Abstract
Many splicing regulators bind to their own pre-mRNAs to induce alternative splicing that leads to formation of unstable mRNA isoforms. This provides an autoregulatory feedback mechanism that regulates the cellular homeostasis of these factors. We have described such an autoregulatory mechanism for two core protein components, U11–48K and U11/U12–65K, of the U12-dependent spliceosome. This regulatory system uses an atypical splicing enhancer element termed USSE (U11 snRNP-binding splicing enhancer), which contains two U12-type consensus 5′ splice sites (5′ss). Evolutionary analysis of the USSE element from a large number of animal and plant species indicate that USSE sequence must be located 25–50 nt downstream from the target 3′ splice site (3′ss). Together with functional evidence showing a loss of USSE activity when this distance is reduced and a requirement for RS-domain of U11–35K protein for 3′ss activation, our data suggests that U11 snRNP bound to USSE uses exon definition interactions for regulating alternative splicing. However, unlike standard exon definition where the 5′ss bound by U1 or U11 will be subsequently activated for splicing, the USSE element functions similarly as an exonic splicing enhancer and is involved only in upstream splice site activation but does not function as a splicing donor. Additionally, our evolutionary and functional data suggests that the function of the 5′ss duplication within the USSE elements is to allow binding of two U11/U12 di-snRNPs that stabilize each others' binding through putative mutual interactions.
Introduction
Alternative splicing is a post-transcriptional regulatory mechanism that creates a multitude of mature mRNAs from a single mRNA precursor (pre-mRNA) and is thought to increase the coding capacity of a genome.1,2 Activation of alternative splicing events is largely dependent on short sequence elements termed splicing enhancers and inhibitors that are located near splice sites, and can be found both in intronic and in exonic locations. These regulatory elements bind to SR and hnRNP class of proteins that can regulate positively or negatively the splice site choice in a context-dependent manner.2 Splicing enhancers are essential for both constitutive and alternative splicing of both the U2-type and U12-type introns.3,4 Additionally, interactions of spliceosome components across an exon on adjacent introns in a process called exon definition influences whether an alternative exon will be included in the mature mRNA product or skipped.5 Exon definition interactions have been shown to predominate over intron definition (i.e. interactions across the intron) when the intron size is larger than ∼250 nt,6 and in addition to the U2-dependent spliceosome, such interactions have also been demonstrated with the U12-dependent spliceosome.7
Besides increasing protein diversity, alternative splicing has also an important role in regulating mRNA levels. The best characterized mechanism uses alternative splicing for inclusion of “poison” cassette exons that introduce premature stop codons (PTC) to mRNA. This triggers the nonsense mediated decay pathway (NMD), and causes clearance of such transcripts from the cellular mRNA pool in a translation dependent manner.8 Many splicing regulators, but also core protein components of the spliceosome use this mechanism in autoregulatory loops, wherein the cellular levels of a particular splicing regulator protein will influence the splicing of its own pre-mRNA and act as a homeostatic mechanism to keep protein levels steady.9-13
Earlier, we have described such a negative feedback loop in two genes encoding for core protein components that are specific to the U12-dependent spliceosome (also called minor spliceosome).12 Both of these proteins, U11–48K and U11/U12–65K (also known as RNPC3), are integral components of the U11/U12 di-snRNP,14-16 which recognizes the 5′ splice site (5′ss) and the branch point sequence (BPS) of U12-type introns.17-19 Both genes contain a novel splicing regulatory element denoted USSE (U11 snRNP-binding splicing enhancer), which is composed of a tandem duplication of 5′ss sequences of U12-type introns that are, however, not used for splicing.12 USSE is recognized by the U11/U12 di-snRNP which activates an alternative upstream U2-type 3′ splice site (3′ss; see Fig. 1A). With the 48K pre-mRNA this leads to the inclusion of an 8 nt alternative exon that disrupts the protein reading frame and leads to alternative splicing–nonsense mediated decay (AS-NMD), while with 65K this leads to formation of mRNA containing a long 3′UTR.11,12 The core USSE element is evolutionarily highly conserved and can be found from both 48K and 65K genes in all mammalian species. Outside the mammalian lineage, the USSE element is widespread, but there is an interesting evolutionary dynamics, in which the USSE element has been lost from either one, or sometimes from both genes. In bird lineage (Sauropoda), and in some species of teleost fish the USSE element has been lost from the 48K gene, but retained in the 65K gene. In contrast, insects have lost USSE from the 65K gene, but retained it in the 48K gene (excluding Diptera that have apparently lost the element from both genes). In plants, the USSE element is located in the 48K gene, but in the 3′UTR instead of an internal intron in the coding region. Thus, the overall USSE architecture in plant 48K genes resembles that of the mammalian 65K gene.11,12
Figure 1.
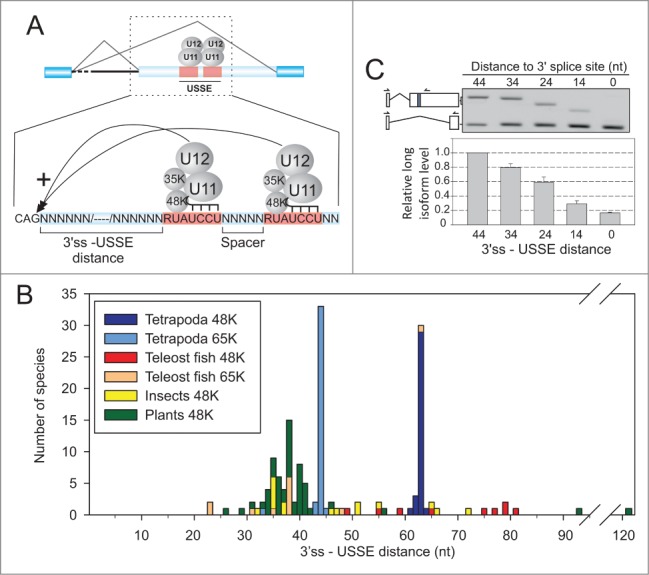
Evolutionary conservation of the 3′ss-USSE distance. (A) Schematic representation of the USSE regulatory module. (B) Distribution of 3′ss-USSE distances in animal and plant species. The different phylogenetic groups are colored differentially as indicated in the figure. (C) RT-PCR analysis of 65K 3′UTR splicing reporter with different 3′ss-USSE distances. Isoforms are labeled on the left side of the gel picture and the isoform ratio (long isoform/short isoform) quantified below the gel picture. Arrows point to the location of RT-PCR primers. Forward primer is shared between the two isoforms.
The need for a tight regulation of minor spliceosome activity and the role of U12-type introns as regulatory elements has been highlighted in several recent reports. The removal of U12-type introns has been shown to be slower or less efficient and to constitute a rate-limiting step for the maturation of mRNAs containing such introns.20-22 The minor spliceosome is essential for fruit fly and zebrafish development23-25 and the activity of the minor spliceosome has been proposed to be regulated through the levels of U6atac snRNA, which can be controlled via the p38MAPK signaling pathway.26 Furthermore, two congenital human diseases have been linked with specific components of the minor spliceosome: partial loss-of-function mutations in the U4atac snRNA gene lead to microcephalic osteodysplastic primordial dwarfism type 1/Taybi-Linder syndrome (MOPD1/TALS),27,28 while mutations in the U11/U12–65K protein component lead to isolated growth hormone deficiency type 1 (IGHD1).29 Additionally, somatic mutations in the minor spliceosome component ZRSR2 involved in 3′ss recognition have been detected in patients suffering from myelodysplastic syndrome (MDS).30 Interestingly, IGHD1 mutations in the 65K protein component appear to be compensated by upregulation of U4atac snRNA levels, suggesting that homeostasis in the levels of specific components of the minor spliceosome is necessary for its proper cellular function.29
The key difference between a typical splicing enhancer and the USSE element is that the latter binds to the U11/U12 di-snRNP, a core component of the spliceosome which functions in the splice site recognition of minor introns. This suggests that the molecular interactions needed for activation of the upstream 3′ss resemble standard exon definition interactions, where U1 snRNP bound to the 5′ss communicate, directly or indirectly, across the exon with U2AF65/35 or U2 snRNP bound to the 3′ end of the upstream intron. In the case of standard exon definition system, this leads to an activation of the 5′ss and splicing of the downstream intron. With the USSE, such 5′splice site activation is prevented because the downstream intron does not contain a suitable U12-type BPS. Therefore, USSE shares features from both exon definition and standard alternative splicing regulation via exonic splicing enhancers (ESE). Here, we asked if this would be reflected in the phylogenetic conservation of the overall architecture of the element, and the mechanism of activation of the upstream 3′ss in 48K and 65K genes. We show that both the 3′ss - USSE distance and the 5′ss - 5′ss distance within the USSE element are evolutionarily constrained and that these constraints are consistent with the model that USSE uses exon definition interactions for splice site activation. Furthermore, we identify the U11–35K protein within the U11/U12 di-snRNP, which is a paralog of the U1–70K protein, as an activator of the upstream 3′ss. Finally, we provide evidence that the function of 5′ss duplication within the USSE element is to allow simultaneous binding of two U11/U12 di-snRNPs that stabilize each other's binding through mutual interactions.
Results
Distance constraints for the alternative splicing activation via USSE
We have earlier described negative feedback regulation of U11–48K and U11/U12–65K transcripts via a U11 snRNP binding splicing enhancer (USSE) that is composed of a duplicated U12-type 5′ss consensus sequence motif (Fig. 1A). The individual 5′ss motifs within USSE are conserved over great evolutionary distance, and are present in species as distant as mammals and plants, but the short spacer sequence separating the two motifs is conserved at the sequence level only in mammals and is more diverse in other species.12 With both the 48K and 65K transcripts, U11 snRNP binding to the USSE leads to the activation of an upstream 3′ splice site (Fig. 1A). While both 5′ss motifs within the USSE are needed for 3′ss activation,12 the mechanistic significance of splice site duplication and the characteristics needed for upstream 3′ss activation have not been explored.
To ask if the spacing between the 3′ss and USSE elements is evolutionary constrained, we analyzed elements from 195 USSE sequences of 116 evolutionarily distant species where appropriate sequence information was available and plotted the distance between 3′ss and USSE, grouped according to the host gene (48K or 65K) and phylogenetic group (Fig. 1B). We found that in terrestrial vertebrates (Tetrapoda) the distance between the two 5′ss motifs was strictly constrained as the observed distances were exactly 63 nt or 43 nt in the 48K and 65K genes, respectively, with few species showing 1 to 2 nt deviations. Fish, insect and plant species show wider variation; in each case the main peak was around 34–41 nt from the upstream 3′ss. It is notable that there are only few species showing shorter 3′ss - USSE distances than 30 nt.
We attempted to address the significance of the observed phylogenetic distribution using an experimental approach. We used a 65K 3′UTR reporter construct (see Figure 3A, a construct containing the USSE element) and modified the distance between 3′ss and USSE, followed by scoring of the upstream 3′ss activation by RT-PCR. We found that for optimal activity, the distance should be at least 30 nt (Fig. 1C). The level of 3′ss activation showed a linear reduction down to 10–20% of the wt value with progressive deletions. These data suggest that, in the mammalian 65K gene, 43 nt is the minimum distance that supports full 3′ss activation, thus providing a putative experimental explanation to the phylogenetic conservation.
Figure 3.
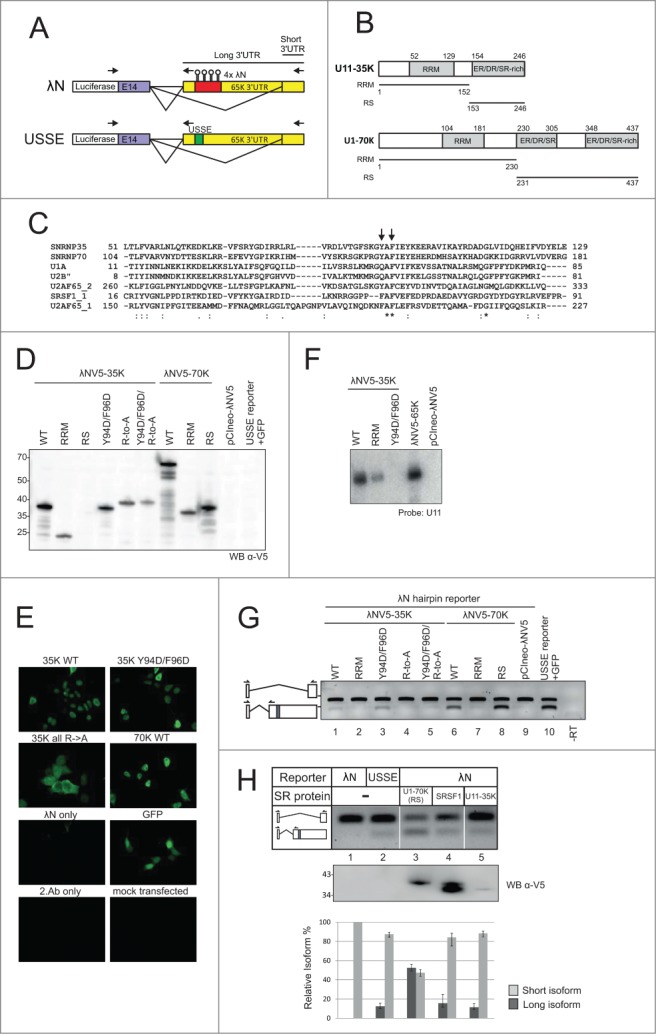
Tethering of 35K and 70K RS domains activates alternative splicing. (A) Splicing reporter constructs used in this study. In the 4 λN hairpin construct, the 65K 3′UTR USSE element is replaced with the hairpins. Arrows point to the location of RT-PCR primers. Forward primer is shared between the two isoforms. (B) Domain structures of U11–35K and U1–70K. Numbers refer to amino acids. Lines below domain structure diagrams indicate the expression constructs called “RRM” and “RS.“ (C) Alignment of amino acid sequence of selected RRM domains from human splicing-related proteins. Numbers indicate amino acids, asterisk indicates identical amino acid residues conserved between members of the protein family, colon indicates residues with highly similar and period with somewhat similar properties. Arrows point to conserved or similar aromatic residues that have been implicated in RNA binding (see text for details). (D) Western blot showing the expression levels of different tagged proteins. (E) α-V5 immunostaining of HEK293 cells expressing the tagged proteins. (F) Northern blot of RNAs coimmunoprecipitating with λNV5-tagged proteins using a probe against U11 snRNA. (G) RT-PCR analysis of 65K 3′UTR splicing reporter cotransfected with the indicated tagged proteins. Isoforms are labeled on the left side of the gel image, with primers as indicated. Note that the long isoform produces the shorter amplicon as the reverse primer had to be placed upstream of the hairpin site. (H) RT-PCR analysis of 65K 3′UTR splicing reporter cotransfected with a set of RS domain containing tagged proteins. The ability of upstream 3′ss activation was assayed by gel electrophoresis (top panel) and qPCR (bottom panel). Error bars indicate standard deviation in a triplicate experiment. Western blot (middle panel) using the α-V5 shows the expression level of different tagged proteins. Positions of the molecular weight markers are indicated on left. In Western blot analyses each lane contained equal μg amounts of protein.
A similar phylogenetic analysis of the spacer length distribution between the two 5′ss-like motifs within a USSE element revealed that virtually all spacers were 3–9 nt in length, with a notable preference for spacers of 3, 6, 7, and 9 nt (Fig. 2A). Testing these distances experimentally, we found that spacers deviating +/− 3 nt from the 6 nt found in the wt 65K gene resulted in essentially wt levels of 3′ss activation (Fig. 2B and C), thus recapitulating the observed spacer length distribution in evolutionarily distant organisms. In contrast, shortening the spacer down to 0 nt led to an almost complete loss the alternative splicing, while spacer extension beyond 9 nt up to 30 nt led to a gradual reduction in the level of 3′ss activation. With short spacers, the experimental results agree with phylogenetic distribution as there are only 3 cases where spacer length was less than 3 nt (Fig. 2A). Interestingly, we also note that although spacer lengths of 4, 5 and 8 nt are rarely present in our phylogenetic sample, at least the 4 and 5 nt long spacers function in vivo at similar levels as the more common spacers (Fig. 2B). In contrast, while none of the spacers in the phylogenetic sample exceeded 10 nt, our experimental results revealed only very mild reduction in the 3′ss activation with spacers up to 30 nt in length (Fig. 2C).
Figure 2.
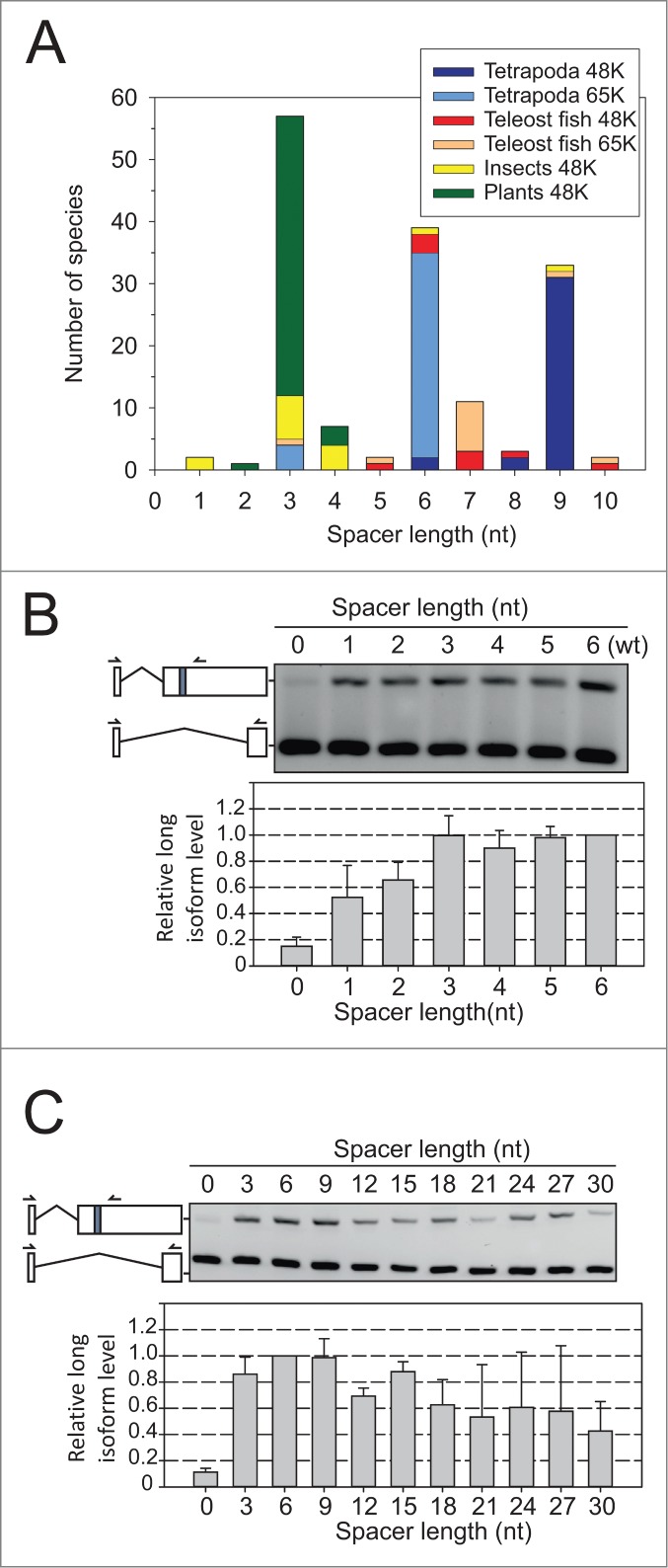
Evolutionary conservation of the USSE spacer length. (A) Distribution of the spacer length between two 5′ss-like motifs within the USSE elements in animal and plant species. The color coding the different phylogenetic groups is the same as in Figure 1B and is indicated in the figure. (B) RT-PCR analysis of 65K 3′UTR splicing reporter with short (0–6 nt) spacer lengths, as in Figure 1C. (C) RT-PCR analysis of 65K 3′UTR splicing reporter with long (up to 30 nt) spacer lengths.
Together, the results revealed a strong correlation between the evolutionary data on 3′ss-USSE and USSE spacer length distributions and experimentally measured 3′ss activation using the 65K 3′UTR reporter. This suggests that the evolutionary constraints observed with the phylogenetic data reflect an evolutionary purifying selection process that removes variants resulting in suboptimal 3′ss activation. Additionally, the phylogenetic aversion of short spacers and the observed reduction in alternative splicing activity provide a strong support to the model where a simultaneous binding of two U11 snRNPs (or U11/U12 di-snRNPs)is needed for upstream 3′ss activation. Most likely, a short spacer leads to a steric clash of the closely spaced U11 snRNPs. Similarly, the observation that a long spacer can support relatively high level of alternative splicing, suggests that the distantly located U11 snRNPs can interact by looping out the intervening long spacer, further supporting the suggestion that both 5′ss elements within the USSE need to be recognized for upstream 3′ss activation.
RS-domain of the U11–35K protein mediates exon definition interactions
U11–35K is the most likely protein component within the U11/U12 di-snRNP to mediate the activation of an upstream 3′ss. This protein shares the overall domain structure with the U1–70K protein, including a short RS-domain. In Arabidopsis, it has been shown to interact with various SR-proteins31 and, in human cells, knockdown of the U11–35K protein leads to a loss of USSE activity.12 Here, we used an RNA tethering assay based on a bacteriophage λ-antiterminator protein N (λN) binding hairpin sequence (BoxB)32 to ask if U11–35K is necessary and sufficient for the activation of the upstream splice site. We replaced the USSE element with four λN hairpin sequences and fused U11–35K and U1–70K proteins (either full-length protein or isolated protein domains) to a V5 tagged λN peptide (Fig. 3A and B). To ensure that potential activation of the upstream 3′ss is not due to tethering the whole U11 or U11/U12 snRNP, we used mutated U11–35K protein in which either the RRM or the RS domain was disabled. The RRM mutation Y94D/F96D targets residues that are conserved in several RRM domains of proteins involved in splicing, and have been shown to be critical for the RNA binding activity with other RRMs (Fig. 3C).33,34 The RS domain was disabled by replacing all arginine residues in the RS domain to alanines (R-to-A mutant).
Transient transfections of the fusion protein constructs together with the λN hairpin reporter into HEK293 cells resulted in roughly equal expression levels for all fusion proteins, with the exception of the 35K RS domain, which could not be expressed, possibly due to instability of the single RS domain (Fig. 3D). Immunostaining with V5 antibody confirmed near complete nuclear localization of the expressed proteins, with the exception of the RS domain mutant which showed a roughly uniform distribution between nucleus and cytoplasm (Fig. 3E). The results are consistent with previous reports on the localization of 35K31 and minor spliceosome components.35 The mislocalization of the RS domain mutant may be due to a failure to shuttle between nucleus and cytoplasm.36,37 We also confirmed that U11 snRNA coimmunoprecipitated with the λNV5-tagged 35K and 35K-RRM, while the Y94D/F96D mutation abolished the binding completely (Fig. 3F).
RT-PCR analysis revealed that replacement of USSE with the λN hairpin sequences resulted in complete inactivation of splicing at the upstream site (Fig. 3G, lane 9). However, tethering of the wild type 35K and 70K proteins led to a partial restoration of the long isoform levels (Fig. 3G, c.f. lanes 1 and 6 to lane 10). While the RRM domains of neither protein were able to activate the upstream 3′ss (lanes 2 and 7), tethering of the U1–70K RS domain resulted in strong activation of the upstream 3′ss and near wt levels of the long isoform (lane 8). The 35K RS domain could not be tested alone due to problems with expression or stability, but activation with the full-length 35K protein containing a mutated RRM (to abolish binding to U11 snRNA; see Fig. 3F) and loss of activity with 35K containing mutated RS domain suggest that the U11–35K protein, and more specifically, its RS domain mediates the activation of the upstream 3′ss.
To compare the ability of 35K in activating the upstream 3′ss to other well-known proteins involved in exon definition interactions that also contain RS domains, we used the same tethering assay followed by qPCR analysis. We compared the 35K activity to that of U1–70K RS domain and the SRSF1 full-length protein in CHO cells. In this assay, the tethered 35K protein was able to activate the upstream 3′ss to a similar level as in the wt USSE reporter (Fig. 3H). SRSF1 also provided a similar activation level. In contrast, U1–70K resulted in strong activation of the upstream 3′ss. The strong 3′ss activation seen with the U1–70K is unlikely to result from high expression levels of the tethered protein. SRSF1 and 35K both show similar 3′ss activation levels despite that SRSF1 shows the highest and 35K the lowest expression levels. Rather, it is likely that even the lowest level of protein expression is already sufficient to saturate binding to the λN hairpin reporter. Together, our results suggest that the 35K protein is able to support similar 3′ss activation as other RS-domain containing proteins.
Discussion
In this study, we have investigated an alternative splicing program that regulates the levels of two minor spliceosome specific proteins, 48K and 65K. These proteins are integral components of the U11/U12 di-snRNP and necessary for the initial recognition of U12-type introns.38 We have earlier shown that the core USSE sequence (i.e., the duplicated 5′ss consensus sequence), needed for activation of alternative splicing within these genes, is extraordinarily conserved in plant and animal kingdoms.12 Here, we show that evolutionary constraints have shaped the overall architecture of the regulatory module consisting of an upstream 3′ss and the USSE element. The distance between the upstream 3′ss and USSE falls to a narrow range between ∼30 and 65 nt in different species. Experimental shortening of this distance in mammalian cells leads to a progressive loss of upstream 3′ss use. A similar constraint was also found from the spacer between the two 5′ss sequences within the USSE sequence suggesting that the precise minimal spacing of the 5′ss elements was also necessary for the activity of the USSE element.
The strict phylogenetic conservation of the 3′ss - USSE distance, particularly in tetrapods, supports the conclusion that U11/U12 di-snRNP bound to the USSE sequence uses exon definition interactions to activate the upstream 3′ss. The observed 3′ss-USSE distances are identical in nearly all tetrapods, specifically 44 nt with the 65K genes and 63 nt with 48K the genes and are similar to the earlier reported minimal distance of ∼50 nt for exon-bridging interactions mammalian system.39-41 Consistently, experimental shortening of the 3′ss-USSE distance using human 65K 3′UTR reporter construct leads to progressive loss of the upstream 3′ss activation (Fig. 1C). This observation can be explained by the classical exon definition model where the shortening of the exon size below 50 nt can lead to a steric clash with the spliceosomal complexes binding to the 3′ and 5′ ends of exon.42,43 Together, these result suggest that the position of the USSE element in respect to upstream 3′ss is dictated by the physical constraints imposed by exon definition interactions. In contrast, similar distance constraints are not apparent for canonical splicing activators (i.e. SR-proteins) acting through ESE sequences, as indicated by transcriptome-wide profiles of ESE hexamers within exons44-46 and the SR-protein binding profiles obtained by CLIP-seq for several mammalian47,48 and Drosophila49 SR-proteins.
It is interesting to note that in plants, and to some extend also in fish and insects, there was substantially more variation with the 3′ss-USSE distance distribution compared to the tetrapod species, and more significantly, a clear shift to shorter distances (Fig. 1B). This may reflect the differences in exon definition interactions in these species as compared to mammalian species. Exon definition has been demonstrated in plants,50 but genomic data shows that plant introns are typically shorter than mammalian introns.51 Therefore, it is possible that plants rely more on intron-definition interactions5 and therefore, the number of components involved in the few exon definition interactions may be smaller, which in turn may allow a closer spacing of the spliceosomal complexes at the exon ends.
The most likely candidate within the U11/U12 di-snRNP to mediate the regulatory exon definition interactions is the U11–35K protein, which alone is sufficient to activate the upstream 3′ss when tethered to the canonical USSE element within the mRNA (Fig. 3). Sequence-level analysis of the mammalian and plant system and functional analysis in plants has earlier identified U11–35K as the functional paralog of U1–70K, which, in mammals, is known to mediate exon definition interactions within the major spliceosome. Furthermore, functional analysis of the 35K protein in plants indicated that the 35K protein is able to interact with various SR-proteins.15,31,52,53 Here, our data indicate that the activation of the upstream 3′ss was dependent on the presence of a functional RS domain in the 35K protein, while the 35K RRM alone failed to activate the upstream 3′ss. Similar results were also obtained with the U1–70K protein, with the difference that the U1–70K RS domain was clearly a more potent activator than the 35K protein. Together, the results further indicate that the exon definition interactions between major and minor spliceosome components are similar to those between components of the major spliceosome only. The activation could take place via a direct interaction between 35K protein and U2AF and/or indirectly, mediated by SR proteins as suggested by the data with the Arabidopsis 35K.31 The difference in splicing activation between 35K and U1–70K is not clear. U1–70K has two RS domains with a higher level of SR/ED/DR dipeptide content compared to the 35K protein, both in mammals and in plants,31 but that appears not to be the only determinant of activity, as SRSF1 has a longer and more pronounced RS domain compared to 35K, yet it does not show a similar increase in 3′ss activation as U1–70K.
In addition to the 3′ss-USSE distance, the other conserved distance was the spacer length within the USSE element. We have earlier suggested that the duplication of the U12-type 5′ss sequence in the USSE element may be a compensation mechanism to allow stable recognition of the USSE by the U11/U12 complex.12 Earlier work has shown that the U11/5′ss interaction is significantly stimulated by a simultaneous U12/BPS interaction.17 Because the USSE elements in both the 48K and 65K genes lack a downstream BPS sequence, the duplication of 5′ss-like motifs may be a compensation to allow stable binding of U11 snRNP to the 5′ss-like motifs.12 This could take place by two different mechanisms. Either the duplicated 5′ss sequences provide a higher affinity platform for a binding of a single U11/U12 complex, or alternatively, the two U11/U12 complexes are simultaneously bound to the two 5′ss sequences within the USSE and their binding is stabilized by a yet to be discovered mutual interaction. The experimental and phylogenetic data support the latter possibility. Specifically, spacer lengths of less than 3 nt are not able to fully support the upstream 3′ss activation, suggesting that there is a minimal distance of 3 nt between the two 5′ss-like motifs needed to accommodate the binding of two snRNP complexes to the USSE element. Accordingly, shortening the distance below 3 nt leads to a steric clash or inability of the two RNA-bound complexes to orient themselves in a way that allows mutual stabilizing interactions. Interestingly, given that the minimal distance of 3 nt is much less than the apparent need of ∼40nt for exon definition interactions, these data also suggest that a potential steric clash in exon definition is not due to the size of the U11/U12 di-snRNP complex. Consistently, work on isolated exon definition complexes has demonstrated the association of U4/U6.U5 tri-snRNP with U2 bound to the upstream 3′ss,54 which may explain the observed distance constraints observed in exon definition.
Finally, our experiments show that lengthening of the spacer sequence up to 30 nt leads to only gradual loss of the 3′ss activation. This observation provides a strong support to the hypothesis that two U11/U12 di-snRNPs are needed for stable mRNA binding in the absence of U12/BPS interaction, and further suggests that the two distantly bound snRNPs can maintain the mutual stabilizing interactions, most likely by looping the intervening RNA segment. The apparent contradiction with the phylogenetic data showing relatively strict spacer length distribution can be explained by the requirement for optimal binding activity with the regulatory cassette. However, our data also suggest that there may be additional USSE-like regulatory sequences besides the ones found from the 48K and 65K genes. Earlier genome-wide bioinformatics searches with rather conservative spacer length cutoff failed to identify additional phylogenetically conserved elements,12 but now more such regulatory elements may be awaiting for discovery.
Materials and Methods
Phylogenetic analysis
The USSE sequences were identified from genomic databases by BLAST searches that initially identified the genes encoding for 48K and 65K proteins in these species. These sequences were subsequently subjected to a detailed sequence searcher to identify the USSE elements as described earlier.12 The obtained sequences containing USSE elements were aligned using the MUSCLE algorithm,55 and the lengths of the 3′ss-USSE element and the USSE spacer were determined manually. The list of the species used in this study is presented in Table S1.
Cell culture
HEK293 (Fig. 3D, E, F and G) and CHO (for characterization of USSE distance constraints in Fig. 1 and 2, and for RS domain activity analysis in Fig. 3H) cells were maintained in DMEM supplemented with FBS, penicillin and streptomycin. Transfections were performed using Lipofectamine 2000 following the manufacturer's recommendations. Transfected splicing reporter amounts were kept low so as not to saturate splicing pathways. 100 ng was transfected in 12-well format or 200 ng in 6-well format, and total DNA amounts were adjusted to those recommended in the standard protocol; for the RNA tethering assay, the remaining DNA amounts was made up of λNV5-fusion constructs to ensure sufficient protein expression levels. 24 h after transfection, cells were harvested by trypsinization and each sample was divided into RNA and protein preparations. Trizol reagent was used for RNA extraction. For protein analysis, the cells were suspended in lysis buffer (25 mM Tris-HCl pH 7.5, 100 mM KCl, 1 mM EDTA, 0.5 mM DTT, protease inhibitor cocktail), sonicated 3 × 30 sec using Bioruptor Twin sonicator, and cleared by 10 min 13 000 rpm centrifugation in an Eppendorf centrifuge.
Plasmids
For USSE mutational studies, the pGL4.13-F65K vector was used as a starting point.12 In wt plasmid, the distance between 3′ss and USSE motif is 44 nt, and the distance between 2 U12-type 5′ss of USSE is 6 nt. PCR based mutagenesis was used to modify distances between upstream 3′ss and USSE, and between the two U12-type 5′ss motifs within the USSE.
To manipulate distance between 3′ss and USSE motifs into 34, 24, 14 and 0 nt, plasmids pGL4.13–65K-del10nt, -del20nt, -del30nt, and -del44nt were constructed by deletion of 10, 20, 30, and 44 bp, respectively, in the sequence between 3′ss and USSE element. Plasmids pGL4.13–65K-insUSSE-3nt, −6nt, −9nt, −12nt, −15nt, −18nt, −21nt, −24nt and pGL4.13–65K-delUSSE-1nt, −2nt, −3nt, −4nt, −5nt and −6nt plasmids were constructed by inserting 3, 6, 9, 12, 15, 18, 21, 24 and deleting 1, 2, 3, 4, 5, and 6 nt, respectively, in the sequence between two U12-type 5′ss motifs of USSE. After deletion and insertion, the final distances between two U12-type 5′ss of USSE were 0, 1, 2, 3, 4, 5, 6 (wt), 9, 12, 15, 18, 21, 24, 27 and 30 nt.
For hairpin tethering studies, the USSE element in pGL4.13-F65K vector was replaced with four BoxB hairpin sequences.32 U11–35K, SRSF1 and U1–70K proteins were tagged with V5 and λN epitopes32 and deletion mutants were constructed as follows: U11–35K RRM contained amino acids 1–152; RS, 153–246; U1–70K RRM, 1–230; RS, 231–437. Y94D/F96D mutant was constructed by PCR mutagenesis by changing U11–35K residues Y94 and F96 to aspartates, and R-to-A mutant was created by replacing all arginine residues in the above defined RS domain by alanines using gene synthesis services (GenScript).
Antibodies
Western blotting, immunoprecipitation and immunocytochemistry were performed with monoclonal mouse anti-V5 antibody (R96025 from Life Technologies).
RT-PCR and qPCR
RNA was treated with RQ DNaseI (Promega) and cDNA synthesis was carried out with random primers and RevertAid reverse transcriptase (Fermentas). Primers used in multiplex PCR and qPCR are listed in Table S2. RT-PCR products were resolved on a 3% Metaphor agarose gel. Images were taken by a LAS3000 imager (Fuji) and analyzed by Aida image analyzing software (Raytest). qPCR was performed in a LightCycler 480 Real-Time PCR System (Roche) in 384- well format. PCR efficiencies (E) of the different primer pairs were taken into account and the isoform ratios were calculated using the formula: Short/Long = E(long)Ct(long)/E(short)Ct(short).
Coimmunoprecipitation
Cells were transfected and harvested as above, except IP lysis buffer was 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA and protease inhibitors. Lysate was precleared by incubation with Protein G Dynabeads (Life Technologies) for 1 h, then incubated with fresh beads and mouse anti-V5 (Life Technologies) for 2 h, and washed three times with IP lysis buffer. Bound material was released with Proteinase K treatment at 60 °C for 45 min, phenol-chloroform extracted and run on 8% urea-PAGE. Northern blotting with U11 specific probe was done as described earlier.12
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Marja-Leena Peltonen for technical assistance during this work and the members of Frilander laboratory for stimulating discussions.
Funding
This work was supported by Academy of Finland [140087 to MJF], Biocentrum Helsinki and Sigrid Jusélius Foundation [both to MJF]. EHN was supported by Helsinki Graduate School in Biotechnology and Molecular Biology.
References
- 1.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010; 463:457-63; PMID:20110989; http://dx.doi.org/ 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem 2015; 84:291-323; PMID:25784052; http://dx.doi.org/ 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hastings ML, Krainer AR. Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA 2001; 7:471-82; PMID:11333026; http://dx.doi.org/ 10.1017/S1355838201002552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ke S, Chasin LA. Context-dependent splicing regulation: Exon definition, co-occurring motif pairs and tissue specificity. RNA Biology 2011; 8:384-8; PMID:21444999; http://dx.doi.org/ 10.4161/rna.8.3.14458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA 2013; 4:49-60; PMID:23044818; http://dx.doi.org/ 10.1002/wrna.1140 [DOI] [PubMed] [Google Scholar]
- 6.Fox-Walsh KL, Dou Y, Lam BJ, Hung S-p, Baldi PF, Hertel KJ. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci USA 2005; 102:16176-81; PMID:16260721; http://dx.doi.org/ 10.1073/pnas.0508489102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Q, Krainer AR. U1-mediated exon definition interactions between AT-AC and GT-AG introns. Science 1996; 274:1005-8; PMID:8875927; http://dx.doi.org/ 10.1126/science.274.5289.1005 [DOI] [PubMed] [Google Scholar]
- 8.Hamid FM, Makeyev EV. Emerging functions of alternative splicing coupled with nonsense-mediated decay. Biochemcal Society Transactions 2014; 42:1168-73; http://dx.doi.org/ 10.1042/BST20140066 [DOI] [PubMed] [Google Scholar]
- 9.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 2007; 446:926-9; PMID:17361132; http://dx.doi.org/ 10.1038/nature05676 [DOI] [PubMed] [Google Scholar]
- 10.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O´Brien G, Shiue L, Clark TA, Blume JE, Ares M. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 2007; 21:708-18; PMID:17369403; http://dx.doi.org/ 10.1101/gad.1525507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turunen JJ, Verma B, Nyman TA, Frilander MJ. HnRNPH1/H2, U1 snRNP and U11 snRNP co-operate to regulate the stability of the U11-48K pre-mRNA. RNA 2013; 19:380-9; PMID:23335637; http://dx.doi.org/ 10.1261/rna.036715.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verbeeren J, Niemelä EH, Turunen JJ, Will CL, Ravantti JJ, Lührmann R, Frilander MJ. An ancient mechanism for splicing control: U11 snRNP as an activator of alternative splicing. Mol Cell 2010; 37:821-33; PMID:20347424; http://dx.doi.org/ 10.1016/j.molcel.2010.02.014 [DOI] [PubMed] [Google Scholar]
- 13.Rösel-Hillgärtner TD, Hung L-H, Khrameeva E, Le Querrec P, Gelfand MS, Bindereif A. A Novel Intra-U1 snRNP Cross-Regulation Mechanism: Alternative Splicing Switch Links U1C and U1-70K Expression. PLoS Genet 2013; 9:e1003856; http://dx.doi.org/ 10.1371/journal.pgen.1003856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turunen JJ, Will CL, Grote M, Lührmann R, Frilander MJ. The U11-48K protein contacts the 5′ splice site of U12-type introns and the U11-59K protein. Mol Cell Biol 2008; 28:3548-60; PMID:18347052; http://dx.doi.org/ 10.1128/MCB.01928-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, Elbashir S, Tuschl T, Lührmann R. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 2004; 10:929-41; PMID:15146077; http://dx.doi.org/ 10.1261/rna.7320604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benecke H, Lührmann R, Will CL. The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11-59K protein. EMBO J 2005; 24:3057-69; PMID:16096647; http://dx.doi.org/ 10.1038/sj.emboj.7600765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frilander MJ, Steitz JA. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev 1999; 13:851-63; PMID:10197985; http://dx.doi.org/ 10.1101/gad.13.7.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarn W-Y, Steitz JA. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 1996; 84:801-11; PMID:8625417; http://dx.doi.org/ 10.1016/S0092-8674(00)81057-0 [DOI] [PubMed] [Google Scholar]
- 19.Hall SL, Padgett RA. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 1996; 271:1716-8; PMID:8596930; http://dx.doi.org/ 10.1126/science.271.5256.1716 [DOI] [PubMed] [Google Scholar]
- 20.Niemelä EH, Oghabian A, Staals RHJ, Pruijn GJM, Frilander MJ. Global analysis of the nuclear processing of unspliced U12-type introns by the exosome. Nucleic Acids Res 2014; 42:7358-69; http://dx.doi.org/ 10.1093/nar/gku391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel AA, McCarthy M, Steitz JA. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J 2002; 21:3804-15; PMID:12110592; http://dx.doi.org/ 10.1093/emboj/cdf297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemelä EH, Frilander MJ. Regulation of gene expression through inefficient splicing of U12-type introns. RNA Biol 2014; 11:1325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markmiller S, Cloonan N, Lardelli RM, Doggett K, Keightley M-C, Boglev Y, Trotter AJ, Ng AY, Wilkins SJ, Verkade H, et al.. Minor class splicing shapes the zebrafish transcriptome during development. Proc Natl Acad Sci USA 2014; 111:3062-7; PMID:24516132; http://dx.doi.org/ 10.1073/pnas.1305536111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otake LR, Scamborova P, Hashimoto C, Steitz JA. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol Cell 2002; 9:439-46; PMID:11864616; http://dx.doi.org/ 10.1016/S1097-2765(02)00441-0 [DOI] [PubMed] [Google Scholar]
- 25.Pessa HKJ, Greco D, Kvist J, Wahlström G, Heino TI, Auvinen P, Frilander MJ. Gene expression profiling of U12-type spliceosome mutant Drosophila reveals widespread changes in metabolic pathways. PLoS ONE 2010; 5:e13215; PMID:20949011; http://dx.doi.org/ 10.1371/journal.pone.0013215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Younis I, Dittmar K, Wang W, Foley SW, Berg MG, Hu KY, Wei Z, Wan L, Dreyfuss G. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife 2013; 2:e00780; PMID:23908766; http://dx.doi.org/ 10.7554/eLife.00780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, Touraine R, Tubacher E, Senni F, Bober MB, Nampoothiri S, et al.. Association of TALS Developmental Disorder with Defect in Minor Splicing Component U4atac snRNA. Science 2011; 332:240-3; PMID:21474761; http://dx.doi.org/ 10.1126/science.1202205 [DOI] [PubMed] [Google Scholar]
- 28.He H, Liyanarachchi S, Akagi K, Nagy R, Li J, Dietrich RC, Li W, Sebastian N, Wen B, Xin B, et al.. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 2011; 332:238-40; PMID:21474760; http://dx.doi.org/ 10.1126/science.1200587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Argente J, Flores R, Gutiérrez-Arumí A, Verma B, Martos-Moreno GA, Cuscó I, Oghabian A, Chowen JA, Frilander MJ, Pérez-Jurado LA. Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency. EMBO Mol Med 2014; 6:299-306; PMID:24480542; http://dx.doi.org/ 10.1002/emmm.201303573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, Sanada M, Grossmann V, Sundaresan J, Shiraishi Y, et al.. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nature Communications 2015; 6:6042; PMID:25586593; http://dx.doi.org/ 10.1038/ncomms7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorković ZJ, Lehner R, Forstner R, Barta A. Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA 2005; 11:1095-107; PMID:15987817; http://dx.doi.org/ 10.1261/rna.2440305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gehring NH, Hentze MW, Kulozik AE. Chapter 23 Tethering Assays to Investigate Nonsense‐Mediated mRNA Decay Activating Proteins. In: Lynne EM, Megerditch K, eds. Methods Enzymol: Academic Press, 2008:467-82. [DOI] [PubMed] [Google Scholar]
- 33.Phelan MM, Goult BT, Clayton JC, Hautbergue GM, Wilson SA, Lian L-Y. The structure and selectivity of the SR protein SRSF2 RRM domain with RNA. Nucleic Acids Res 2012; 40:3232-44; PMID:22140111; http://dx.doi.org/ 10.1093/nar/gkr1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Screaton GR, Cáceres JF, Mayeda A, Bell MV, Plebanski M, Jackson DG, Bell JI, Krainer AR. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J 1995; 14:4336-49; PMID:7556075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pessa HKJ, Will CL, Meng X, Schneider C, Watkins NJ, Perälä N, Nymark M, Turunen JJ, Lührmann R, Frilander MJ. Minor spliceosome components are predominantly localized in the nucleus. Proc Natl Acad Sci USA 2008; 105:8655-60; PMID:18559850; http://dx.doi.org/ 10.1073/pnas.0803646105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delestienne N, Wauquier C, Soin R, Dierick J-F, Gueydan C, Kruys V. The splicing factor ASF/SF2 is associated with TIA-1-related/TIA-1-containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression. FEBS Journal 2010; 277:2496-514; PMID:20477871; http://dx.doi.org/ 10.1111/j.1742-4658.2010.07664.x [DOI] [PubMed] [Google Scholar]
- 37.Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Cáceres JF. Nuclear Export and Retention Signals in the RS Domain of SR Proteins. Mol Cell Biol 2002; 22:6871-82; PMID:12215544; http://dx.doi.org/ 10.1128/MCB.22.19.6871-6882.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turunen JJ, Niemelä EH, Verma B, Frilander MJ. The significant other: splicing by the minor spliceosome. Wiley Interdiscip Rev RNA 2013; 4:61-76; PMID:23074130; http://dx.doi.org/ 10.1002/wrna.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berget SM. Exon Recognition in Vertebrate Splicing. J Biol Chem 1995; 270:2411-4; PMID:7852296; http://dx.doi.org/ 10.1074/jbc.270.6.2411 [DOI] [PubMed] [Google Scholar]
- 40.Dominski Z, Kole R. Cooperation of pre-mRNA sequence elements in splice site selection. Mol Cell Biol 1992; 12:2108-14; PMID:1569943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol 1991; 11:6075-83; PMID:1944277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arias MA, Lubkin A, Chasin LA. Splicing of designer exons informs a biophysical model for exon definition. RNA 2015; 21:213-29; PMID:25492963; http://dx.doi.org/ 10.1261/rna.048009.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Black DL. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev 1991; 5:389-402; PMID:2001841; http://dx.doi.org/ 10.1101/gad.5.3.389 [DOI] [PubMed] [Google Scholar]
- 44.Ke S, Shang S, Kalachikov SM, Morozova I, Yu L, Russo JJ, Ju J, Chasin LA. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res 2011; 21:1360-74; PMID:21659425; http://dx.doi.org/ 10.1101/gr.119628.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 2006; 15:2490-508; PMID:16825284; http://dx.doi.org/ 10.1093/hmg/ddl171 [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res 2005; 33:5053-62; PMID:16147989; http://dx.doi.org/ 10.1093/nar/gki810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo Gene W, Ares M Jr, Fu X-D. Genome-wide Analysis Reveals SR Protein Cooperation and Competition in Regulated Splicing. Mol Cell 2013; 50:223-35; PMID:23562324; http://dx.doi.org/ 10.1016/j.molcel.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Änkö M-L, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer K. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 2012; 13:R17; http://dx.doi.org/ 10.1186/gb-2012-13-3-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradley T, Cook ME, Blanchette M. SR proteins control a complex network of RNA-processing events. RNA 2015; 21:75-92; PMID:25414008; http://dx.doi.org/ 10.1261/rna.043893.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson CG, Clark GP, Lyon JM, Watters J, McQuade C, Brown JWS. Interactions between introns via exon definition in plant pre-mRNA splicing. Plant J 1999; 18:293-302; http://dx.doi.org/ 10.1046/j.1365-313X.1999.00463.x [DOI] [Google Scholar]
- 51.Alexandrov N, Troukhan M, Brover V, Tatarinova T, Flavell R, Feldmann K. Features of Arabidopsis Genes and Genome Discovered using Full-length cDNAs. Plant Mol Biol 2006; 60:69-85; PMID:16463100; http://dx.doi.org/ 10.1007/s11103-005-2564-9 [DOI] [PubMed] [Google Scholar]
- 52.Will CL, Schneider C, Reed R, Lührmann R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 1999; 284:2003-5; PMID:10373121; http://dx.doi.org/ 10.1126/science.284.5422.2003 [DOI] [PubMed] [Google Scholar]
- 53.Lorković ZJ, Lopato S, Pexa M, Lehner R, Barta A. Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleoprotein-specific proteins suggest their involvement in pre-mRNA splicing. J Biol Chem 2004; 279:33890-8; PMID:15166240; http://dx.doi.org/ 10.1074/jbc.M400270200 [DOI] [PubMed] [Google Scholar]
- 54.Schneider M, Will CL, Anokhina M, Tazi J, Urlaub H, Lührmann R. Exon Definition Complexes Contain the Tri-snRNP and Can Be Directly Converted into B-like Precatalytic Splicing Complexes. Mol Cell 2010; 38:223-35; PMID:20417601; http://dx.doi.org/ 10.1016/j.molcel.2010.02.027 [DOI] [PubMed] [Google Scholar]
- 55.Edgar R. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004; 5:113; PMID:15318951; http://dx.doi.org/ 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


