ABSTRACT
Circular RNAs (circRNAs) are widely expressed in animal cells, but their biogenesis and functions are poorly understood. CircRNAs have been shown to act as sponges for miRNAs and may also potentially sponge RNA-binding proteins (RBPs) and are thus predicted to function as robust posttranscriptional regulators of gene expression. The joint analysis of large-scale transcriptome data coupled with computational analyses represents a powerful approach to elucidate possible biological roles of ribonucleoprotein (RNP) complexes. Here, we present a new web tool, CircInteractome (circRNA interactome), for mapping RBP- and miRNA-binding sites on human circRNAs. CircInteractome searches public circRNA, miRNA, and RBP databases to provide bioinformatic analyses of binding sites on circRNAs and additionally analyzes miRNA and RBP sites on junction and junction-flanking sequences. CircInteractome also allows the user the ability to (1) identify potential circRNAs which can act as RBP sponges, (2) design junction-spanning primers for specific detection of circRNAs of interest, (3) design siRNAs for circRNA silencing, and (4) identify potential internal ribosomal entry sites (IRES). In sum, the web tool CircInteractome, freely accessible at http://circinteractome.nia.nih.gov, facilitates the analysis of circRNAs and circRNP biology.
KEYWORDS: CircRNA-miRNA, CLIP-Seq, circRNA siRNA, circRNA IRES, divergent primer design, RNA-binding proteins, sponge circRNAs, transcriptome
Introduction
Circular RNAs (circRNAs) are widely expressed RNAs lacking 5′ and 3′ ends, forming instead covalently closed RNA loops. In eukaryotes, circRNAs arise most often through backsplicing, a process in which the 5' and 3' ends of a spliced RNA are covalently linked to form a closed RNA molecule that contains exon and/or intron sequences.1-3 Although a few examples of circRNAs have been known for several decades, only with the recent widespread use of high-throughput RNA sequencing and bioinformatics have we learned that circRNAs constitute a vast class of stable RNAs expressed endogenously in cells, often with tissue-specific patterns.1-5 The functional roles of circRNAs are not as well established as those of other noncoding (nc)RNAs such as microRNAs (miRNAs). However, one prominent mechanism whereby circRNAs are believed to function is by sponging miRNAs, sequestering them away from protein-coding mRNAs.2 It has also been postulated that circRNAs could serve as sponges for RNA-binding proteins (RBPs), platforms for assembly of RBPs, and protein-coding templates for translation.6
RBPs control all stages of post-transcriptional gene expression, including the splicing, export, turnover, translation, and localization of mRNAs.7-9 By modulating gene expression, RBPs play key roles in virtually all cellular processes – proliferation, differentiation, motility, senescence, apoptosis, as well as the cellular responses to stresses, mitogens, and immune triggers.10,11 Consequently, RBPs have been implicated in a wide range of human diseases such as cancer, muscle pathologies, and neurodegenerative conditions.12-16 Recent developments in the analysis in RNA-protein (RNP) interactions using cross-linking techniques such as CLIP (cross-linking immunoprecipitation), PAR-CLIP (photoactivatable-ribonucleoside-enhanced CLIP), HITS-CLIP (high-throughput sequencing CLIP), and iCLIP (individual-nucleotide-resolution CLIP) have identified with unprecedented precision the binding sequences of RBPs on target RNAs.17-20
MicroRNAs (miRNAs) comprise another major class of posttranscriptional regulators. They bind thousands of human transcripts through partial complementarity and generally reduce their expression by suppressing mRNA translation and/or stability.21-23 In some cases miRNAs may enhance mRNA translation.24,25 MicroRNAs are initially synthesized as primary (Pri)-microRNA transcripts, which are processed by the microprocessor complex into microRNA precursors (Pre)-microRNAs.26-29 Pre-microRNAs are then exported to the cytoplasm where they are further processed by DICER1 and loaded onto the RNA-inducible silencing complex (RISC) to be directed to target mRNAs.28-31
RBPs and miRNAs can bind the same mRNA in a variety of functional manners; for example, they can compete, cooperate, or bind sequentially to a given mRNA.32-34 Such corregulatory effects by RBPs and miRNAs influence physiologic processes as well as disease.35,36 Thus, it is important to understand in detail their interaction with mRNAs. Several databases are available which facilitate research on RBPs (e.g., starBase) and miRNAs (e.g., TargetScan) interacting with specific mRNAs. Here, we have developed a new computational resource named ‘CircInteractome’ which enables researchers to search systematically for possible interactions of circRNAs with RBPs and miRNAs. This tool will greatly facilitate the identification of circRNAs that can sponge miRNAs and/or RBPs and will enhance the search for circRNAs with potential function as platforms for the assembly of RBPs. Additional features such as the ability to display the mRNA counterpart and the genomic and mature sequences of the circRNA, as well as the ability to design specific circRNA divergent primers and circRNA-directed siRNAs, are aimed at further facilitating other aspects of circRNA research. The CircInteractome tool currently searches against Homo sapiens databases but will be expanded to include other species in the future.
Results and discussion
CircRNA-wide mapping of RBPs and RBP ‘super-sponges’
Published CLIP datasets do not specify if the RBP binding sites are present in linear or circular RNAs (unless the CLIP hit spans a junctional sequence). Thus, we utilized CLIP datasets as indicated in the workflow (Fig. 1) to create a comprehensive binding map of RBPs to circRNAs (Fig. 2). We integrated 93 independently reported CLIP datasets from various RBPs (Table S1) obtained from different tissues and cell lines.37 Computational analyses revealed that for select RBPs there were large numbers of binding sites in circRNA sequences (Table S2); for instance, we identified ˜117,000 circRNAs that could potentially associate with the RBP EIF4A3 (Fig. S1A). Analysis of other RBPs using CircInteractome indicated that they could also potentially interact with numerous circRNAs (Fig. S1A). For example, the mature circRNA hsa_circ_0000020 hosts multiple binding sites for several RBPs like HuR (6 sites) and FMRP (10 sites) (Fig. 2A, B). Thus, we hypothesized that circRNAs with relatively high density of binding sites for any single RBP could potentially act as a ‘sponge’ or a ‘decoy’ for that RBP. This sponging function would be enhanced by the long half-lives of circRNAs. By extension, circRNAs with exceptionally high density of binding sites for a given RBP might be considered to be ‘super-sponges’; for example, hsa_circ_0024707 (428 nt long) could function as a super-sponge for AGO2, since AGO2 can potentially bind this relatively short circRNA at 85 predicted positions (Table S2). In sum, this tool facilitates the search for potential RBPs interacting with circRNAs, and can identify possible RBP sponges as indicated in Figure 2 and Figure S3.
Figure 1.
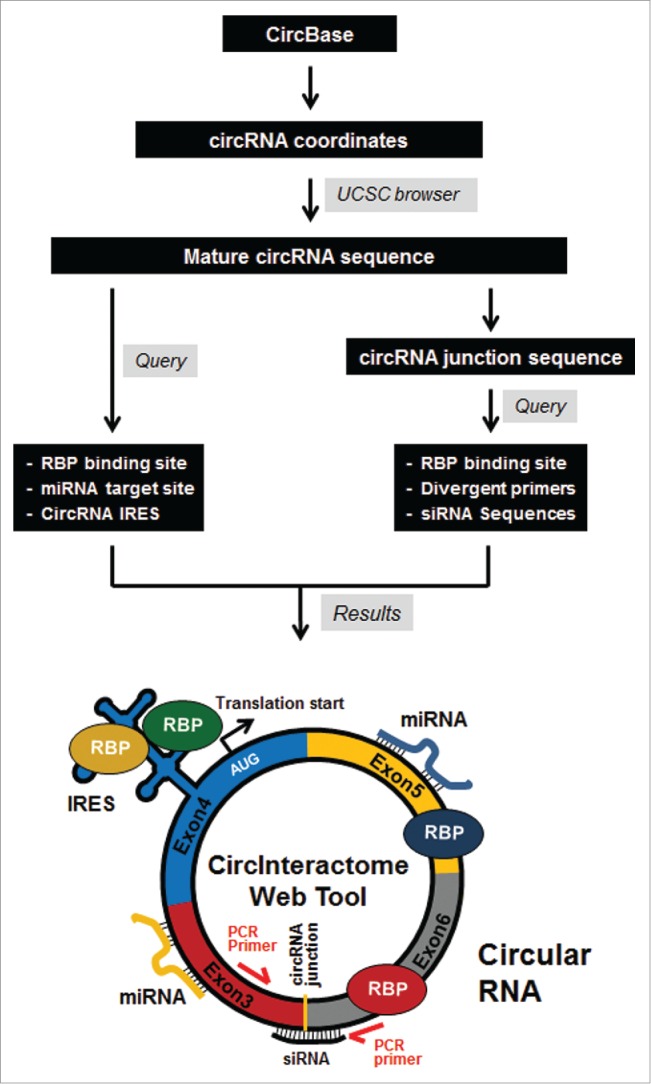
Workflow of the web tool Circular RNA Interactome or ‘CircInteractome’.
Figure 2.
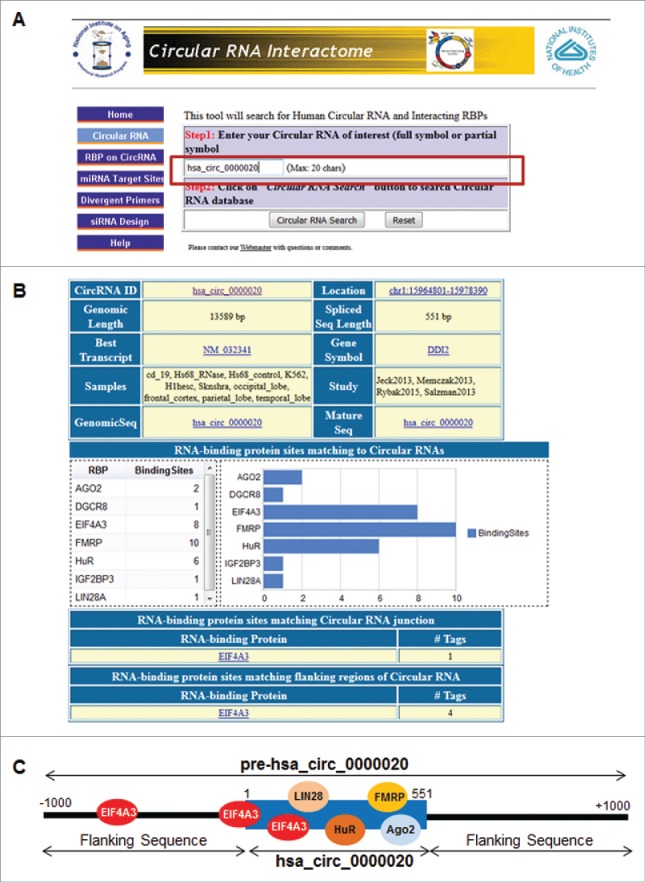
View of CircInteractome input and output pages. A. Example the ‘Circular RNA’ page exhibiting the input parameters needed for a CircInteractome run. B. Screenshot of ‘Circular RNA search’ for has_circ_0000020, showing RBPs binding in different regions of this circRNA. C. Schematic representation of the potential binding sites of different RBPs to mature and pre-hsa_circ_0000020.
RBPs on circRNA junctions
RBPs may also interact with circRNA junctions and play a role in circRNA splicing, processing, folding, stabilization, and localization. To test this possibility, we queried CircInteractome for possibly binding sites for RBPs at select sequences spanning 100 nt upstream and downstream from the junction site (Fig. 2B). Computational analysis of the RBP binding sites from various datasets (Table S1) revealed that RBPs may indeed interact with circRNA junctions. For instance, EIF4A3 targets junction sequences with much higher frequency than the frequency seen for targeting the body of mature circRNAs (Fig. S1B). This suggests that EIF4A3 could have a preference for binding to circRNA junctions compared to other RBPs (Fig. S1B). An example of this type of search for hsa_circ_0000020 is shown in Figure 2B and Figure S4A.
Mapping binding sites of RBPs on pre-circRNA
The splicing machinery can generate circRNAs through non-linear back-splicing, which joins 5′ and 3′ ends covalently to make a circRNA.38 Splicing events are tightly regulated by RBPs and snRNAs binding near the splice sites.39 Thus, we used CircInteractome to search all datasets (Table S1) in order to identify the binding sites of RBPs in the flanking sequences upstream and downstream of the mature circRNA. For instance, the flanking sequences on both sides of the hsa_circ_0000020 junction indicated 4 binding sites for EIF4A3 (Fig. 2C, bottom; Fig. S4B), suggesting a possible role for EIF4A3 in the biogenesis of hsa_circ_0000020. Our analysis also uncovered a relatively higher frequency of putative binding sites for splicing factors like TDP43 at the flanking sequences of circRNAs, which point to a possible role for these RBPs in circRNA splicing/biogenesis (Fig. S1C).
Potential circRNA translation though IRES
CircRNAs are believed to not be translated. While most circRNAs do not appear to be associated with polyribosomes, the possibility exists that some circRNAs might be translated into protein products.3 In this regard, linear long noncoding (lnc)RNAs are not generally translated into proteins, but a subset of them appear to be a source of functional small or micropeptides due to the presence of short open reading frames (ORFs).40 For example, a micropeptide encoded by a noncoding RNA was recently found to regulate muscle performance.41 In addition, the viral circRNA CCC (covalently closed circular, 220 nt long) was recently found to be fully translated into a 16-kDa highly basic protein in infected rice plants.42 Bioinformatic analysis revealed the presence of IRES (internal ribosome entry sites) and sites for RBPs that regulate IRES-mediated translation (IRES trans-acting factors or ITAFs) in several circRNA sequences (Table S3).43 For example, we found that hsa_circ_0041407 contains an IRES (underlined sequence) and partial coding sequences of MAX network transcriptional repressor (MNT; Fig. 3A, B) and postulated that hsa_circ_0041407 could give rise to a small chimeric protein of ∼31 kDa (highlighted sequence, Fig. 3B). To our surprise, IRES regions of circRNAs are predicted sites for many RBPs, including HuR and PTB, 2 proteins reported to modulate IRES-driven translation.43 Together, these findings suggest that circRNAs could be translated through IRES sequences, and thus IRES-bearing circRNAs detected in association with polysomes warrant future exploration.
Figure 3.
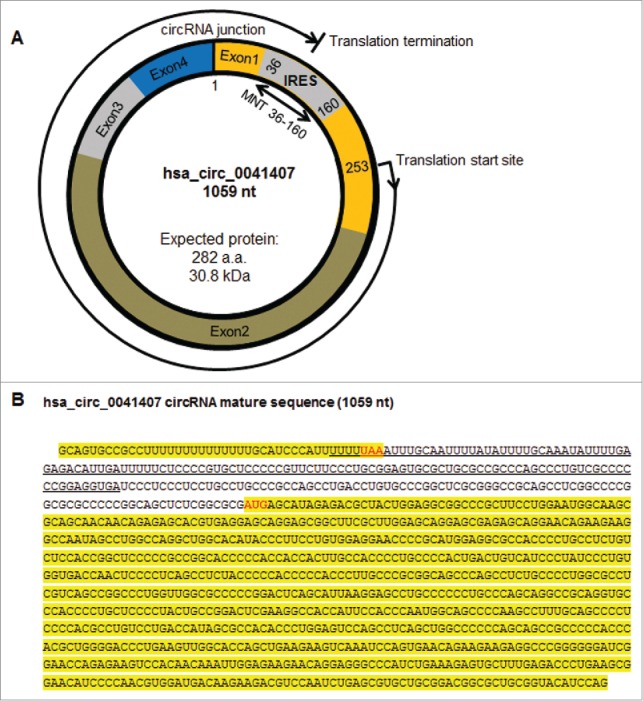
Predicted IRES possibly mediating circRNA translation. A. Schematic representation of hsa_circ_0041407, which contains the MNT 36-160 IRES that may potentially drive translation. B. The sequence of hsa_circ_0041407 highlighted can potentially be translated via the IRES underlined.
miRNA ‘super-sponge’ circRNAs
The main function described for circRNAs thus far in the literature is that of miRNA sponges, as shown for the circRNA ciRS-7, which has multiple binding sites for miR-7.5 To characterize miRNA-circRNA interactions (Fig. 4, Fig. S5), we incorporated into CircInteractome the ability to search using the TargetScan algorithm, which predicts miRNAs that target circRNA by surveying for 7-mer or 8-mer complementarity to the seed region as well as the 3′ end of each miRNA.46 A survey of miRNA target sites in circRNAs revealed the presence of numerous target sites for a specific miRNA. In cases of exceptionally high hit numbers, a circRNA could be considered a ‘super-sponge’ for microRNAs. Strikingly, over 3,000 circRNAs were found to have at least 20 miRNA target sites in a single circRNA, and most of them had AGO2 binding sites (Table S4). For example, hsa_circ_0139850, which is only 437 nt long, has 22 sites for miR-7 and thus might act a super-sponge for miR-7 (Fig. 4). The general finding that circRNAs often have more miRNA target sites than those predicted by chance lends further support to the idea that binding microRNAs could be one of the key functions of circRNAs.
Figure 4.
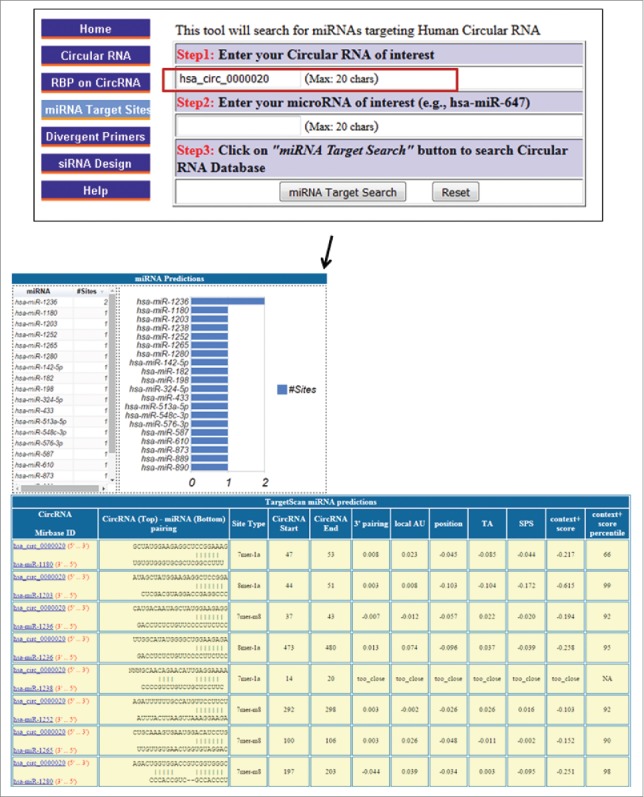
Potential miRNA-circRNA interactions. For a given circRNA ID entered, the miRNAs potentially targeting that circRNA are identified.
Divergent primer design
Designing specific primers for quantification of circRNA using qPCR amplification can be challenging and prone to errors, since the mature circRNA sequences after splicing are not readily available in many cases and the primers must be divergent and must span the junction. At present, there is no software or web server that enables direct design of divergent primers specific to circRNAs. Thus, we incorporated primer design tools into CircInteractome (Primer345 or NCBI primer design tool) and used as template the sequence around the circRNA junction to ensure that the circRNA was amplified specifically by reverse transcription (RT) followed by real-time quantitative (q)PCR analysis (Fig. 5A). To illustrate this feature of CircInteractome, we show the design of divergent primers at the junction sequence of hsa_circ_0000020 (Fig. 5B; Fig. S6A,B).
Figure 5.
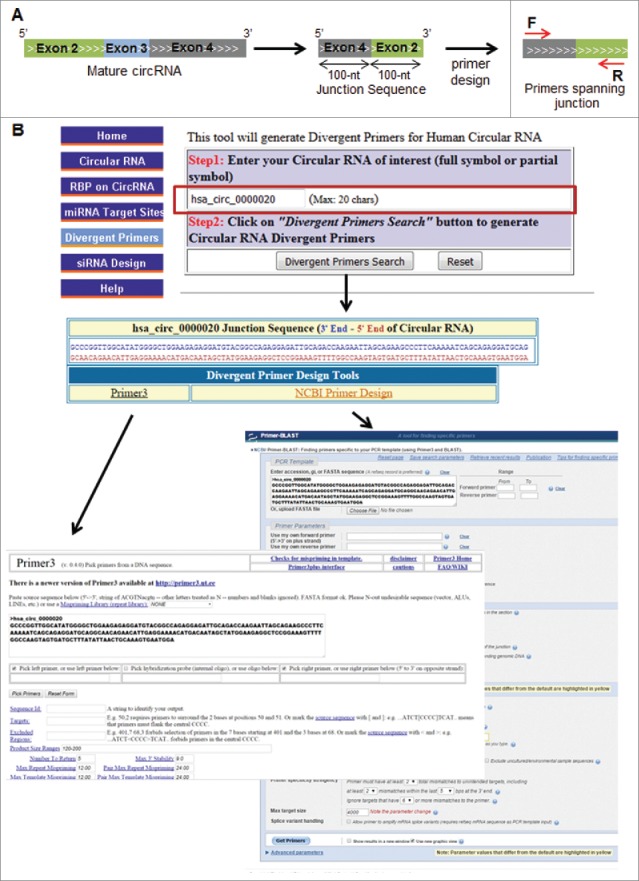
CircRNA primer design. A. Schematic representation of the design of divergent primers for circRNA detection by RT-qPCR analysis. B. Input webpage for divergent primer design. C. Screenshot of the output page showing the junction sequence of circRNA and links of primer design tools (Primer3 and NCBI).
siRNA design
Designing siRNAs selectively directed at circRNAs is challenging due to the limited knowledge of the junctional sequences and the lack of appropriate software tools to help design them. Thus, we incorporated into CircInteractome the ability to generate 21-nt siRNAs targeting junctional sequences spanning 5–16 nt on either side of the junction. Using standard siRNA design criteria, we filtered junctional sequences to find the best siRNA target sequence46,47; users need to avoid siRNAs with long stretches rich in G/C, and confirm using BLAST that the siRNAs are specific. For better silencing effects, the output siRNAs may be ordered with an additional 2 nucleotides (dTdT) as 3′ DNA overhangs (Fig. S7). Users may then purchase them from any source; links to forms to complete siRNA orders using IDT or Dharmacon are provided. Examples of the 10 best possible siRNA target sequences for hsa_circ_0000020 are shown (Fig. 6). In short, CircInteractome can be used to predict siRNA sequences targeting the circRNA junctions.
Figure 6.
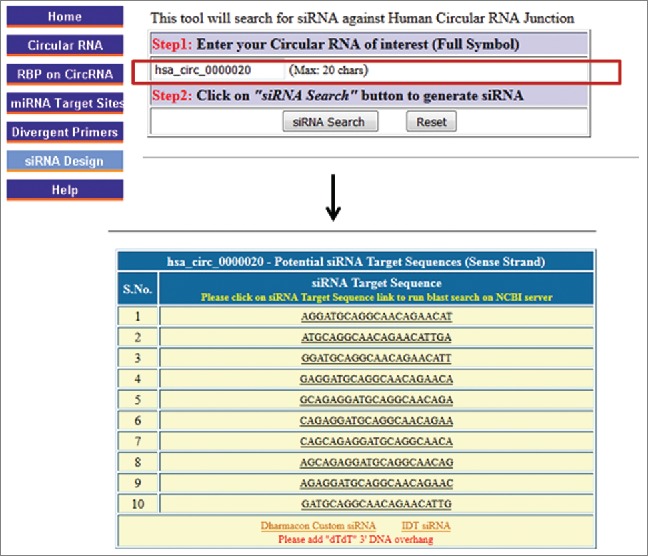
CircRNA siRNA design. Screenshot of the CircInteractome siRNA design page, including an example of output siRNAs targeting the junction sequence of a given circRNA.
Conclusions and future directions
This freely available web service can facilitate the study of circular RNAs and their interactions with other binding factors, mainly RBPs and miRNAs. CircInteractome provides researchers with valuable detail about circular RNAs and their possible role in sequestering RBPs and/or miRNAs and thereby reducing their availability for mRNAs. CircInteractome also facilitates the design of primers for studying circRNA by RT-qPCR analysis. CircInteractome can be used to predict RBP binding to upstream and downstream sequences of the pre-spliced transcript, thus potentially shedding light into the biogenesis of circRNAs. The interaction of translation regulators (miRNAs, RBPs, ITAFs) and the presence of IRESs in circRNAs can help to decipher whether circRNAs have protein/peptide-coding capabilities.
CircInteractome incorporates several features from other freely available web resources, such as circBase, StareBase 2.0, TargetScan 7.0, and Primer3. By integrating these resources, CircInteractome enables the user to find out the genomic and mature circRNA sequences (Fig. S2), circRNA-binding partners (RBPs, miRNAs), siRNAs, and primers to study circRNA levels, localization, and function.
Although CircInteractome has many user-friendly features for circRNA researchers, it is limited in its ability to predict RBP and miRNA interactions when circRNAs form secondary or tertiary structures. Given that all of the data provided here are predicted based on sequence matches and the presence of secondary or tertiary structures in circRNA cannot be considered systematically, experimental validation is essential to verify RBP and miRNA functional sites.
We plan to maintain, update and curate CircInteractome in the foreseeable future, and will include additional RBPs, microRNAs, and circRNAs as they become available. Since RBP and miRNA sites may be masked or revealed depending on whether the RNA is single- or double-stranded, we will also include information on secondary structure as it is reported. In addition, experimental validation of circRNAs interacting with RBPs and microRNAs will also be included in CircInteractome. We will also integrate circRNAs from other species such as mouse and monkey, and will include predictions of RNA hybrids including circRNA:mRNA and circRNA:lncRNA. Collectively, this resource will accelerate our efforts to understand the roles of circRNAs in biological processes relevant to health and disease.
Methods
Acquisition of circular RNA sequences
CircBase (http://www.circbase.org) is a public database that was developed to gather unified datasets of circRNA IDs, genomic coordinates and best transcripts.48 The mature sequences of all of the reported circRNAs were downloaded from the UCSC browser mirror (http://genome.mdc-berlin.de) at the Max-Delbrück-Centrum für Molekulare Medizin (MDC), Berlin, Germany.49A complete list of all human circRNAs used in this study is available in the CircInteractome website. We also retrieved 1000 bp upstream and downstream of circRNA from the UCSC genome browser mirror as circRNA flanking regions. We used 100 nucleotides from the circRNA 3′ and joined them to 100 nucleotides at the 5′ to identify the junctional sequence. For circRNAs shorter than 200 nt, we divided the RNA sequence in to two equal halves and the first half was joined at the 3′ of the second half to generate the template for primer design (Fig. 5A). This dataset was used for sequence alignment with RBP binding sites.
Acquisition of binding sites of RBPs on circRNAs
Datasets including the binding sites of 35 RBPs (Table S1) identified by PAR-CLIP, HITS-CLIP, or iCLIP were retrieved from starBase v2.0 (http://starbase.sysu.edu.cn/),37 UCSC browser, and published datasets. A complete list of all datasets used in the study is shown in Table S2. Data were previously analyzed and thus we used these datasets for our analysis without modification. The sequences for the RBP CLIP clusters were downloaded from the UCSC browser mirror (http://genome.mdc-berlin.de)49 and were analyzed against circRNA mature, flanking, and junction sequences of circRNAs with 95% sequence homology and maximum of 2 mismatches to find their binding sites (Fig. 1). When more than one tag was predicted at the same site for an RBP, only one tag was considered as the binding sequence.
IRES dataset
The sequences of reported IRES were downloaded from the IRESite (http://iresite.org/), which contains information about experimentally validated IRES sequences.50 These IRES sequences were analyzed against mature circRNA sequences to find out the presence of IRES in circRNAs; 99% similarity between the IRES sequence and the mature circRNA sequence was allowed (Fig. 1). IRES sequences present in circRNAs were further analyzed against the RBP CLIP cluster sequences to identify binding sites of different RBPs in the IRES sequences of circRNAs (Fig. 1).
miRNA Target ciRNAs
Mature sequences of circRNAs were used in the TargetScan Perl Script to predict the miRNAs which have sequence complementarity with circRNA.44 The complete miRNA list and sequences were taken from the microRNA database (http://www.mirbase.org/).51
Divergent primer design
The circRNA junction sequences were retrieved from the mature circRNA sequences and exported to primer design tools (Primer3 or NCBI) to give maximum of 5 sets of primer pairs for PCR products of sizes ranging from 120–200 bp and spanning the junction (Fig. S6A, B). For circRNAs smaller than 200 nt, users are given a choice for the length of PCR amplicon based on circRNA length.
CircRNA siRNA design
siRNAs spanning circRNA junction were designed using criteria for 21-nt siRNAs described previously.46,47 To design the 21-nt siRNA target sequence spanning the junction, we used a minimum of 5 nt on either side of the junction.
Availability
The database is freely accessible through the web server at http://circinteractome.nia.nih.gov.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
This research was supported in full by the National Institute on Aging Intramural Research Program of the National Institutes of Health.
References
- 1.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell type-specific features of circular RNA expression. PLoS Genet 2013; 9:e1003777; PMID:24039610; http://dx.doi.org/ 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; http://dx.doi.org/ 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 3.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141-57; PMID:23249747; http://dx.doi.org/ 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7:e30733; PMID:22319583; http://dx.doi.org/ 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; http://dx.doi.org/ 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 6.Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. EMBO J 2013; 32:923-5; PMID:23463100; http://dx.doi.org/ 10.1038/emboj.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 2004; 24:5534-47; PMID:15169913; http://dx.doi.org/ 10.1128/MCB.24.12.5534-5547.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007; 131:174-87; PMID:17923096; http://dx.doi.org/ 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 9.Blencowe BJ. Alternative splicing: New insights from global analyses. Cell 2006; 126:37-47; PMID:16839875; http://dx.doi.org/ 10.1016/j.cell.2006.06.023 [DOI] [PubMed] [Google Scholar]
- 10.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008; 582:1977-86; PMID:18342629; http://dx.doi.org/ 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 2008; 389:243-55; PMID:18177264; http://dx.doi.org/ 10.1515/BC.2008.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. WIRES RNA 2010; 1:214-29; PMID:21935886; http://dx.doi.org/ 10.1002/wrna.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao FB, Taylor JP. RNA-binding proteins in neurological disease. Brain Res 2012; 1462:1-2; PMID:22682432; http://dx.doi.org/ 10.1016/j.brainres.2012.05.038 [DOI] [PubMed] [Google Scholar]
- 14.Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Syst Biol RNA Binding Proteins 2014; 825:1-55; PMID:25201102; http://dx.doi.org/23415593 10.1007/978-1-4939-1221-6_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet 2013; 29:318-27; PMID:23415593; http://dx.doi.org/ 10.1016/j.tig.2013.01.004 [DOI] [PubMed] [Google Scholar]
- 16.Abdelmohsen K, Gorospe M. RNA-binding protein nucleolin in disease. RNA Biol 2012; 9:799-808; PMID:22617883; http://dx.doi.org/ 10.4161/rna.19718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science 2003; 302:1212-5; PMID:14615540; http://dx.doi.org/ 10.1126/science.1090095 [DOI] [PubMed] [Google Scholar]
- 18.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010; 141:129-41; PMID:20371350; http://dx.doi.org/ 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al.. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008; 456:464-U22; PMID:18978773; http://dx.doi.org/ 10.1038/nature07488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 2010; 17:909-U166; PMID:20601959; http://dx.doi.org/ 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15-20; PMID:15652477; http://dx.doi.org/ 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 22.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science 2004; 304:594-6; PMID:15105502; http://dx.doi.org/ 10.1126/science.1097434 [DOI] [PubMed] [Google Scholar]
- 23.Abdelmohsen K, Hutchison ER, Lee EK, Kuwano Y, Kim MM, Masuda K, Srikantan S, Subaran SS, Marasa BS, Mattson MP, et al.. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol Cell Biol 2010; 30:4197-210; PMID:20584986; http://dx.doi.org/ 10.1128/MCB.00316-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasudevan S, Tong YC, Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007; 318:1931-4; PMID:18048652; http://dx.doi.org/ 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- 25.Panda AC, Sahu I, Kulkarni SD, Martindale JL, Abdelmohsen K, Vindu A, Joseph J, Gorospe M, Seshadri V. miR-196b-mediated translation regulation of mouse insulin2 via the 5 ' UTR. PLoS ONE 2014; 9:e101084; PMID:25003985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004; 23:4051-60; PMID:15372072; http://dx.doi.org/ 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 2006; 13:1097-101; PMID:17099701; http://dx.doi.org/ 10.1038/nsmb1167 [DOI] [PubMed] [Google Scholar]
- 28.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009; 10:126-39; PMID:19165215; http://dx.doi.org/ 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- 29.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev 2010; 24:1086-92; PMID:20516194; http://dx.doi.org/ 10.1101/gad.1919710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Ann Rev Biochem 2010; 79:351-79; PMID:20533884; http://dx.doi.org/ 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Prot Pept Sci 2012; 13:372-9; PMID:22708488; http://dx.doi.org/ 10.2174/138920312801619394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol 2011; 31:4219-31; PMID:21859890; http://dx.doi.org/ 10.1128/MCB.05955-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Gen Dev 2009; 23:1743-8; PMID:19574298; http://dx.doi.org/ 10.1101/gad.1812509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciafre SA, Galardi S. microRNAs and RNA-binding proteins A complex network of interactions and reciprocal regulations in cancer. RNA Biol 2013; 10:935-43; PMID:23696003; http://dx.doi.org/ 10.4161/rna.24641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agami R. microRNAs, RNA binding proteins and cancer. Eur J Clin Invest 2010; 40:370-4; PMID:20486997; http://dx.doi.org/ 10.1111/j.1365-2362.2010.02279.x [DOI] [PubMed] [Google Scholar]
- 37.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nuc Acids Res 2014; 42:D92-D7; PMID:24297251; http://dx.doi.org/ 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014; 20:1829-42; PMID:25404635; http://dx.doi.org/ 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet 2011; 27:89-97; PMID:21232811; http://dx.doi.org/ 10.1016/j.tig.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al.. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 2014; 33:981-93; PMID:24705786; http://dx.doi.org/ 10.1002/embj.201488411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, et al.. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015; 160:595-606; PMID:25640239; http://dx.doi.org/ 10.1016/j.cell.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.AbouHaidar MG, Venkataraman S, Golshani A, Liu BL, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci USA 2014; 111:14542-7; PMID:25253891; http://dx.doi.org/ 10.1073/pnas.1402814111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Salas E, Lozano G, Fernandez-Chamorro J, Francisco-Velilla R, Galan A, Diaz R. RNA-binding proteins impacting on internal initiation of translation. Intl J Mol Sci 2013; 14:21705-26; PMID:24189219; http://dx.doi.org/ 10.3390/ijms141121705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007; 27:91-105; PMID:17612493; http://dx.doi.org/ 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3-new capabilities and interfaces. Nuc Acids Res 2012; 40; PMID:22730293; http://dx.doi.org/15215364 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngoc BT, Ho TB, Saori K. A Descriptive method for generating siRNA design rules. Lect Notes Comput Sc 2013; 7803:196-205; http://dx.doi.org/ 10.1007/978-3-642-36543-0_21 [DOI] [Google Scholar]
- 47.Naito Y, Yamada T, Ui-Tei K, Morishita S, Saigo K. siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nuc Acids Res 2004; 32:W124-W9; PMID:15215364; http://dx.doi.org/ 10.1093/nar/gkh442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014; 20:1666-70; PMID:25234927; http://dx.doi.org/ 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al.. The UCSC genome browser database: update 2009. Nuc Acids Res 2009; 37:D755-D61; PMID:18996895; http://dx.doi.org/ 10.1093/nar/gkn875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mokrejs M, Masek T, Vopalensky V, Hlubucek P, Delbos P, Pospisek M. IRESite-a tool for the examination of viral and cellular internal ribosome entry sites. Nuc Acids Res 2010; 38:D131-D6; PMID:19917642; http://dx.doi.org/ 10.1093/nar/gkp981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nuc Acids Res 2014; 42:D68-D73; PMID:24275495; http://dx.doi.org/ 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


