ABSTRACT
The generation of mature mRNA in the protozoan parasite Trypanosoma brucei requires coupled polyadenylation and trans splicing. In contrast to other eukaryotes, we still know very little on components, mechanisms, and dynamics of the 3′ end-processing machinery in trypanosomes. To characterize the catalytic core of the polyadenylation complex in T. brucei, we first identified the poly(A) polymerase [Tb927.7.3780] as the major functional, nuclear-localized enzyme in trypanosomes. In contrast, another poly(A) polymerase, encoded by an intron-containing gene [Tb927.3.3160], localizes mainly in the cytoplasm and appears not to be functional in general 3′ end processing of mRNAs. Based on tandem-affinity purification with tagged CPSF160 and mass spectrometry, we identified ten associated components of the trypanosome polyadenylation complex, including homologues to all four CPSF subunits, Fip1, CstF50/64, and Symplekin, as well as two hypothetical proteins. RNAi-mediated knockdown revealed that most of these factors are essential for growth and required for both in vivo polyadenylation and trans splicing, arguing for a general coupling of these two mRNA-processing reactions.
KEYWORDS: CPSF, mRNA processing, polyadenylation, poly(A) polymerase, trans splicing, Trypanosoma brucei
Introduction
The expression of polycistronic protein-coding genes in the protozoan parasite Trypanosoma brucei requires coupling of the two major mRNA processing steps, trans splicing and polyadenylation, to produce mature mRNAs. The protein components, the mechanisms and dynamics of both processes and in particular their mechanistic linkage are still not well characterized. Although studies in the last decade have identified numerous spliceosomal components, we still know very little about the composition and functioning of the polyadenylation machinery in trypanosomes.1,2,3
In higher eukaryotes the polyadenylation complex consists of a multitude of proteins, which assemble in an orchestrated manner on their target RNAs. The specificity of 3′ end processing of newly transcribed RNA is driven by two cis-acting elements: the highly conserved AAUAAA hexanucleotide, located 10–30 nucleotides upstream of the pre-mRNA cleavage site, and a U- or GU-rich sequence around 30 nucleotides downstream of the cleavage site.4,5 First, the cleavage and polyadenylation specificity factor (CPSF) recognizes the polyadenylation signal, followed by binding of the cleavage stimulation factor (CstF) to the U/GU-rich sequence. Recruitment of poly(A) polymerase as well as other factors such as cleavage factor I and II (CFI and CFII) completes complex formation. After cleavage, mediated by the CPSF73 subunit, which acts as the endonuclease, the poly(A) tail is elongated by the poly(A) polymerase, first in a distributive, and after binding of the poly(A) binding protein (PABP) to the nascent mRNA, in a processive manner, resulting in poly(A) tail extension to approximately 250 nucleotides.6,7
In trypanosomes most of the individual components of the polyadenylation machinery remain elusive. Only three subunits of the CPSF complex, CPSF30, CPSF73, and Fip1, have been identified as orthologues of their mammalian counterparts.3,8,9 However, even for the poly(A) polymerase, in other eukaryotes probably the best delineated component, definitive identification and biochemical characterization in trypanosomes are still missing. In T. brucei, at least two genes code for putative poly(A) polymerases [Tb927.3.3160 and Tb927.7.3780], the first of which is peculiar in that it represents one of the two genes in trypanosomes that require cis splicing: Its two exons are interrupted by an intronic sequence.10 However, recent studies support the hypothesis that in trypanosomes cis splicing might not be essential for survival, questioning whether the poly(A) polymerase from the intron-containing gene in fact represents a functional enzyme.11
In addition, the polyadenylation signal itself is poorly defined in trypanosomes, since the two canonical cis-acting elements, AAUAAA and the U/GU-rich sequence, are not conserved. Previous studies indicated that a polypyrimidine tract upstream of a trans splice site affects polyadenylation of the upstream gene and trans splicing of the downstream gene.12-14 Regulation of both individual RNA-processing reactions through a shared element may underline the close coupling of the two mechanisms. Recently, initial suggestive evidence of a physical linkage of the trans splicing and polyadenylation machineries was obtained, based on copurification of the polyadenylation factor CPSF73 with the spliceosomal U1 snRNP protein U1A.3 However, a biochemical linkage of both mechanisms has not been fully described yet.
Here we report the identification and initial characterization of a major functional poly(A) polymerase in procyclic trypanosomes. By assaying for non-specific in vitro polyadenylation activity, only the putative, nuclear-localized poly(A) polymerase Tb927.7.3780 showed in vitro activity, whereas the cytoplasmic poly(A) polymerase Tb927.3.3160, expressed from an intron-containing gene, does not appear to be functional under these conditions. Moreover, by measuring the in vivo polyadenylation status of mRNAs, knockdown of only the nuclear-localized poly(A) polymerase [Tb927.7.3780], but not of the other putative enzyme [Tb927.3.3160], resulted in a significant shortening of poly(A) tails. In addition, searching for the individual factors of the trypanosomatid polyadenylation complex, we identified by mass spectrometry ten components: CPSF160/100/73/30, Fip1, CstF50/64, Symplekin, and two hypothetical proteins. RNAi-mediated knockdowns of the individual factors identified in our study revealed that most of them are essential for growth and for achieving normal poly(A) tail length as well as for trans splicing, strongly supporting a general coupling mechanism of both mRNA-processing reactions.
Results
The trypanosomatid poly(A) polymerases
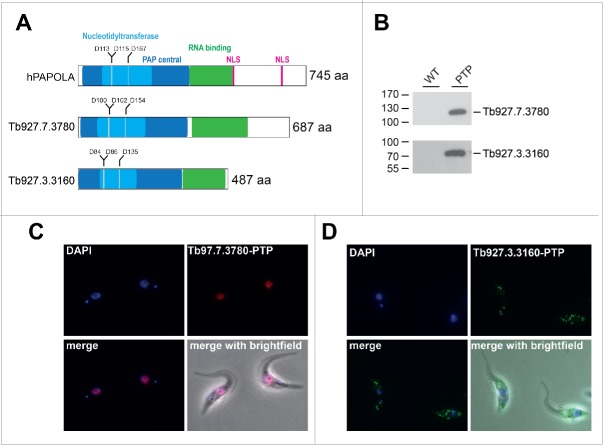
To identify the major functional poly(A) polymerase in T. brucei, we focus here on two putative poly(A) polymerases [Tb927.3.3160 and Tb927.7.3780] and initially compared the sequences with their human homolog, PAPOLA (Fig. 1A and Fig. S1). Based on the Needleman-Wunsch alignment algorithm, the human PAPOLA protein shows 17.4% and 24% identity with the trypanosomatid Tb927.3.3160 and Tb927.7.3780 proteins from T. brucei, respectively. Although there are regions of low sequence similarity, structure predictions of these two T. brucei poly(A) polymerases suggest a strong structure conservation of the two proteins. Moreover, by domain structure prediction of these two T. brucei poly(A) polymerases, the highly conserved domains of the poly(A) polymerase described in higher eukaryotes, such as the central domain harboring the nucleotidyltransferase activity including the three catalytic aspartic acid residues, are present in both trypanosomatid proteins.15-17 Following the central domain, an RNA-binding domain was predicted for all three proteins. However, both trypanosomatid proteins lack a recognizable, classical nuclear localization signal at their C-terminus.
Figure 1.
Expression and cellular localization of two poly(A) polymerases (PAPs) in T. brucei. (A) Domain organization of the PAPs, comparing the human poly (A) polymerase α (hPAPOLA) and two putative trypanosomatid PAPs [Tb927.7.3780 and Tb927.3.3160]. The PAP central domain, harboring the nucleotidyltransferase domain and the conserved catalytic aspartic acid residues (in blue), the RNA-binding domain (in green), and the known nuclear localization signals (NLS; in magenta) are illustrated.(B) The expression of PTP-tagged putative poly(A) polymerase [Tb927.7.3780] and [Tb927.3.3160] in T. brucei cells was detected by Western blotting, using polyclonal anti-protein A antibodies (lane PTP). As a control, T. brucei 427 wildtype (lane WT) cells were included. Protein size markers in kDa. (C,D) Cell lines stably expressing PTP-tagged versions of the two poly(A) polymerases were fixed and stained with DAPI. The localization of tagged proteins was assessed by indirect immunofluorescence, using anti-protein A antibodies followed by alexa-594 and alexa-488-coupled secondary antibodies for Tb927.7.3780 and Tb927.3.3160, respectively. Superimpositions of DAPI and PTP-tagged proteins (merge) as well as with brightfield are shown (merge with brightfield).
To compare the two putative enzymes, we next generated two clonal procyclic cell lines, which stably express either Tb927.3.3160 or Tb927.7.3780, each with a C-terminal PTP-tag (protein C epitope / TEV cleavage site / 2x protein A epitope). Expression of both proteins was monitored by Western blot analysis (Fig. 1B) and in addition, the subcellular distribution of both proteins was visualized by indirect immunofluorescence (Fig. 1C-D). Interestingly, the PTP-tagged version of the Tb927.7.3780 predominantly localizes to the nucleus, whereas the putative poly(A) polymerase Tb927.3.3160 clearly localizes mainly in distinct cytoplasmic spots, suggesting differential functions. However, one has to consider that the large C-terminal tag may influence the localization within the cell.
Only the putative poly(A) polymerase Tb927.7.3780 shows in vitro activity
To assess and compare the enzymatic activity of both proteins, we expressed His-tagged versions of the two poly(A) polymerases in Sf9 insect cells, followed by HPLC purification (Fig. 2A). The purified recombinant proteins were assayed for unspecific polyadenylation activity, using an RNA substrate derived from the 3′ end of the T. brucei α-tubulin mRNA. Interestingly, only the Tb927.7.3780 protein showed in vitro activity under these conditions, in contrast to the Tb927.3.3160 protein (Fig. 2B). The RNA substrate was efficiently elongated by Tb927.7.3780 (approximately 150 nucleotides within 10 min), suggesting a high processivity of the enzyme (Fig. 2B). As previously shown for the human poly(A) polymerase, non-specific activity is very inefficient in the absence of specificity factors, such as CPSF or CFI; however, if Mg2+ is replaced by Mn2+, the unspecific activity is strongly enhanced.18,19 This is also the case for the poly(A) polymerase Tb927.7.3780 in trypanosomes (Fig. 2B).
Figure 2.
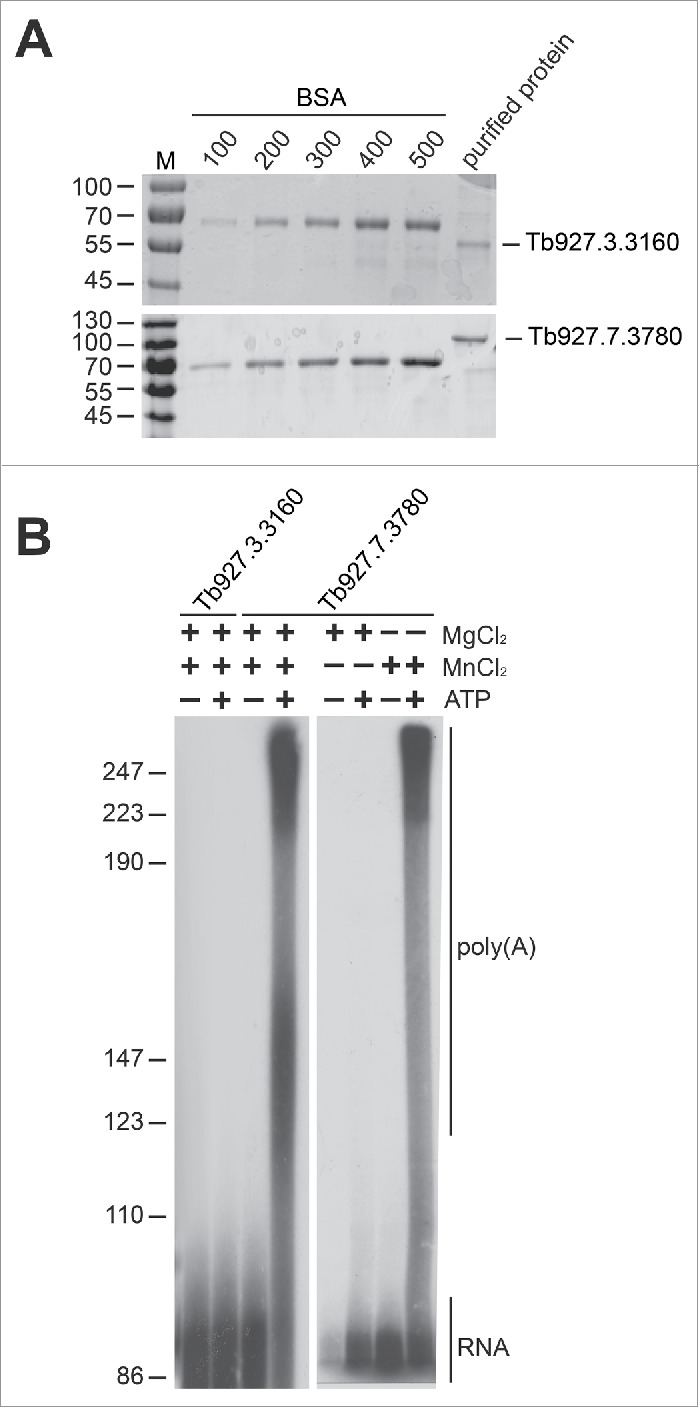
Recombinant putative poly(A) polymerase Tb927.7.3780 polyadenylates in vitro.(A) Quantification of recombinant baculovirus-expressed poly(A) polymerases. Recombinant proteins were expressed in baculovirus-infected Sf9 cells, HPLC-purified, and analyzed by SDS-PAGE and Coomassie staining. BSA standards (100–500 ng) were used for quantification. Protein size markers in kDa. (B) Only the putative poly(A) polymerase Tb927.7.3780 shows in vitro activity and requires manganese as a cofactor for unspecific polyadenylation. In vitro transcribed 32P-labeled RNA (derived from the T. brucei α-tubulin 3′ UTR) was incubated at 37°C with baculovirus-expressed and purified putative poly(A) polymerase [Tb927.3.3160 or Tb927.7.3780]. Reactions were performed in the presence of MnCl2 or MgCl2 or a mixture of both (1.5 mM), and either in presence (+) or absence (−) of ATP. Reactions were analyzed after 30 minutes on a denaturing 12.5% polyacrylamide gel. Marker sizes in nucleotides.
In vivo activity of the putative poly(A) polymerases
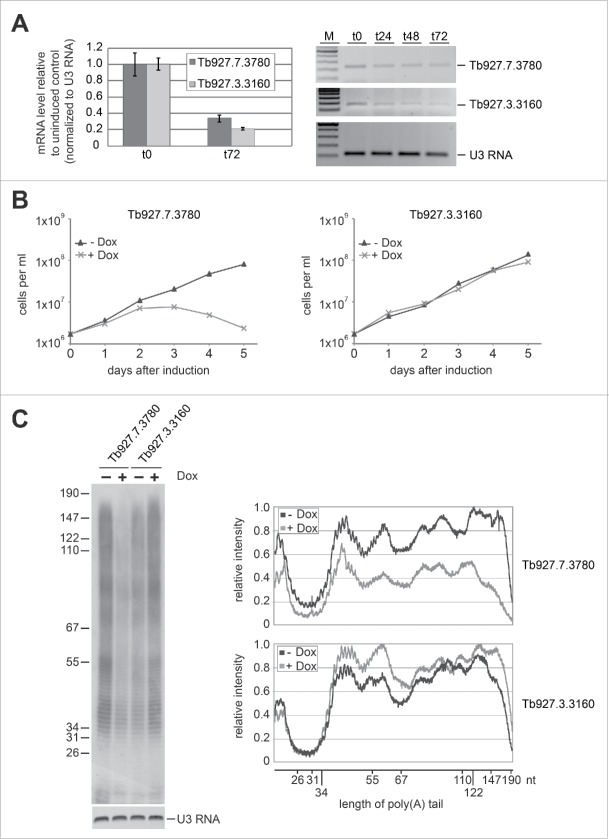
To compare the activities of the two trypanosomatid poly(A) polymerases in vivo, we silenced them individually in procyclic T. brucei by doxycycline-inducible RNAi. The knockdown efficiency was confirmed by RT-qPCR, using primers detecting the respective Tb927.3.3160 and Tb927.7.3780 mRNAs (Fig. 3A, left side). After three days of RNAi induction, we observed a knockdown efficiency of approximately 80% and 70%, respectively. In addition, we performed semiquantitative RT-PCR using the same primer pairs, confirming the knockdown (Fig. 3A, right side). When we monitored the cell viability upon knockdown, only after silencing Tb927.7.3780 expression, cell growth was rapidly affected; in contrast, knockdown of Tb927.3.3160 did not result in a phenotypic growth defect (Fig. 3B).
Figure 3.
Only the putative poly(A) polymerase Tb927.7.3780 is active in polyadenylation in vivo. (A) RNAi-mediated knockdown of the two trypanosomatid poly(A) polymerases. mRNA levels were analyzed by real-time RT-PCR (left) or semiquantitative RT-PCR (right) from uninduced cells (t0) and after 1, 2, and 3 d of RNAi induction (t24-t72, as indicated). U3 RNA served for normalization and as a control. M, markers (100, 200, 300, 400, and 500 bp). (B) Only the putative poly(A) polymerase Tb927.7.3780, but not Tb927.3.3160, is essential for cell viability. Growth curves of representative cell lines are shown, in which RNAi was induced by doxycycline, (+ Dox, lines with asterisks) and respective uninduced controls (−Dox, lines with triangles).(C) Depletion of poly(A) polymerase Tb927.7.3780, but not of Tb927.3.3160, impairs polyadenylation in vivo. RNA from uninduced (− Dox) and induced (+ Dox) cells was splint-labeled, digested with RNases A/T1, followed by separation of the labeled tails on a 15% denaturing gel. U3 RNA detected by RT-PCR served as a loading control. Marker sizes are indicated in nucleotides. The relative intensities of both signals were measured with ImageJ and plotted. The black and gray lines represent the tail length distributions in uninduced and induced cells, respectively.
To investigate whether individual knockdown of the two poly(A) polymerases affects the global mRNA polyadenylation status in vivo, total RNA was analyzed after three days, and the poly(A) tail length was examined according to Tkcaz et al.3 In brief, poly(A) tails were labeled at their 3′ end by splint ligation, using an oligo(dG)-oligo(dT) primer to allow subsequent fill-in with 32P-dCTP. Total RNA was then treated with RNase A and T1, leaving intact poly(A) tails, which were analyzed by gel electrophoresis. The relative band intensities of the poly(A) tails were measured by ImageJ and normalized against signals of U3 RNA (Fig. 3C). The results clearly indicate that Tb927.7.3780 depletion severely reduced relative amounts and lengths of the poly(A) tails, unlike Tb927.3.3160 depletion, where an opposite effect of a slight increase in activity was observed.
Taken together, our results indicate that the poly(A) polymerase Tb927.7.3780 (annotated as putative) represents the major functional nuclear poly(A) polymerase in trypanosomes. In contrast, the poly(A) polymerase from the intron-containing gene Tb927.3.3160 localizes mainly in the cytoplasm and appears not to be functional in general 3′ polyadenylation.
Identification of the constituents of the polyadenylation complex
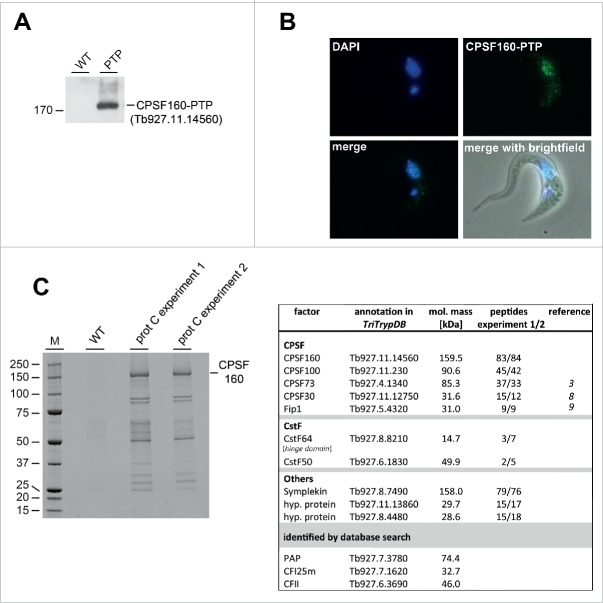
To systematically investigate the composition of the trypanosomatid polyadenylation machinery, we used tandem affinity purification (TAP), based on the largest subunit of the CPSF complex, the putative CPSF160 [Tb927.11.14560]. We generated a clonal procyclic cell line, which stably expresses CPSF160 with a C-terminal PTP tag (for Western blot analysis, see Fig. 4A; for its cellular distribution, Fig. 4B). As expected, CPSF160-PTP predominantly localizes to the nucleus, with only minor staining of the cytoplasm.
Figure 4.
Identification of components of the polyadenylation complex, based on TAP-tag purification of the nuclear localized trypanosomatid CPSF160 homolog. (A) Expression of PTP-tagged CPSF160 homolog [Tb927.11.14560; lane PTP] in T. brucei cells was detected by Western blotting using polyclonal anti-protein A antibodies. As a control, T. brucei 427 wildtype (lane WT) cells were analyzed. Protein size marker in kDa. (B) Nuclear localization of CPSF160-PTP. Cell lines stably expressing PTP-tagged T. brucei CPSF160 were fixed and stained with DAPI (DAPI). The localization of tagged protein was assessed by indirect immunofluorescence, using anti-protein A antibodies (CPSF160-PTP). In addition, superimpositions of DAPI and PTP-tagged protein (merge) as well as with brightfield are shown (merge with brightfield). (C) CPSF160-associated proteins identified by mass spectrometry. Extracts were prepared from T. brucei cell lines that stably express tagged CPSF160 (in two biological replicates; experiment 1/2), followed by TAP-tag affinity purification. Proteins after the second purification step (protein C elution) were analyzed by SDS-PAGE and Coomassie staining (lanes prot C experiment 1/2). As control, T. brucei wildtype cells were used (lane WT). Protein size markers in kDa (M). Putative protein constituents of the T. brucei polyadenylation complex were identified by mass spectrometry. Each protein is described by name (factor), the TriTrypDB annotation number (http://tritrypdb.org/tritrypdb/), its molecular mass (in kDa), the number of exclusive unique peptide counts obtained by mass spectrometry, and, if applicable, by literature reference. Below, the three proteins identified only by database search (PAP, CFI25m, and CFII) are listed by name, TriTrypDB annotation number, and molecular mass.
Based on CPSF160-PTP, two independent tandem affinity purifications (TAP) were performed (prot C experiment 1 and 2); as a control, wild-type T. brucei cells (WT) were used (Fig. 4C). Factors co-purifying with CPSF160 were analyzed by mass spectrometry and are listed in Fig. 4C (right panel; for raw data, see Table S1): Eight proteins were identified by homology search as putative factors of the polyadenylation complex in trypanosomes, including five CPSF subunits (CPSF160; CPSF100; CPSF73; CPSF30; Fip1), two constituents of CstF [CstF64 hinge domain (see below); CstF50], and the scaffold protein Symplekin. Two additional proteins were found [Tb927.11.13860; Tb927.8.4480], which are likely associated with the polyadenylation complex, since they are clearly above the peptide counts of the background control (WT). In addition to mass spectrometry-based homology search, we further confirmed our data by comparative InterProScan protein domain identification of the individual trypanosomatid factors and their respective human counterparts (Fig. S2).20 Some proteins display only low sequence similarity to their human counterparts. This applies in particular to the putative CstF64, which was classified only through a putative hinge domain present in both the trypanosomatid and human protein (see Discussion).
In addition, we did homology-based databank searches for trypanosomatid homologs of other eukaryotic polyadenylation factors, which had not appeared in our mass spectrometry analysis. As a result, we identified potential homologs of the 25 kDa subunit of cleavage factor I (CFIm25) [Tb927.7.1620] and cleavage factor II (CFIIm) [Tb927.6.3690] (Fig. S2).
Polyadenylation factors are required for both polyadenylation and trans splicing
To assess the functional relevance of the proteins identified in the polyadenylation complex, we depleted each of them by inducible RNAi: Each of the individual knockdowns showed a severe growth defect in procyclic trypanosomes, with the exception of one of the two hypothetical proteins, Tb927.11.13860 (Fig. S3).
As described above for the two poly(A) polymerases, the in vivo polyadenylation status was examined after knockdown of the individual factors for three days (Fig. S4, summarized in Fig. 5A). Knockdown of CPSF100, CPSF73, and CPSF30 strongly reduced the overall poly(A) tail lengths. In contrast, depletion of CPSF160 and Fip1 -as part of the CPSF complex- showed no significant effect. CstF50 depletion strongly affected polyadenylation, in contrast to CstF64 depletion. Note that the putative CstF64 analyzed here exhibited only a very low sequence similarity to its mammalian counterpart (see above); it may represent a truncated homolog functionally different from the mammalian factor or a trypanosomatid-specific factor.
Figure 5.
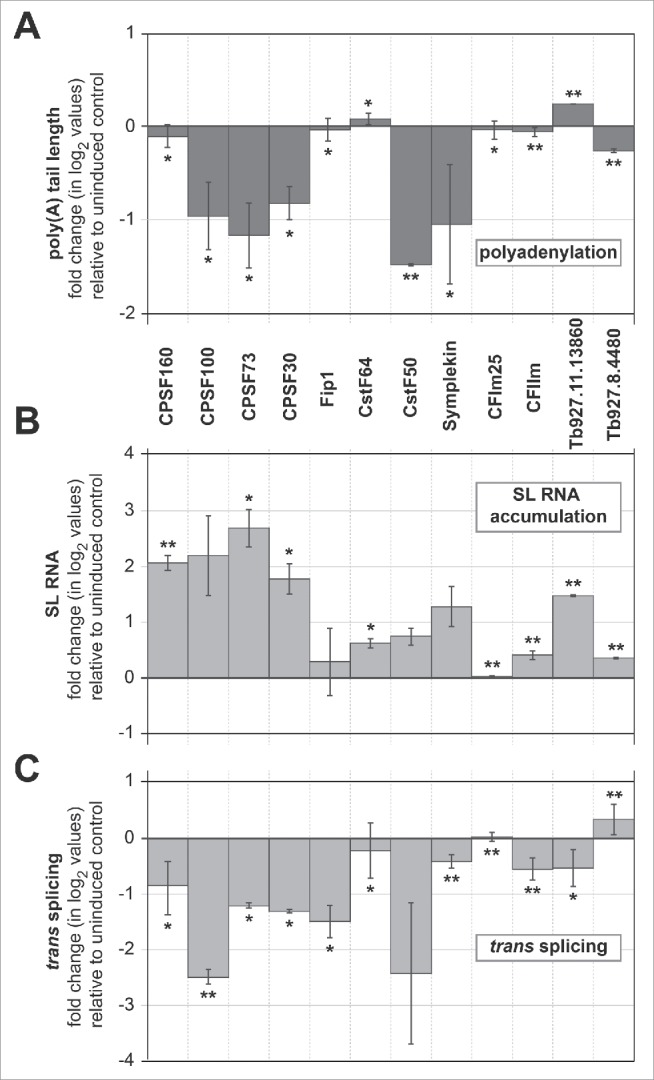
Polyadenylation factors required for both polyadenylation and splicing in vivo. The following factors were subjected to RNAi knockdown (TriTrypDB numbers in parenthesis): CPSF160 [Tb927.11.14560]; CPSF100 [Tb927.11.230]; CPSF73 [Tb927.4.1340]; CPSF30 [Tb927.11.12750]; Fip1 [Tb927.5.4320]; CstF50 [Tb927.6.1830]; CstF64 [Tb927.8.8210]; CFIm25 [Tb927.7.1620]; CFIIm [Tb927.6.3690]; Symplekin [Tb927.8.7490]; Tb927.11.13860; Tb927.8.4480. Changes in the overall poly(A) tail length (A), in SL RNA accumulation (B), and in trans splicing of PPIase mRNA (C) upon depletion of individual polyadenylation factors are graphically shown. Poly(A) tail lengths were analyzed and quantitated by the ‘Riemann's sum’, comparing induced and uninduced cells (for raw data, see Fig. S4). SL RNA accumulation and trans-splicing activities were determined by RT-qPCR on SL RNA and trans spliced PPIase mRNA, respectively, and normalized to U3 RNA. For both polyadenylation, SL RNA accumulation, and trans splicing, fold changes (in log2 values) of uninduced and induced levels were calculated, with standard deviations derived from three biological replicates (*for P values < 0 .05; ** for P values < 0 .01).
Depletion of the scaffold factor Symplekin resulted in a severe polyadenylation phenotype. In contrast, depletion of the two cleavage factors CFIm25 and CFIIm did not significantly affect poly(A) tail length. Finally, when analyzing the two un-annotated proteins [Tb927.8.4480 and Tb927.11.13860], small, but significant stimulatory and inhibitory effects were observed, respectively.
As several previous studies have indicated, both major pre-mRNA processing steps, trans splicing and polyadenylation, are interconnected. Therefore we also analyzed trans splicing activities to compare them directly with the corresponding polyadenylation effects. Both spliced-leader (SL) RNA (Fig. 5B), which is known to accumulate upon inhibition of trans splicing (for example, see reference 2), and a specific trans-spliced mRNA (PPIase) (Fig. 5C) were measured by quantitative RT-PCR after knockdown of the individual polyadenylation factors. Depletion of the CPSF complex subunits 160, 100, 73, and 30 resulted in spliced-leader accumulation and severe splicing deficiencies. However, for Fip1 only blocking of splicing was observed, but no effect on SL RNA accumulation (Fig. 5C). Knockdown of both CstF subunits 64 and 50, Symplekin, CFIIm, and the two hypothetical proteins Tb927.11.13860 and Tb927.8.4480 resulted in moderate effects on spliced-leader accumulation and in most cases also on inhibition of trans splicing. However, depletion of CFIm25 had neither an effect on SL RNA accumulation nor on trans splicing. These effects on PPIase trans splicing were largely confirmed with another gene, PRP8, analyzed by semiquantitative RT-PCR (Fig. S5).
We conclude that the corresponding knockdown effects of most core polyadenylation factors on both polyadenylation and trans splicing, as well as splicing deficiencies after CPSF160 and Fip1 depletion, argue for a general coupling of these two major RNA processing steps.
Discussion
Regulation of gene expression in trypanosomes depends mainly on post-transcriptional control.21 Unravelling the mechanisms of mRNA processing therefore is a prerequisite to understand the post-transcriptional regulation mechanisms in these parasites. In this study we focused on the 3′ end processing machinery. In particular, we characterized for the first time the major functional nuclear poly(A) polymerase in T. brucei [Tb927.7.3780], which so far was annotated as a putative enzyme. Another poly(A) polymerase paralog [Tb927.3.3160], which is encoded by an intron-containing gene in trypanosomes, does not represent the major enzyme responsible for nuclear mRNA 3′ end processing. However, both proteins share strong structure similarities with other eukaryotic poly(A) polymerases (Fig. 1A). The tripartite domain architecture of classical poly(A) polymerases consisting of the N-terminal catalytic domain, the central domain (also referred as PAP domain), and the C-terminal RNA-binding domain, is conserved in both trypanosomatid paralogs.22 Both Tb927.3.3160 and Tb927.7.3780 lack a clearly defined nuclear localization signal (NLS); however, some basic residues can be found near the C-terminus of the Tb927.7.3780 protein, which may act as an NLS (Fig. S1). Of particular interest is the observed cytoplasmic localization of the putative poly(A) polymerase paralog Tb927.3.3160, specifically within distinct spots inside the cytosol. Despite the fact that only the Tb927.7.3780 protein showed in vitro as well as in vivo polyadenylation activity, this argues for functions of the Tb927.3.3160 protein other than 3′ end processing of pre-mRNAs. Despite our negative results in in vitro polyadenylation, we cannot rule out that Tb927.3.3160 may represent a functional poly(A) polymerase, for example, with a limited, specialized set of targets, which our method used here did not detect. For example, cytoplasmic polyadenylation or uridylation may regulate translation or stability of a subset of mRNAs; alternatively, additional factors might be required for activity.23,24
Another basic question that arose is the biological role of cis splicing in trypanosomes. A recent study from our group supports the hypothesis that cis splicing may be an evolutionary relic.11 The findings presented here further support this provocative idea. In trypanosomes only two genes contain intron sequences and require cis splicing for proper mRNA maturation: Tb927.3.3160 and Tb927.8.1510, the latter of which codes for an ATP-dependent DEAD box helicase. However, neither RNAi depletion of either of these two factors (this study and reference 25) nor inhibition of cis splicing result in a severe growth phenotype.11 Nevertheless, this does not exclude that the two introns may fulfill an important, still unknown function.
Our identification and initial characterization of the individual components of the trypanosomatid polyadenylation complex provides an important initial step in understanding the evolution of mRNA processing mechanisms. The considerable differences of cis-acting polyadenylation elements between trypanosomes, yeast, and mammals raises the question whether the protein factors involved are correspondingly divergent in trypanosomes. For example, for some mammalian polyadenylation factors, such as CstF50, no counterpart could be identified in yeast.26
In trypanosomes, only Fip1 and CPSF30 had previously been identified and functionally described so far.8,9 Specifically, CPSF30 is required for polycistronic mRNA processing.8 In addition, the CPSF73 ortholog associates with the U1 snRNP-specific U1A protein in trypanosomes.3 Here we identified eight additional factors as part of the trypanosomatid polyadenylation machinery, based on mass spectrometry and database search. However, the poly(A) polymerase was not copurified, which is in line with previous observations, suggesting that the poly(A) polymerase is not tightly associated with other processing factors; instead, there may be a dynamic or transient association with the core polyadenylation complex.27 Even if the sequence similarity between these trypanosome polyadenylation factors and their human counterparts is low, they share for the most part the domain architecture. This becomes particularly obvious when comparing the conservation of the subunits of the CPSF complex (Fig. S2). For example, the CPSF73 subunit, which attracted special attention as the functional endonuclease in the mammalian polyadenylation reaction, shares specific, catalytically important residues with the trypanosome homolog: The Zn-binding residues (mainly histidines) are conserved in the trypanosomatid CPSF73, indicating that CPSF73 likely also act as the endonuclease during polyadenylation in trypanosomes (Fig. S2).6
Some of the new trypanosome factors display only low sequence conservation. The putative trypanosome CstF64 strikingly differs from the human counterpart (compare the molecular masses of 14 kDa vs. 64 kDa for the T. brucei and the human CstF64, respectively) and could only be identified through a putative hinge domain required for CstF64 interaction with CstF77 and Symplekin.28,29 The otherwise conserved N-terminal RRM domain, however, is completely missing in trypanosomes. Therefore the trypanosomatid CstF64 may have retained only and function as a scaffolding protein required for the recruitment of other 3′ end processing factors.
Finally, some known mammalian polyadenylation factors could neither be identified in our mass spectrometry approach nor through extensive database homology search: As an example, we did not find a trypanosome homolog of WDR33, a subunit of the CPSF complex. Two independent studies had recently identified WDR33 as the factor that together with CPSF30 recognizes the AAUAAA polyadenylation signal.30,31 Since trypanosomes lack the classical hexameric polyadenylation signal, this appears plausible.
Depletion of the individual factors identified in our study showed severe growth defects and inhibition of polyadenylation in most cases. Interestingly, the two newly identified hypothetical proteins, Tb927.11.13860 and Tb927.8.4480, of which no known homolog or conserved domain structure could be identified, activate or repress polyadenylation, respectively. Such effects have been described in higher eukaryotes for factors that are not constituents of the core polyadenylation complex, but are required for general regulation of polyadenylation.32,33 In addition, we observed trans splicing defects consistent with close coupling of both mRNA processing reactions in trypanosomes. Interestingly, we did not detect for all factors effects on polyadenylation and trans splicing: For example, knockdown of cleavage factor I m25 (CFIm25) neither affected poly(A) tail length nor splicing. In higher eukaryotes CFIm25 is involved in the regulation of alternative polyadenylation, especially in the repression of proximal polyadenylation sites leading to longer 3′UTRs.34 Since alternative polyadenylation is common in trypanosomes as well, CFIm25 identified here is a potential regulator of this process.35,36
In other eukaryotes, a direct linkage between spliceosomal and 3′ end processing components had been demonstrated by several studies.37-39 In trypanosomes, so far only the polypyrimidine tract is known to affect trans splicing and polyadenylation of two adjacent genes.12-14 Until recently no factor could be identified, which directly links the polyadenylation complex and the trans-spliceosome in trypanosomes. This is in line with our result that no known spliceosomal factor copurified with CPSF160. However, a recent study provided first evidence that the polyadenylation factor CPSF73 copurifies with the U1A protein, a specific component of the spliceosomal U1 snRNP.3 Note that the spliceosomal U1 snRNP was thought to be required only for cis splicing, not for trans splicing. However, we recently reported that the U1 snRNP may also associate with the trans-spliceosome, indicating a physical linkage between cis- and trans-splicing.11 Taken together this suggests that the U1 snRNP functions in both splicing and polyadenylation, likely linking both processes.
Unravelling the individual constituents of the polyadenylation complex in combination with what is already known about the components of the trypanosomatid spliceosome will help us to further understand the principle mechanisms of mRNA processing in trypanosomes.
Materials and methods
Cell culture, extract preparation, and immunofluorescence
For the generation of cell lines expressing PTP-tagged Tb927.3.3160, Tb927.7.3780, and CPSF160, the pC-PTP-Neo vector was used.40 The T. brucei open reading frames for Tb927.3.3160 (nts 672–1461), Tb927.7.3780 (nts 1172–2061), and CPSF160 (nts 3601–4356) were PCR-amplified and inserted in-frame into the pC-PTP-NEO vector upstream of the PTP tag sequence, using the ApaI and NotI restriction sites. For genomic integration, 10 µg of linearized constructs were transfected into procyclic T. brucei 427 cells and cloned by limiting dilution in the presence of G418 (40 µg/ml Geneticin; Gibco-BRL).
Cell culture of T. brucei 427 and 29–13 was described previously.1,41 Cell lysates were prepared in extraction buffer [150 mM KCl, 20 mM Tris-Cl pH 7.7, 3 mM MgCl2, 0.5 mM DTT, containing a Complete Mini, EDTA-free protease inhibitor cocktail tablet (Roche)], and using a Dounce homogenizer, followed by sonication. Cell lysates were supplemented with 0.1% Tween-20, and centrifuged twice at 20,000 x g for 15 min to remove aggregates.
The cellular distribution of Tb927.3.3160-PTP, Tb927.7.3780-PTP, and CPSF160-PTP was analyzed by indirect immunofluorescence as described.1
Database analysis
Protein sequence alignments were performed by the EMBOSS (http://www.ebi.ac.uk/Tools/psa/emboss_needle/) program. Pattern and profile search were done by InterProScan.20 Protein structure prediction was carried out via the GeneSilico metaserver.42
Expression and purification of recombinant poly(A) polymerases
The open reading frames of Tb927.3.3160 and Tb927.7.3780 were cloned into the vector pFastBac HTb (Life Technologies) replacing the first methionine with the N-terminal His-tag of the vector. Recombinant baculovirus for protein expression in Sf9 cells was obtained, using the Bac-to-Bac® baculovirus expression system (Life Technologies). For purification of recombinant proteins, extracts from infected SF9 cells were prepared, and the proteins were purified by Ni-nitrilotriacetic acid affinity chromatography, followed by gel filtration on an ÄKTApurifier high-pressure liquid chromatography system (GE Healthcare).
In vitro polyadenylation assays
For standard in vitro polyadenylation assays 4 nM in vitro transcribed 32P-labeled RNA, derived from the 3′ end of T. brucei α-tubulin mRNA, was assembled with poly(A) buffer (25 mM Tris-Cl pH 7.4, 50 mM KCl, 1.5 mM MgCl2, 1.5 mM MnCl2, 0.05 mM EDTA, 0.5 mM DTT, 10% glycerol, 2.6% polyvinyl alcohol [PVA]). 32 nM recombinant purified poly(A) polymerase was added on ice. The reaction was started by the addition of 1 mM ATP and incubation at 37°C. RNAs were ethanol-precipitated and analyzed by denaturing polyacrylamide gel electrophoresis.
RNA analysis, RNA interference (RNAi) silencing, and real-time RT-PCR
Total RNA was isolated from procyclic T. brucei cells using Trizol reagent (Life Technologies) and further purified by the RNeasy Mini Kit (Qiagen).
The RNAi constructs were made, using the stem-loop vector pLEW100 according to an established cloning strategy.41 The resulting construct was linearized with SacII, and 10 µg were transfected into T. brucei 29–13 cells by electroporation. Transformants were cloned by limiting dilution in the presence of G418 (15 µg/ml), hygromycin (50 µg/ml), and phleomycin (2.5 µg/ml). RNAi was induced by the addition of 1 mg/ml of doxycycline. Cells were counted every day and diluted to 2 × 106 cells/ml. Semiquantitative as well as quantitative real-time RT-PCR were performed as described.1 Sequences of oligonucleotides used are available upon request.
Poly(A) tail length measurement
Labeling of polyadenylated RNAs was done as described by Tkcaz et al.3 Quantitation was performed by measuring the relative intensities of 32P-labeled product bands, using ImageJ.43
Fold changes were obtained by calculating the middle ‘Riemann sum’, which can be understood as a weighted average of the polyadenylation efficiency:
in which S is the 'Riemann sum' of the function f(x) over the total measured intensities (n) divided into separate sections (i).
Mass spectrometric analysis
Tandem affinity purification of PTP-tagged proteins was done as described by Schimanski et al,40 with minor modifications: Briefly, T. brucei cells were collected from 3-liter cultures (corresponding to ∼4 ml packed cell volume) and lysed in 7 ml extraction buffer (150 mM KCl, 20 mM Tris-Cl pH 7.7, 3 mM MgCl2, 0.5 mM DTT, containing a Complete Mini, EDTA-free protease inhibitor cocktail tablet [Roche]). For IgG affinity chromatography, 300 μl of IgG Sepharose 6 Fast Flow beads (packed bead volume; GE Healthcare) were incubated with extract for two hours at 4°C. Beads were washed extensively with PA-150 buffer (150 mM KCl, 20 mM Tris-Cl pH 7.7, 3 mM MgCl2, 0.5 mM DTT, 0.1% Tween20), followed by TEV protease buffer (150 mM KCl, 20 mM Tris-Cl pH 7.7, 3 mM MgCl2, 0.5 mM EDTA; 0.5 mM DTT, 0.1% Tween20). Tagged proteins were eluted in 2 ml of TEV protease buffer containing 250 units of AcTEV protease (Life Technologies). For anti-ProtC affinity purification, CaCl2 was added to the eluate to a final concentration of 2 mM. The eluate was incubated for 2 hours at 4°C with 300 μl of anti-protein C affinity matrix (packed bead volume; Roche). The beads were washed with PC-150 buffer (150 mM KCl, 20 mM Tris-Cl pH 7.7, 3 mM MgCl2,1 mM CaCl2, 0.1% Tween20), and the ProtC-tagged proteins were eluted at room temperature with 0.5 ml EGTA elution buffer (5 mM Tris-Cl pH 7.7, 10 mM EGTA, 5 mM EDTA). Eluted proteins were concentrated, using a vacuum concentrator and StrataClean resins (Stratagene), analyzed on 4–12% NuPAGE gels (Life Technologies), and stained with Coomassie brilliant blue G-250.
Proteins were in-gel digested with endoproteinase trypsin and analyzed by LC-MS/MS under standard conditions. Data were searched against a T. brucei protein sequence database (NCBInr v.11.09.2013, 38057 entries). Data were evaluated with scaffold software (Proteome Software, Version 4.0.4), and proteins were ranked by their unique peptide counts in LC-MS/MS analysis, using a peptide and protein threshold of 95.0% and a minimum of three peptides.
Supplementary Material
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Georges Martin for discussions as well as members of our laboratory for critical comments and advice. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG Bi 316/17–1 and IRTG1384, to A.B.).
References
- 1.Palfi Z, Jaé N, Preußer C, Kaminska KH, Bujnicki JM, Lee JH, Günzl A, Kambach C, Urlaub H, Bindereif A. SMN-assisted assembly of snRNP-specific Sm cores in trypanosomes. Genes Dev 2009; 23:1650-64; PMID:19605687; http://dx.doi.org/ 10.1101/gad.526109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luz Ambrosio D, Lee JH, Panigrahi AK, Nguyen TN, Cicarelli RMB, Günzl A. Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors. Eukaryot Cell 2009; 8:990-1000; PMID:19429779; http://dx.doi.org/ 10.1128/EC.00075-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tkacz ID, Gupta SK, Volkov V, Romano M, Haham T, Tulinski P, Lebenthal I, Michaeli S. Analysis of spliceosomal proteins in trypanosomatids reveals novel functions in mRNA processing. J Biol Chem 2010; 285:27982-99; PMID:20592024; http://dx.doi.org/ 10.1074/jbc.M109.095349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proudfoot NJ, Brownlee GG. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature 1976; 263:211-14; PMID:822353; http://dx.doi.org/ 10.1038/263211a0 [DOI] [PubMed] [Google Scholar]
- 5.Gil A, Proudfoot NJ. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit β-globin mRNA 3′ end formation. Cell 1987; 49:399-406; PMID:3568131; http://dx.doi.org/ 10.1016/0092-8674(87)90292-3 [DOI] [PubMed] [Google Scholar]
- 6.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 2006; 444:953-56; PMID:17128255; http://dx.doi.org/ 10.1038/nature05363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahle E. 3′-end cleavage and polyadenylation of mRNA precursors. Biochim Biophys Acta 1995; 1261:183-94; PMID:7711061; http://dx.doi.org/ 10.1016/0167-4781(94)00248-2 [DOI] [PubMed] [Google Scholar]
- 8.Hendriks EF, Abdul-Razak A, Matthews KR. tbCPSF30 depletion by RNA interference disrupts polycistronic RNA processing in Trypanosoma brucei. J Biol Chem 2003; 278:26870-78; PMID:12746436; http://dx.doi.org/ 10.1074/jbc.M302405200 [DOI] [PubMed] [Google Scholar]
- 9.Bercovich N, Levin MJ, Vazquez MP. The FIP-1 like polyadenylation factor in trypanosomes and the structural basis for its interaction with CPSF30. Biochem Biophys Res Commun 2009; 380:850-55; PMID:19338765; http://dx.doi.org/10688355 10.1016/j.bbrc.2009.01.182 [DOI] [PubMed] [Google Scholar]
- 10.Mair G, Shi H, Li H, Djikeng A, Aviles HO, Bishop JR, Falcone FH, Gavrilescu C, Montgomery JL, Santori MI, et al.. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA 2000; 6:163-69; PMID:10688355; http://dx.doi.org/ 10.1017/S135583820099229X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preußer C, Rossbach O, Hung LH, Li D, Bindereif A. Genome-wide RNA-binding analysis of the trypanosome U1 snRNP proteins U1C and U1-70K reveals cis/trans-spliceosomal network. Nucleic Acids Res 2014; 42:6603-15; PMID:24748659; http://dx.doi.org/8504937 10.1093/nar/gku286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev 1993; 7:996-1007; PMID:8504937; http://dx.doi.org/ 10.1101/gad.7.6.996 [DOI] [PubMed] [Google Scholar]
- 13.Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev 1994; 8:491-501; PMID:7907303; http://dx.doi.org/ 10.1101/gad.8.4.491 [DOI] [PubMed] [Google Scholar]
- 14.Benz C, Nilsson D, Andersson B, Clayton C, Guilbride DL. Messenger RNA processing sites in Trypanosoma brucei. Mol Biochem Parasitol 2005; 143:125-34; PMID:15993496; http://dx.doi.org/ 10.1016/j.molbiopara.2005.05.008 [DOI] [PubMed] [Google Scholar]
- 15.Zhelkovsky AM, Kessler MM, Moore CL. Structure-function relationships in the Saccharomyces cerevisiae poly(A) polymerase. Identification of a novel RNA binding site and a domain that interacts with specificity factor(s). J Biol Chem 1995; 270:26715-20; PMID:7592899; http://dx.doi.org/ 10.1074/jbc.270.44.26715 [DOI] [PubMed] [Google Scholar]
- 16.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J 1996; 15:2593-603; PMID:8665867 [PMC free article] [PubMed] [Google Scholar]
- 17.Martin G, Jenö P, Keller W. Mapping of ATP binding regions in poly(A) polymerases by photoaffinity labeling and by mutational analysis identifies a domain conserved in many nucleotidyltransferases. Protein Sci 1999; 8:2380-91; PMID:10595540; http://dx.doi.org/ 10.1110/ps.8.5.1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christofori G, Keller W. 3′ cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell 1988; 54:875-89; PMID:2842067; http://dx.doi.org/ 10.1016/S0092-8674(88)91263-9 [DOI] [PubMed] [Google Scholar]
- 19.Wahle E, Martin G, Schiltz E, Keller W. Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J 1991; 10:4251-57; PMID:1756732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al.. InterProScan 5: genome-scale protein function classification. Bioinformatics 2014; 30:1236-40; PMID:24451626; http://dx.doi.org/ 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol 2007; 156:93-101; PMID:17765983; http://dx.doi.org/ 10.1016/j.molbiopara.2007.07.007 [DOI] [PubMed] [Google Scholar]
- 22.Martin G, Keller W, Doublié S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J 2000; 19:4193-4203; PMID:10944102; http://dx.doi.org/ 10.1093/emboj/19.16.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rissland OS, Norbury J. The Cid1 poly(U) polymerase. Biochim Biophys Acta 2008; 1779:286-94; PMID:18371314; http://dx.doi.org/ 10.1016/j.bbagrm.2008.03.003 [DOI] [PubMed] [Google Scholar]
- 24.Kashiwabara S, Nakanishi T, Kimura M, Baba T. Non-canonical poly(A) polymerase in mammalian gametogenesis. Biochim Biophys Acta 2008; 1779:230-38; PMID:18294465; http://dx.doi.org/ 10.1016/j.bbagrm.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 25.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 2011; 21:915-24; PMID:21363968; http://dx.doi.org/ 10.1101/gr.115089.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takagaki Y, Manley JL. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol 1997; 17:3907-14; PMID:9199325; http://dx.doi.org/ 10.1128/MCB.17.7.3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 2009; 33:365-76; PMID:19217410; http://dx.doi.org/ 10.1016/j.molcel.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 2000; 20:1515-25; PMID:10669729; http://dx.doi.org/ 10.1128/MCB.20.5.1515-1525.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruepp MD, Schweingruber C, Kleinschmidt N, Schümperli D. Interactions of CstF-64, CstF-77, and Symplekin: implications on localization and function. Mol Biol Cell 2011; 22:91-104; PMID:21119002; http://dx.doi.org/ 10.1091/mbc.E10-06-0543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schönemann L, Kühn U, Martin G, Schäfer P, Gruber AR, Keller W, Zavolan M, Wahle E. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev 2014; 28:2381-93; PMID:25301781; http://dx.doi.org/ 10.1101/gad.250985.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR 3rd, Ule J, Manley JL, Shi Y. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 2014; 28:2370-80; PMID:25301780; http://dx.doi.org/ 10.1101/gad.250993.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H, Lee JH, Lee Y. Regulation of poly(A) polymerase by 14-3-3ϵ. EMBO J 2003; 22:5208-19; PMID:14517258; http://dx.doi.org/ 10.1093/emboj/cdg486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell 2011; 41:409-18; PMID:21329879; http://dx.doi.org/ 10.1016/j.molcel.2011.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruber AR, Martin G, Keller W, Zavolan M. Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol 2012; 9:1405-12; PMID:23187700; http://dx.doi.org/ 10.4161/rna.22570 [DOI] [PubMed] [Google Scholar]
- 35.Siegel TM, Hekstra DR, Wang X, Dewell S, Cross GAM. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 2010; 38:4946-57; PMID:20385579; http://dx.doi.org/ 10.1093/nar/gkq237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog 2010. 6:e1001090; PMID:20838601; http://dx.doi.org/ 10.1371/journal.ppat.1001090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev 2000; 14:403-14; PMID:10691733; http://dx.doi.org. 10.1101/gad.14.4.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 2006; 23:195-205; PMID:16857586; http://dx.doi.org/ 10.1016/j.molcel.2006.05.037 [DOI] [PubMed] [Google Scholar]
- 39.Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J 2006; 25:4854-64; PMID:17024186; http://dx.doi.org/ 10.1038/sj.emboj.7601331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schimanski B, Nguyen TN, Günzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell 2005; 4:1942-50; PMID:16278461; http://dx.doi.org/ 10.1128/EC.4.11.1942-1950.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem 2000; 275:40174-79; PMID:11013266; http://dx.doi.org/ 10.1074/jbc.M008405200 [DOI] [PubMed] [Google Scholar]
- 42.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res 2003; 31:3305-07; PMID:12824313; http://dx.doi.org/ 10.1093/nar/gkg557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9:671-75; PMID:22930834; http://dx.doi.org/ 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.