, prediction of target genes, analysis of related biological processes and metabolic pathways will be very meaningful to further protect and develop this valuable marine crop in the future.46,47
Results and discussion
HiSeq of small RNAs of E. denticulatum
As shown in the overview in Fig. 1, a high-quality small RNA population (its concentration was about 42 ng/L) isolated from total RNA of E. denticulatum was subjected to HiSeq, thereby generating 11,444,177 clean reads (Table 1). The most frequent small-RNA length was 19 nt (Fig. 2). The numbers and proportions of unique and total small RNAs mapped to expressed sequence tags (ESTs) can be seen in Table 2. After removing RNAs such as rRNAs, tRNAs, snoRNAs and snRNAs identified by BLASTn searching, the remaining sequences were annotated as miRNAs and classified in this study as either known or conserved. Among the various small RNA categories shown in Fig. S1A and S1B, 267,018 reads (7,919 unique reads) were annotated as conserved miRNAs. Another 11,073,638 reads (267,018 unique reads) could not be annotated; these novel miRNAs were further analyzed.
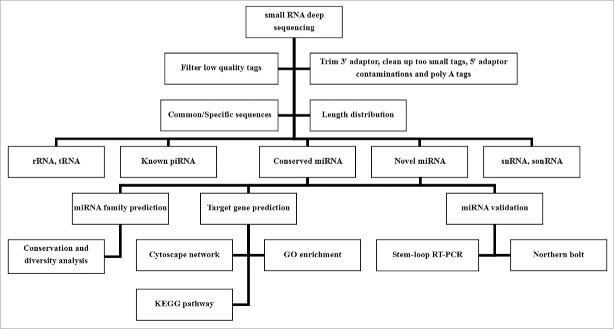
Figure 1.
Overview of experiments and bioinformatics analyses applied to Eucheuma denticulatum.
Table 1.
Summary of reads generated from sequencing of Eucheuma denticulatum small-RNA libraries.
| Type | Reads | Percent (%) |
|---|---|---|
| Total reads | 11556963 | |
| High quality | 11536642 | 100% |
| 3′adapter null | 1392 | 0.01% |
| Insert null | 2306 | 0.02% |
| 5′adapter contaminants | 26879 | 0.23% |
| Smaller than 18nt | 61785 | 0.54% |
| PolyA | 103 | 0.00% |
| Clean reads | 11444177 | 99.20% |
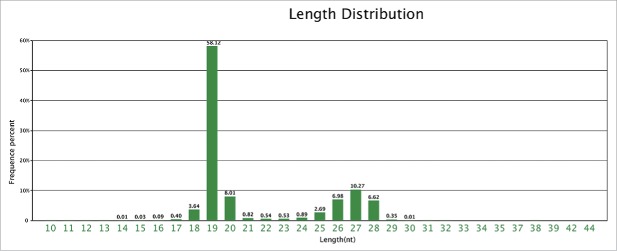
Figure 2.
Length distribution of small RNAs in Eucheuma denticulatum. As shown by the indicated frequency percentages, 19-nt-long reads were the most abundant.
Table 2.
Summary of small RNAs mapped to transcripts and ESTs of Eucheuma denticulatum.
| Unique reads | Percent (%) | Total reads | Percent (%) | |
|---|---|---|---|---|
| Total small RNAs | 2997135 | 100% | 11444177 | 100% |
| Mapped to transcripts and ESTs | 66052 | 2.2% | 1477411 | 12.91% |
Identification of novel miRNAs in E. denticulatum
To predict novel miRNAs in E. denticulatum, we aligned the unannotated sequences to ESTs of this red alga. After screening the candidates based on stringent identification criteria described in Materials and Methods, only one novel miRNA (temporarily named as ede-miR1) could be identified. The secondary structure of this novel miRNA precursor can be seen in Fig. 3. In this study, we therefore identified a total of 134 miRNAs (133 conserved and 1 novel one) in E. denticulatum (Table S1). The length distribution of identified miRNAs ranged from 18 to 27 nt, with 19 nt the most frequent length (Fig. S2A). These sizes are typical of plant miRNAs. According to the distribution of nucleotides in different positions of the detected miRNAs, the least common nucleotide was C (Fig. S2B), and U was the most biased first-position nucleotide (Fig. S2C), which is consistent with the basic characteristics of most identified miRNA sequences.48
Figure 3.
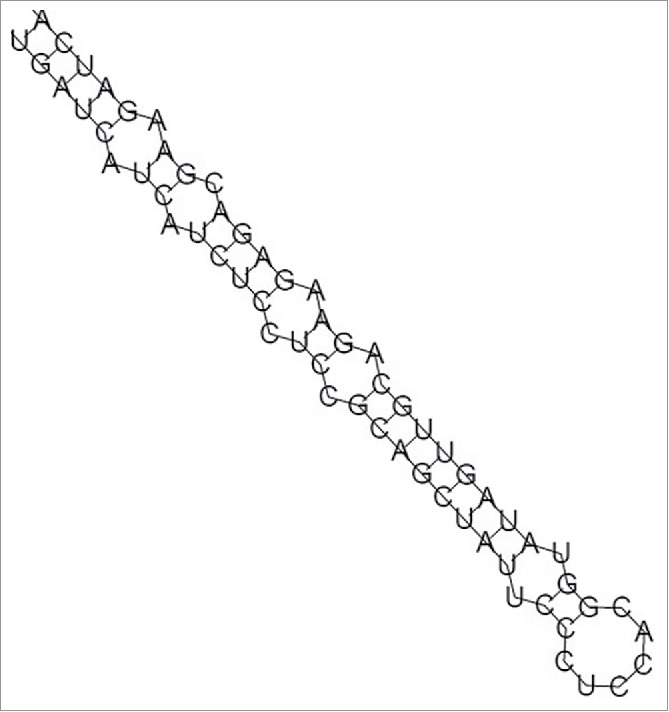
Secondary structure of predicted pre-miRNAs in Eucheuma denticulatum. The stem-loop structure of the novel miRNA precursor ede-miR1 in E. denticulatum is indicated.
Conservation and diversity of miRNA families
Potential miRNA families can be predicted on the basis of sequence homology between predicted and known miRNAs stored in miRBase. Altogether, 126 miRNA families with 133 members were predicted in this study (Table S2). As shown in Table 3, most families had only one representative in E. denticulatum, with miR164, miR166, miR169, miR319, miR397, miR398 and miR5562 having 2 members. To investigate the conservation and diversity of these predicted miRNA families across plants, we additionally performed a BLAST search of 21 randomly selected new predicted miRNA families in E. denticulatum against miRNA families in other plant species (Table S3). As shown in Fig. 4, the 21 families were found in 11 different plant species. Among them, 8 and 6 families had homologs in Oryza sativa and Arabidopsis thaliana, respectively. Moreover, miR6463 homologs were found in Populus trichocarpa, A. thaliana, Medicago truncatula, Glycine max and Brachypodium distachyon, indicating that these RNA sequences may originate from a common sequence in an ancestral species. Finally, these identified miRNA families were found in both bryophytes and angiosperms, revealing the diversity of these miRNA families in different plant species. Whether there is a potential relationship among conservation and diversity of miRNA families, small RNAs evolution as well as species evolution remains to be explored in the future.
Table 3.
Sizes of miRNA families identified in Eucheuma denticulatum.
| Size of family member | Number of miRNA family | Percent of miRNA family (%) |
|---|---|---|
| 1 | 126 | 94.73% |
| 2 | 7 | 5.27% |
Figure 4.
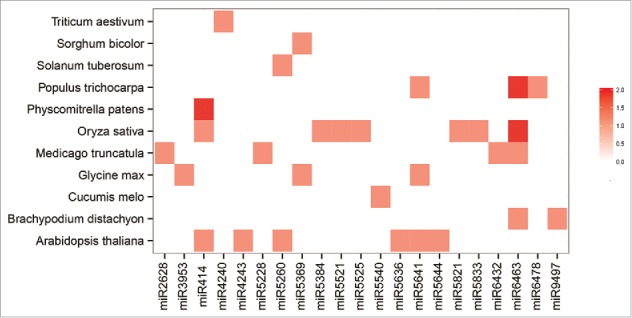
Distribution of predicted conserved miRNA families in different plant species. Data for mature miRNAs in different plant species are from miRBase21.0. Color coding is used to indicate the number of miRNA members in each family, with dark red corresponding to the highest number and white the lowest.
Validation of miRNAs in E. denticulatum
To validate the credibility of identified miRNAs, 4 miRNAs were first randomly selected for RNA gel blot detection49 using the digoxigenin-labeled oligonucleotide probes listed in Table S4. Second, 13 miRNAs (12 conserved and one novel) were randomly chosen and subjected to stem-loop reverse transcription (RT)-PCR validation50 using the primer sequences given in Table S5. As shown in Fig. 5A, 3 of the 4 miRNAs could be detected by probe hybridization, while all 13 miRNAs were detectable by stem-loop RT-PCR (Fig. 5B). The results of these 2 validation approaches demonstrate that the majority of miRNAs identified in this study are credible. As is well known, some miRNAs are expressed at low levels.51 In addition, some genes are expressed in different tissues or at different stages.52 This spatial and temporal variation in miRNAs expression may be the main factor hindering miRNA validation, such as the negative result seen for miR7734-5p and the low expression of miR2667a observed in our northern blot analysis (Fig. 5A). In spite of this fact, the 2 validation rates in this study (75% and 100%) are still high based on the principle of random sampling, which indicates that our applied methods were effective. Furthermore, the spatial and temporal expression analysis on some miRNAs will be conducted.
Figure 5.
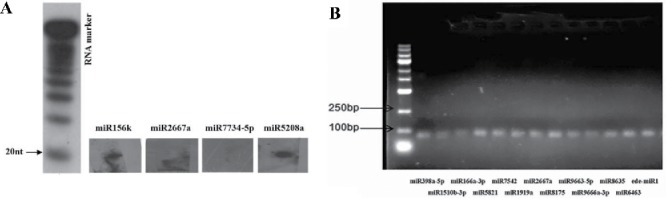
Gel-based detection of Eucheuma denticulatum miRNAs. (A) RNA gel blot hybridization of digoxigenin-labeled probes for 4 E. denticulatum miRNAs. (B) Agarose gel of stem-loop reverse transcription PCR products based on 13 E. denticulatum miRNAs.
Prediction of E. denticulatum miRNA target genes
Plant miRNAs can bind nearly perfectly with their targets by complementary matching to regulate the mRNA post-transcriptional process by mRNA cleavage or translational inhibition.53 To predict as many target genes as possible in E. denticulatum, both psRobot54 and TargetFinder 55 were used to predict putative targets. As shown in Table 4, 542 and 621 target genes were predicted based on 627 targets predicted by psRobot and 813 targets predicted by TargetFinder, respectively. After removing duplicates, 871 target genes were identified (Fig. S3). Moreover, 155 and 214 targets respectively obtained by psRobot and TargetFinder were predicted as potential translational inhibition locations based on psRNATarget analysis 56 (Table S6). The observed percentages of translational inhibition (32.8% and 35.7%) demonstrate that cleavage may be the main miRNA inhibition pattern during the process of mRNA post-transcription in E. denticulatum.
Table 4.
Summary of predicted conserved miRNA targets in Eucheuma denticulatum.
| Software | miRNA number | Target gene number | Count of targets | Target location number |
|---|---|---|---|---|
| psRobot | 67 | 542 | 626 | 627 |
| TargetFinder | 92 | 621 | 810 | 813 |
| Total | 100 | 871 | 1079 |
Gene ontology (GO) analysis
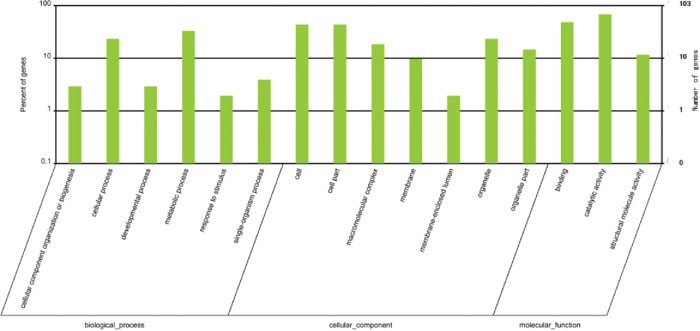
Statistical results of GO classification demonstrate that the majority of putative target genes were related to BP and CC categories (Fig. 6).57 Directed acyclic graphs of the top 10 enriched GO terms based on the 3 ontologies (BP, CC and MF) can be seen in Fig. S4A–S4C. As shown in Table S7, the 3 most highly enriched GO terms in BP, CC and MF categories were related to porphyrin-containing compound biosynthetic process, plastid stroma, and catalytic activity, respectively. Notably, some enriched GO terms may provide reference information for protection and utilization of this red algal resource.58 For example, the enriched target genes (GO: 0050896) regulated by miR535d were associated with E. denticulatum response to stimulus (Tables S6 and S7), possibly providing some clues for future study of the adaption of eucheuma to various environmental stresses.
Figure 6.
Gene Ontology (GO) classification of target genes. The x-axis shows the diverse biological functions of target genes based on 3 GO categories (biological process, cellular component and molecular function). The y-axis shows the percentage and number of these target genes.
Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis
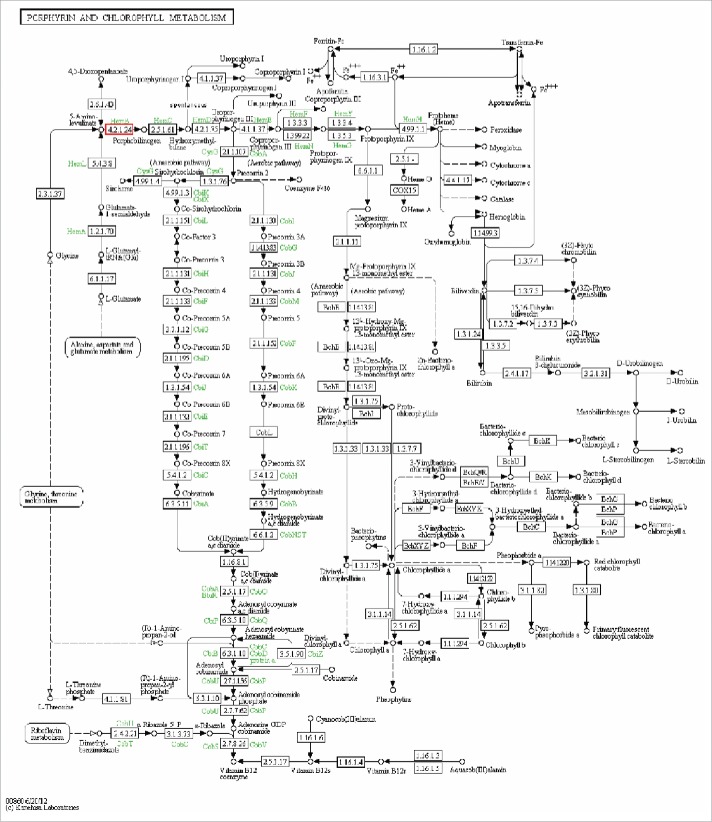
A KEGG reference pathway analysis was carried out based on the functional hierarchy of target genes in the KEGG orthology system.59 As shown in Table S8, the most significantly enriched pathways were biosynthesis of secondary metabolites (pathway ID: ko01110) and metabolic pathways (pathway ID: ko01100). Because these metabolic categories are so broad,60 however, no pathway maps could be identified in the KEGG database. Nevertheless, the porphyrin and chlorophyll metabolism category (pathway ID: ko000860) attracted our attention because of its essential role in plant growth and development (Fig. 7).61 The potential target gene, HemB (including 2 types: HemB1 and HemB2), is involved in the synthesis of chlorophyll precursor and plays an essential role in plant early embryonic development.62 Research has demonstrated that transcription factors FHY3/FAR1 can regulate HemB1 gene expression via 2 regulatory modes: 63 either through binding of FHY3/FAR1 to the promoter of HemB1 to activate its expression, or by direct interaction of the regulation factor PIF1 with FHY3 or FAR1 to partly repress the activation of HemB1 expression by those transcription factors. As the mechanism underlying post-transcriptional regulation of HemB1 expression is still unclear, however, further study is needed in conjunction with future explorations of the eucheuma growth and development process.
Figure 7.
Porphyrin and chlorophyll metabolic pathway. The porphyrin and chlorophyll metabolic pathway was revealed by KEGG analysis. Small boxes represent proteins or enzymes (with EC nos.), with red boxes corresponding to the candidate target genes encoding them. Specific genes or enzymes are indicated in green for some species. Small circles indicate metabolites, and arrows represent the different metabolic pathways.
Cytoscape network analysis
Cytoscape networks were constructed to illustrate the relationships of miRNAs, miRNA-target gene and target genes in E. denticulatum.64 As shown in Fig. S5 the most enriched and cross-linked networks were revealed to be miR5260-110 target genes and miR398b-3p-miR398a-5p-miR418-miR1919a-miR159g-3p-miR5565e-target genes. The number of miRNAs and target genes and the comprehensive co-regulatory networks among them demonstrate that some important biological processes in the organism may be regulated in a complicated, indirect fashion.65
Our described findings, which correspond to the first reported identification of miRNAs in E. denticulatum, narrow the existing research gap and lay a foundation for more detailed study of eucheuma small RNAs. Future investigations that involve mining of some key miRNAs, validation of target genes and their functions, and exploration of their regulatory mechanisms and potential metabolic pathways will be necessary for future protection and utilization of this marine alga.
Materials and methods
Plant material
Three samples of E. denticulatum at the sporophyte stage were collected on 1 June, 2014 from the southeastern coast of Hainan near the South China Sea (about 19° N latitude and 110° E longitude) by researchers from the College of Marine Life, Ocean University of China (Qingdao, China). Six tissue portions from different locations were removed from each sample and sheared into approximately 1.5–2-cm cubes. The fresh tissues from the different samples were pooled together, immediately frozen in liquid nitrogen and stored at −75°C.
Small RNA library construction and deep sequencing
After extraction of high-quality total RNA (its concentration was about 450ng/uL, and its value of OD260/280 was between 1.8 and 2.2) from approximately 80 mg of the collected tissue pool with Trizol reagent (Invitrogen, Carlsbad, California, USA), about 10 μg of enriched small RNA was isolated using an Illumina TruSeq Small RNA Sample Prep kit (Illumina, San Diego, California, USA) and used for small-RNA library construction. Small-RNA library quality assessment was performed on an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, California, USA) and a StepOnePlus Real-Time PCR system (ABI, Carlsbad, California, USA). Deep sequencing was performed on an Illumina HiSeq 2000 system (Illumina).
Standard bioinformatics analysis of small RNAs
As shown in Fig. 1, raw sequencing reads were filtered to remove unclean reads, including low-quality tags, insert null tags, 3′ adaptor null tags, and tags with 5′ adaptor contaminants, poly-A tails or lengths less than 18 nt. The remaining clean reads were annotated by alignment against E. denticulatum ESTs in GenBank (July 2014) using SOAP (http://soap.genomics.org.cn/)66 and against all plant miRNAs in miRBase 21.0 (July 2014) using BLAST. After removing tRNAs, rRNAs, snRNAs, snoRNAs and known miRNAs identified in Rfam 11.0 (http://rfam.sanger.ac.uk),67 GenBank and miRBase 21.0 databases, the remaining unannotated small RNAs were used for novel miRNA prediction.
Identification of novel miRNAs
Novel miRNAs and their precursors were identified by aligning the unannotated reads against eucheuma ESTs using Mireap (http://sourceforge.net/projects/mireap/).68 We filtered out unreasonable reads that did not meet the following criteria:69 (i) the precursor could form a perfect stem-loop structure; (ii) the miRNA and miRNA* formed a duplex with no more than 2 nucleotides on 5′- and 3′-end overhangs; (iii) no loops and bulges larger than 4 nt were located within the miRNA-miRNA* duplex and (iv) the value of the precursor's minimum free energy of folding (MFE) was no more than −18 kcal/mol.70 The remaining high-confidence sequences were retained as potential novel miRNAs, and the secondary structures of their precursors were obtained using Mireap.
Prediction of miRNA families
After using ClustalX 2.0 71 and MEGA 5 72 to conduct an alignment analysis and BLAST search of sequences of conserved miRNAs combined with their precursors against miRNA sequences deposited in miRBase 21.0, we considered sequences with homology percentages of at least 98% and with no more than 2 mismatches among them to belong to the same family. To explore the conservation and diversity of E. denticulatum miRNA families in different plant species, we also conducted a BLAST comparison of 21 randomly selected miRNA families identified in E. denticulatum vs. plant miRNA families in miRBase.
Northern blot validation
Four miRNAs were randomly selected and subjected to RNA gel blot detection.73 First, approximately 30 μg of total RNA was resolved on a 15% denaturing polyacrylamide gel and electrically transferred to Hybond-N+nylon membrane (Amersham Biosciences, London, UK). Blot hybridization was then carried out with miRNA-complementary oligonucleotides labeled with digoxigenin (Roche, Basel, Switzerland). The oligo sequences are listed in Table S7. In brief, membranes were subjected to the following steps:74 (i) pre-hybridization incubation at 62°C for 2 h followed by hybridization with incubation at 42°C for 18 h; (ii) 2 washings with 2× saline sodium citrate/0.1% sodium dodecyl sulfate for 5 min at room temperature and then 2 washings with 0.5× saline sodium citrate/0.1% sodium dodecyl sulfate for 15 min at 68°C; (iii) incubation in blocking buffer for 30 min followed by addition of 20 mL antibody buffers and incubation for 30 min at room temperature; (iv) immersion in detection buffer for 5 min and then incubation at 25°C for 5 min with 1 mL added CDP-Star (a chemical luminescence material with the molecular formula C18H19Cl2Na2O7P); and (v) incubation at 37°C for 10 min and exposure to X-ray film at 25°C for 15 min.
RT-PCR validation
To further validate our identification results, 13 miRNAs, including 12 conserved miRNAs and one novel miRNA, were randomly selected for stem-loop RT-PCR detection.75 RT primer, forward primer and universal primer sequences used in this study are given in Table S8. First, the RT reaction was carried out according to the HiScript 1st Strand cDNA Synthesis kit protocol (Vazyme Biotech, New York, USA) in 20-μL reaction volumes consisting of 1 μg RNA, 0.5 μL RT primer, 5 μL of 2× RT Mix, 1 μL RT Enzyme Mix and RNase-free double-distilled H2O. Reaction conditions were 25°C for 5 min, 42°C for 20 min, 85°C for 10 min and 4°C for 5 min. PCR amplification was then carried out according to the 2× Taq PCR Master Mix kit instructions (Vazyme Biotech) in reaction mixtures of 1 μL cDNA, 0.5 μL universal primer, 0.5 μL forward primer, 5 μL 2× Taq PCR Master Mix and 3 μL distilled H2O. The PCR cycling program was as follows: 94°C for 5 min, followed by 35 cycles of 94°C for 15 s, 60°C for 30 s and 72°C for 15 s, with a final step of 72°C for 7 min and then 4°C for 2 min. The resulting PCR products were electrophoretically analyzed on a 1.2% agarose gel.
Target gene prediction
Prediction of miRNA target genes was performed by running psRobot (http://omicslab.genetics.ac.cn/psRobot/)76 and TargetFinder (http://carringtonlab.org/).77 All identified miRNAs were used as queries against eucheuma transcripts and ESTs deposited in GenBank. Based on perfect complementary and close homology between miRNAs and target transcripts in plants,78 potential targets were those with the following characteristics:79 (i) no more than 4 mismatches in all, with no more than one mismatch in positions 1–9 and no mismatches at positions 10 and 11; (ii) no deletions or insertions; (iii) a perfect duplex at positions 8–12; (iv) no loops or bulges in either strand; (v) overhangs on 5′ and 3′ ends of no more than one nucleotide and (vi) a MFE value of less than −18 kcal/mol between the miRNA and its complementary sequence. Moreover, potential translational inhibition was predicted based on whether one mismatch could be detected in positions 9–11 by using psRNATarget (http://plantgrn.noble.org/psRNATarget/).56
GO classification and enrichment
We analyzed GO functions using the following equation:80
In this equation, N is the total number of genes with GO annotations, n is number of target gene candidates in N, M is the total number of genes annotated to a certain GO term and m is the number of target gene candidates in M. GO terms with Bonferroni-corrected p-values ≤ 0.05 were defined as significantly enriched in target genes. In addition, GO classification into BP, CC and MF categories was conducted using the AmiGO tool of the Gene Ontology Consortium (http://geneotology.org/).81 Enriched GO terms and their topological structures were obtained using the Goseq package.82 We also constructed directed acyclic graphs of the top 10 enriched terms based on the 3 classifications. Because of the scattered, sparse nature of the gene functional distribution obtained in this study, GO classification and enrichment analysis of target genes regulated by novel miRNAs could not be conducted effectively.
KEGG pathway prediction
We inferred the main referenced metabolic pathways based on the diverse target gene functions uncovered by KEGG enrichment analysis. Target genes with KEGG orthology IDs were obtained based on similar functional products deposited in KOBAS 2.0 (http://kobas.cbi.pku.edu.cn/home.do).83 The same equation used in the GO analysis was applicable to the KEGG pathway analysis, except that N was the total number of genes with KEGG annotations and M was the total number of genes annotated to a certain pathway. Only genes with a false discovery rate ≤ 0.05 were considered as significantly enriched target genes. Based on the results of the KEGG enrichment analysis, reference metabolic pathway maps were obtained using KegSketch software (http://genome.jp/kegg/).84 For the same reason given for the GO analysis, target genes regulated by novel miRNAs could not be effectively subjected to KEGG pathway analysis.
Cytoscape network construction
To reveal correlations among miRNAs, miRNA-target genes and target genes in E. denticulatum, Cytoscape networks were constructed according to the Cytoscape software manual (http://www.cytoscape.org).85,86
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Dr. Barbara Goodson (University of Texas, USA) for her critical reviews of the manuscript and editorial assistance with the English. We thank Professor Tao Liu in College of Marine Life, Ocean University of China for the support of Eucheuma dentticulatum samples, and Beijing genomics of institute (BGI) for assistance with sequencing.
Funding
This work was supported by National Science Foundation of China [Shulian Xie, 31370239, Jia Feng, 31200164].
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 2.Zhu JK. Reconstituting plant miRNA biogenesis. Proc Natl Acad Sci U S A 2008; 105:9851; PMID:18632572; http://dx.doi.org/ 10.1073/pnas.0805207105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of microRNAs from moss. Plant J 2005; 43:837-48; PMID:16146523; http://dx.doi.org/ 10.1111/j.1365-313X.2005.02499.x [DOI] [PubMed] [Google Scholar]
- 4.Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell 2005; 17:1397-411; PMID:15805478; http://dx.doi.org/ 10.1105/tpc.105.031682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo YS, Liu HH, Yang ZX, Chen J, Sun YM, Ren X. Identification and characterization of miRNAome in tobacco (Nicotiana tabacum) by deep sequencing combined with microarry. Gene 2012; 201:24-32;PMID:22575711; http://dx.doi.org/ 10.1016/j.gene.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 6.Jiao Y, Song W, Zhang M, Lai J. Identification of novel maize miRNAs by measuring the precision of precursor processing. B M C Plant Biol 2011; 11:141; PMID:22014170; http://dx.doi.org/ 10.1186/1471-2229-11-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang WW, Luo YP, Gong X, Zeng WH, Li SG. Computational identification of 48 potato microRNAs and their targets. Comput Biol Chem 2009; 33:84-93; PMID:18723398; http://dx.doi.org/ 10.1016/j.compbiolchem.2008.07.006 [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843-54; PMID:8252621; http://dx.doi.org/16736022 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 9.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nature Genetics 2006; 38:S31-6; PMID:16736022; http://dx.doi.org/ 10.1038/ng1791 [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Yu X, Cai Y, Zheng H, Yu D, Liu G, Zhou Q, Hu S, Hu F. Next-generation small RNA sequencing for microRNAs profiling in the honey bee Apis mellifera. Insect Mol Biol 2010; 19:799-805; PMID:20807255; http://dx.doi.org/24667243 10.1111/j.1365-2583.2010.01039.x [DOI] [PubMed] [Google Scholar]
- 11.Lv DK, Ge Y, Bai X, Li Y, Zhu YM. Bioinformatics in plant miRNA research. China J Bioinformatics 2009; 2:113-116. Available: http://en.cnki.com.cn/Article_en/CJFDTotal-XXSW200902007.htm [Google Scholar]
- 12.Su C, Yang XZ, Gao SQ, Tang YM, Zhao CP, Li L. Identification and characterization of a subset of microRNAs in wheat (Triticum aestivum L). Genomics 2014; 103:298-307; PMID:24667243; http://dx.doi.org/ 10.1016/j.ygeno.2014.03.002 [DOI] [PubMed] [Google Scholar]
- 13.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004; 16:2001-19; PMID:15258262; http://dx.doi.org/17991681 10.1105/tpc.104.022830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 2006; 57:19-53; PMID:16669754; http://dx.doi.org/17991681 10.1146/annurev.arplant.57.032905.105218 [DOI] [PubMed] [Google Scholar]
- 15.Jin LG, Wang C, Liu JY. Plant microRNA. Chin J Biochem Mol Biol 2006; 22:609-14. Available: http://cjbmb.bjmu.edu.cn/CN/abstract/abstract19639.shtml [Google Scholar]
- 16.Hunter C, Poethig RS. Missing links: miRNAs and plant development. Curr Opin Genet Dev 2003; 13:372-78; PMID:12888010; http://dx.doi.org/17991681 10.1016/S0959-437X(03)00081-9 [DOI] [PubMed] [Google Scholar]
- 17.Kidner CA, Martienssen RA. Macro effects of microRNAs in plants. Trends Genet 2003; 19:13-6; PMID:12493243; http://dx.doi.org/17991681 10.1016/S0168-9525(02)00011-2 [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Wang W, Sun X, Liang Z, Wang F. Conserved and novel heat stress-responsive microRNAs were identified by deep sequencing in Saccharina japonica (Laminariales, Phaeophyta). Plant Cell Environ 2005; 38:1357-7. Available: http://onlinelibrary.wiley.com/doi/10.1111/pce.12484/full; http://dx.doi.org/ 10.1111/pce.12484 [DOI] [PubMed] [Google Scholar]
- 19.Xia BM, Zhang JP. Records of Chinese seaweeds. Beijing: Science Press; 1999: pp128-32 [Google Scholar]
- 20.Fan X, Han LJ, Zheng NY. Analysis of nutritional components of common edible seaweeds in China. Chin J Mar Drugs 1993; 4:32-7. Available: http://www.cqvip.com/qk/91183X/199304/1237420.html [Google Scholar]
- 21.Ask EI, Azanza RV. Advances in cultivation technology of commercial eucheumatoid species: a review with suggestions for future research. Aquaculture 2002; 206:257-77; http://dx.doi.org/ 10.1016/S0044-8486(01)00724-4 [DOI] [Google Scholar]
- 22.Tye RJ. Industrial and non-food uses for carrageenan. Carbohydrate Polymers 1989; 10:259-80; http://dx.doi.org/ 10.1016/0144-8617(89)90066-0 [DOI] [Google Scholar]
- 23.Li LH, Qi B, Yang SL. Functional effect of dietary fiber from Eucheuma on reducing serum lipids. J Fishery Sci China 2008; 15:943-9. Available: http://lib.cqvip.com/qk/98270X/200806/28730479.html [Google Scholar]
- 24.Jiang ZY, Liu Y, Tang Y, Cen YZ, Wang CJ. Effect of seleno-polysaccharide of Eucheuma on growth and apoptosis of hepatoma carcinoma cell strain. J Sun Yat-sen University (Med Sci) 2008; 29:270-77. Available: http://www.cqvip.com/Main/Detail.aspx?id=27279605 [Google Scholar]
- 25.Liu Y, Jiang ZY, Shi SS, Lin C, Tang Y, Cen YZ, Wang CJ. Effect of Seleno-polysaccharide of eucheuma on the growth and apoptosis of uterinecervix cancer cell strain. J Jinan University (Med Edition) 2007; 28:546-9. Available: http://lib.cqvip.com/qk/94341X/200706/26176624.html [Google Scholar]
- 26.Bai X, Lin C, Jiang ZY, Yuan GX, Shen WZ, Li YQ, Tang Y. Inhibiting effects of Eucheuma polysaccharide selenide on the proliferation of carcinoma cell line HEp-2. Cancer Res Prevention Treatment 2007; 34:249-52. Available: http://www.cnki.com.cn/Article/CJFDTotal-ZLFY200704005.htm [Google Scholar]
- 27.Ye SM, Cen YZ, Zhang MY, Wang YF. Antiviral activities of polysaccharides from Eucheuma gelatinae and Eucheuma striatum in vitro. Chin J Mar Drugs 2007; 26:14-9. Available: http://www.cnki.com.cn/Article/CJFDTotal-HYYW200703002.htm [Google Scholar]
- 28.Liu Y. Study on anti-tumor and anti-virus effect of seleno-polysaeeharide of eueheuma. Jinan University MS Dissertation. 2008. Available: http://cdmd.cnki.com.cn/Article/CDMD-10559-2009011402.htm [Google Scholar]
- 29.Liu JG, Pang T, Wang L, Li J, Lin W. The reasons causing catastropic death in tropical carrageenan producing seaweeds and their difference in resistance to illness. Oceanologia et Limnologia Sinica 2009; 40:235-41. Available: http://www.cnki.com.cn/Article/CJFDTotal-HYFZ200902022.htm [Google Scholar]
- 30.Azanza-Corrales R, Aliaza TT, Montano E. Recruitment of Eucheuma and Kappaphycus on a farm in Tawi-Tawi, Philippines. Hydrobiologia 1999; 327:235-44. Available: http://link.springer.com/chapter/10.1007%2F978-94-009-1659-3_33 [Google Scholar]
- 31.Park S, Yoon JH. Ruegeria arenilitoris sp. nov., isolated from the seashore sand around a seaweed farm. Antonie Van Leeuwenhoek Int J Microbiol 2012; 102:581-89; PMID:22669198; http://dx.doi.org/17991681 10.1007/s10482-012-9753-8 [DOI] [PubMed] [Google Scholar]
- 32.Robertson-Andersson DV, Potgieter M, Hansen J, Bolton JJ, Troell M, Anderson RJ, Halling C, Probyn T. Integrated seaweed cultivation on an abalone farm in South Africa. J Appl Phycol 2008; 20:579-95. Available: http://link.springer.com/chapter/10.1007%2F978-1-4020-9619-8_18; http://dx.doi.org/ 10.1007/s10811-007-9239-7 [DOI] [Google Scholar]
- 33.Kühlmann KJ. Evaluations of marine reserves as basis to develop alternative livelihoods in coastal areas of the Philippines. Aquaculture Int 2002; 10:527-49. Available: http://link.springer.com/article/10.1023%2FA:1023955626357; http://dx.doi.org/ 10.1023/A:1023955626357 [DOI] [Google Scholar]
- 34.Padolina WG. The seaweed industry and R&D: a solid partnership. Hydrobiologia 1999; 399:25-6. Available: http://link.springer.com/article/10.1023%2FA:1017219101607 [Google Scholar]
- 35.Sundström J, Collén J, Abrahamsson K, Pedersén M. Halocarbon production and in vivo brominating activity of Eucheuma denticulatum. Phytochemistry 1996; 42:1527-30. Available: http://www.sciencedirect.com/science/article/pii/0031942296001975; http://dx.doi.org/ 10.1016/0031-9422(96)00197-5 [DOI] [Google Scholar]
- 36.Glenn EP, Doty MS. Growth of the seaweeds Kappaphycus alvarezii, K. striatum and Eucheuma denticulatum as affected by environment in Hawaii. Aquaculture 1990; 84:245-55. Available: http://www.doc88.com/p-4177778843535.html; http://dx.doi.org/ 10.1016/0044-8486(90)90090-A [DOI] [Google Scholar]
- 37.Glenn EP, Doty MS. Photosynthesis and respiration of the tropical red seaweeds, Eucheuma striatum (Tambalang and elkhorn varieties) and E. denticulatum. Aquatic Botany 1981; 10:353-64. Available: http://www.doc88.com/p-1156692733568.html; http://dx.doi.org/ 10.1016/0304-3770(81)90033-4 [DOI] [Google Scholar]
- 38.Li LH, Yang SL, Qi B. Studies of optimization of extraction process of dietery fibre from Eucheuma with biologic enzyme by using uniform design. Food Sci 2006; 27:292-96. Available: http://www.cnki.com.cn/Article/CJFDTotal-SPKX200610070.htm [Google Scholar]
- 39.Guo L, Bao HY, Ye SM, Xing YY, Xu SY, Cen YZ. Antibacterial activities of Eucheuma gelatinae, Eucheuma striatum and their hydrolysates. J Jinan University 2002; 23:79-83. Available: http://www.cnki.com.cn/Article/CJFDTotal-JNDX200203014.htm [Google Scholar]
- 40.Zhang T, Shi X, He JH, Chen QF, Feng ZH, He PM. Preliminary analysis on identification Eucheuma and Kappaphycus (Rhodopyta) by ISSR. Acta Oceanologica Sinica 2011; 33:173-8. Available: http://www.cnki.com.cn/Article/CJFDTotal-SEAC201103020.htm [Google Scholar]
- 41.Othman R, Nor DM, Daud H, Adnan AM. An expressed sequence tag approach for cloning of phytochelatin synthase cDNA clone from Eucheuma denticulatum (Rhodophyta). J Biotechnol 2008; 136:S541-2; http://dx.doi.org/ 10.1016/j.jbiotec.2008.07.1272 [DOI] [Google Scholar]
- 42.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res 2007; 36:D154-8; PMID:17991681; http://dx.doi.org/ 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi S, Gao ZX, Zhao H, Zeng C, Luo W, Chen B, Wang WM. Identification and characterization of microRNAs involved in growth of blunt snout bream (Megalobrama amblycephala) by Solexa sequencing. BMC Genomics 2013; 14:754PMID:23324084; http://dx.doi.org/ 10.1186/1471-2164-14-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Ju Z, Li Q, Hou Q, Wang C, Li JB, Li RL, Wang LL, Sun T, Hang SQ, et al.. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in holstein cattle. Int J Biol Sci 2011; 7:1016-26; PMID:21912509; http://dx.doi.org/17173072 10.7150/ijbs.7.1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genetics 2009; 10:94-108; PMID:19148191; http://dx.doi.org/17173072 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Epping JT, et al.. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-9; PMID:10802651; http://dx.doi.org/17173072 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28:27; PMID:10592173; http://dx.doi.org/17173072 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kluiver J, van den Berg A, de Jong D, Blokzijl T, Harms G, Bouwman E, Jacobs S, Poppema S, Kroesen BJ. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene 2007; 26:3769-76; PMID:17173072; http://dx.doi.org/ 10.1038/sj.onc.1210147 [DOI] [PubMed] [Google Scholar]
- 49.Rio DC. Northern blots for small RNAs and microRNAs. Cold Spring Harb Protoc 2014; 7:793-7; PMID:24987143; http://dx.doi.org/23415324 10.1101/pdb.prot080838 [DOI] [PubMed] [Google Scholar]
- 50.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nquyen JT, Barbisin M, Xu NL, Mahuvakar VR, Anderson MR, et al.. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005; 33:179; PMID:16314309; http://dx.doi.org/23415324 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. B M C Plant Biol 2008; 8:37-42. Available: http://www.biomedcentral.com/1471-2229/8/37; http://dx.doi.org/ 10.1186/1471-2229-8-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallory AC, Vaucheret H. MicroRNAs: something important between the genes. Curr Opin Plant Biol 2004; 2:120-5; PMID:15003210; http://dx.doi.org/23415324 10.1016/j.pbi.2004.01.006 [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Zheng Y, Jagadeeswaran G, Sunkar R. Characterization of small RNAs and their target genes in wheat seedlings using sequencing-based approaches. Plant Science 2013; 203-204:17-24; PMID:23415324; http://dx.doi.org/ 10.1016/j.plantsci.2012.12.014 [DOI] [PubMed] [Google Scholar]
- 54.Wu HJ, Ma YK, Chen T, Wang M, Wang XJ. PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res 2012; 40:W22-8; PMID:22693224; http://dx.doi.org/ 10.1093/nar/gks554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bo X, Wang S. TargetFinder: a software for antisense oligonucleotide target site selection based on MAST and secondary structures of target mRNA. Bioinformatics 2005; 21:1401-2; PMID:15598838; http://dx.doi.org/ 10.1093/bioinformatics/bti211 [DOI] [PubMed] [Google Scholar]
- 56.Dai XB¸, Zhao XP. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 2011; 39:W155-9; PMID:21622958; http://dx.doi.org/15695432 10.1093/nar/gkr319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure, Bioinformatics. 2006; 13:1600-7; PMID:16606683; http://dx.doi.org/15695432 10.1093/bioinformatics/btl140 [DOI] [PubMed] [Google Scholar]
- 58.Sheng X, Song X, Yu Y, Niu L, Li S, Li H, Wei C, Liu T, Zhang L, Du L Characterization of microRNAs from sheep (Ovis aries) using computational and experimental analyses. Mol Biol Rep 2011; 38: 3161-71; PMID:20140706; http://dx.doi.org/15695432 10.1007/s11033-010-9987-3 [DOI] [PubMed] [Google Scholar]
- 59.Qiu C, Wang J, Cui Q. miR2Gene: pattern discovery of single gene, multiple genes, and pathways by enrichment analysis of their microRNA regulators. B M C Systems Biol 2011; PMID:22784580; http://dx.doi.org/15695432 10.1186/1752-0509-5-S2-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su H. Study on the Secondary metabolites from three red algae and two marine actinomycetes. Chinese Acad Sci Ph D Dissertation. May, 2009. Available: http://cdmd.cnki.com.cn/Article/CDMD-80068-2009126134.htm [Google Scholar]
- 61.Nakanishi H, Nozue H, Suzuki K, Kaneko Y, Taguchi G, Hayashida N. Characterization of the Arabidopsis thaliana mutant pcb 2 which accumulates divinyl chlorophylls. Plant Cell Physiol 2005; 46:467-473; PMID:15695432; http://dx.doi.org/ 10.1093/pcp/pci053 [DOI] [PubMed] [Google Scholar]
- 62.Beale S. Green genes gleaned. Trend Plant Sci 2005; 10:309-312; PMID:15951223; http://dx.doi.org/22634759 10.1016/j.tplants.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 63.Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R. Transposase-derived proteins FHY3/FAR1 interact with Phytochrome-Interacting Factor1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 2012; 24:1984-2000; PMID:22634759; http://dx.doi.org/ 10.1105/tpc.112.097022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Politano G, Benso A, Savino A, DiCarlo S. ReNE: A cytoscape plugin for regulatory network enhancement. PLoS One 2014; 9:e115585; PMID:25541727; http://dx.doi.org/ 10.1371/journal.pone.0115585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA. RNA 2005; 11:1753-61; PMID:16314451; http://dx.doi.org/ 10.1261/rna.2248605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li R, Li Y, Kristiansen K, Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008; 24:713-4; PMID:18227114; http://dx.doi.org/ 10.1093/bioinformatics/btn025 [DOI] [PubMed] [Google Scholar]
- 67.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res 2005; 33:D121-4; PMID:15608160; http://dx.doi.org/ 10.1093/nar/gki081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Zhang Z, Liu F, Vongsangnak W, Jing Q, Shen B. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res 2012; 40:4298-305; PMID:22287634; http://dx.doi.org/ 10.1093/nar/gks043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al.. A uniform system for microRNA annotation. RNA 2003; 9:277-9; PMID:12592000; http://dx.doi.org/ 10.1261/rna.2183803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warris S, Boymans S, Muiser I, Noback M, Krijnen W, Nap JP. Fast selection of miRNA candidates based on large-scale pre-computed MFE sets of randomized sequences. BMC Res Notes 2014; 7:34; PMID:24393391; http://dx.doi.org/ 10.1186/1756-0500-7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al.. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947-8; PMID:17846036; http://dx.doi.org/ 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 72.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731-9; PMID:21546353; http://dx.doi.org/ 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Várallyay É, Burgyán J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nature Protocols 2008; 3:190-6; PMID:18274520; http://dx.doi.org/10089203 10.1038/nprot.2007.528 [DOI] [PubMed] [Google Scholar]
- 74.Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucl Acids Res 2004; 22:e175; PMID:15598818; http://dx.doi.org/10089203 10.1093/nar/gnh171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Varkonyi-Gasic E, Hellens RP. Quantitative stem-loop RT-PCR for detection of microRNAs. Meth Mol Biol 2011; 744:145-57; PMID:21533691; http://dx.doi.org/10089203 10.1007/978-1-61779-123-9_10 [DOI] [PubMed] [Google Scholar]
- 76.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 2011; 39:W155-9; PMID:21622958; http://dx.doi.org/10089203 10.1093/nar/gkr319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lavorgna G, Guffanti A, Borsani G, Ballabio A, Boncinelli E. TargetFinder: searching annotated sequence databases for target genes of transcription factors. Bioinformatics 1999; 15:172-3; PMID:10089203; http://dx.doi.org/ 10.1093/bioinformatics/15.2.172 [DOI] [PubMed] [Google Scholar]
- 78.Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res 2005; 5:336-360; PMID:15916721; http://dx.doi.org/17367786 10.1038/sj.cr.7290302 [DOI] [PubMed] [Google Scholar]
- 79.Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett 2007; 581:1464-1474; PMID:17367786; http://dx.doi.org/ 10.1016/j.febslet.2007.02.074 [DOI] [PubMed] [Google Scholar]
- 80.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010; 11:R14; PMID:20132535; http://dx.doi.org/21715386 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 2004; 32:D258-61; PMID:14681407; http://dx.doi.org/21715386 10.1093/nar/gkh066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srivastava P K, Moturu TR, Pandey P, Baldwin IT, Pandey SP. A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. B M C Genomics 2014; 15:348; PMID:24885295; http://dx.doi.org/21715386 10.1186/1471-2164-15-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011; 39:W316-22; PMID:21715386; http://dx.doi.org/ 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Reaearch 2012; 40:D109-14; PMID:22080510; http://dx.doi.org/25541727 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Praneenararat T, Takagi T, Iwasaki W. Integration of interactive, multi-scale network navigation approach with Cytoscape for functional genomics in the big data era. B M C Genomics 2012; 13:S24; PMID:23281970; http://dx.doi.org/25541727 10.1186/1471-2164-13-S7-S24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Politano G, Benso A, Savino A, DiCarlo S. ReNE: A cytoscape plugin for regulatory network enhancement. PLoS One 2014; 9:e115585; PMID:25541727; http://dx.doi.org/ 10.1371/journal.pone.0115585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.