ABSTRACT
U6 small nuclear RNAs are part of the splicing machinery. They exhibit several unique features setting them appart from other snRNAs. Reports of introns in structured non-coding RNAs have been very rare. U6 genes, however, were found to be interrupted by an intron in several Schizosaccharomyces species and in 2 Basidiomycota. We conducted a homology search across 147 currently available fungal genome and identified the U6 genes in all but 2 of them. A detailed comparison of their sequences and predicted secondary structures showed that intron insertion events in the U6 snRNA were much more common in the fungal lineage than previously thought. Their positional distribution across the entire mature snRNA strongly suggests a large number of independent events. All the intron sequences reported here show canonical splice site and branch site motifs indicating that they require the splicesomal pathway for their removal.
KEYWORDS: Fungi; homology search; intron; snRNA; snRNA evolution
Introduction
The removal of introns from mRNA precursors (pre-mRNA) is facilitated by the spliceosome, a multimeric machinery ubiquitous among Eukarya. This complex involves the pre-mRNA, 4 different small nuclear ribonucleoproteins (snRNP) and several other auxiliary proteins.1 The snRNPs are usually composed of a single small nuclear RNA (snRNA) and a set of associated proteins. In eukaryotes, this holds for the snRNAs U1, U2, and U5, while both U4 and U6 snRNA are base-paired with each other and incorporated into a single snRNP.2,3 We refer to an excellent review by Matera and Wang1 for details on the precise role of each snRNA and its protein factors.
U6 is the best conserved snRNA, pointing at a central role in the splicing process.4 It is also exceptional in several other aspects: while Pol II transcribed U1, U2, U4, and U5 snRNAs share a common 2,2,7-trimethylguanosine (TMG) 5′ cap and an internal Sm protein binding site, U6 snRNA lacks these 2 structural features. Instead of a TMG cap, U6 genes possess a γ-monomethyl phosphate ester as 5′ end modification.5 U6 snRNA genes are transcribed by RNA polymerase III in vertebrates,6,7 insects8 and the budding yeast9,10 and encode the common Poly-T tract at their 5′ end, which is a characteristic termination signal of Pol III.
In yeast species, U6 transcription depends on 3 sequence motifs: 1) the TATA-Box upstream of transcription start site (TSS), 2) an internal box A (downstream but close to TSS) and 3) a box B sequence motif that resides ˜120nt downstream of transcription termination site. Minimal identifying sequence elements of the latter 2 motifs were determined as TRGYNNANNNG and GWTCRANNC in 10 yeast species.11 In general, promoter structures of Pol III transcripts are rather dynamic, i.e., they might vary between different Pol III transcripts of the same organism or between the same transcript in different species. In other Pol III transcripts of S.cerevisiae for example, box A and B are located downstream of the TSS but upstream of the mature product. The U6 snRNA in the fission yeast S.pombe, on the other hand, harbors a B box that is shifted from a distant downstream flanking region into a region of the RNA precursor that is later removed by splicing.11
An ∼50nt long intron-like sequence has been detected in the S.pombe U6 snRNA.12 Although U6 constitutes an essential component of the spliceosome, it was demonstrated that the common pre-mRNA splicing procedure is in charge for the removal of the U6 intron here.13 Homologous introns were subsequently found in closely related species of the Schizosaccharomyces genus.14 These introns are inserted at the homologous sequence positions within the U6 precursor and share considerable sequence similarity indicating a common origin. Additional introns encoded by the 2 Basidiomycota Rhodotorula hasegawae and Rhodosporidium dacryoidum were not found to be homologous to the Schizosaccharomyces introns suggesting that the introns arose at unrelated time points during fungal evolution.15
We collected U6 snRNAs from 147 genomes by homology search and analyzed them with respect to potential intron insertions and promoter elements. Our survey covers the complete set of fungal genomes published by summer 2015. We identified a total of 59 introns, which appear to have been inserted in in a few lineage specific insertion events and in a larger number of quite recent, essentially species-specific events.
Materials and Methods
We analyzed the evolution of U6 snRNA genes within 147 fungal organisms whose genomes are available in decent quality. Selected organisms ranged from Microsporidia, Mucoromycotina, Blastocladiomycota, and Basidiomycota to a large group of Ascomycota. A complete taxonomic tree can be found on the supplement page1 .
Detection of U6 snRNA genes
All U6 snRNAs that are annotated in the Rfam database16 for our fungi representatives were used as queries in a BLAST-based homology search. Additional paralogs and new orthologs were retrieved directly. Missing sequences were searched with relaxed BLAST parameters, regarding word size and gap penalties, to retrieve short conserved regions which were then concatenated in a subsequent chaining process. This method enabled us to detect intron interrupted snRNA genes even without a query containing a homologous intron. GotohScan17 was applied to species where no U6 candidate was uncovered with the initial BLAST-based search.
Detection of sequence motifs
To detect the intron characteristic sequence motifs we applied MEME18 on the putative intron sequences to retrieve motifs of length 7nt (5splice site and branch site) and 5nt (3splice site), respectively.
We extended the pre-snRNA U6 by 300nt up- and downstream and used MEME to detect the Pol III characteristic sequence motifs: TATA box, box A, and box B. Box A motifs were searched in the flanking regions and the mature snRNA, with the initial consensus sequence TRGYNNANNNG. Boxes B were searched in the potential introns and the flanking region of the snRNA with the starting consensus sequence GNTCNANNC. Both initial box motifs were retrieved from.11 Since MEME has problems with identifying a given but variable (in length) motif within a highly conserved RNA, we additionally applied FIMO, a motif detection tool that is also part of the MEME-suite2 , with the same consensus sequences to search for potential A box motifs in mature U6 snRNAs for sequences were no other A box was detected. TATA boxes were exclusively searched in the 300nt upstream region with consensus sequence TATAWW.
Cross-validation with RNA-seq data
Published RNA-seq data of total RNA for species with intron interrupted U6 snRNAs is available for Fusarium graminearum,19 Schizosaccharomyces pombe,20 and Trichoderma reesei.21 We blasted the U6 candidates in the referenced Genome Browsers or against the mapped short read archives to evaluate a potential transcription and the excision of our computationally identified introns.
Results
In 145 of the 147 fungal species, we found a total number of 334 U6 snRNA genes by means of BLAST. In the Microsporidia Vittaforma corneae, Edhazardia aedis, and Nematocida parisii U6 seems to be highly diverged. Nevertheless, we were able to annotate a potential U6 snRNA gene in N.parisii using the GotohScan approach leaving merely V.corneae and E.aedis without a detected U6 transcript.
The length of the mature transcript ranges from 100nt in Aspergillus or Schizosaccharomyces to 120nt in Basidiomycota due to an enlarged region directly upstream of the poly-T termination signal. Gene copy numbers are mainly conserved in the respective fungal lineages. In Taphrinomycotina and Saccharomycotina, U6 is commonly present in a single copy. The exception of this rule is given by Metschnikowia bicuspidata, whose genome harbors 14 nearly identical copies. Organisms in other lineages like Leotiomycetes or Dothidiomycetes typically encompass one, two, or three paralogous U6 snRNA genes, while Agaricomycetes harbor 4 to 9 different genes on average, see Fig. 1.
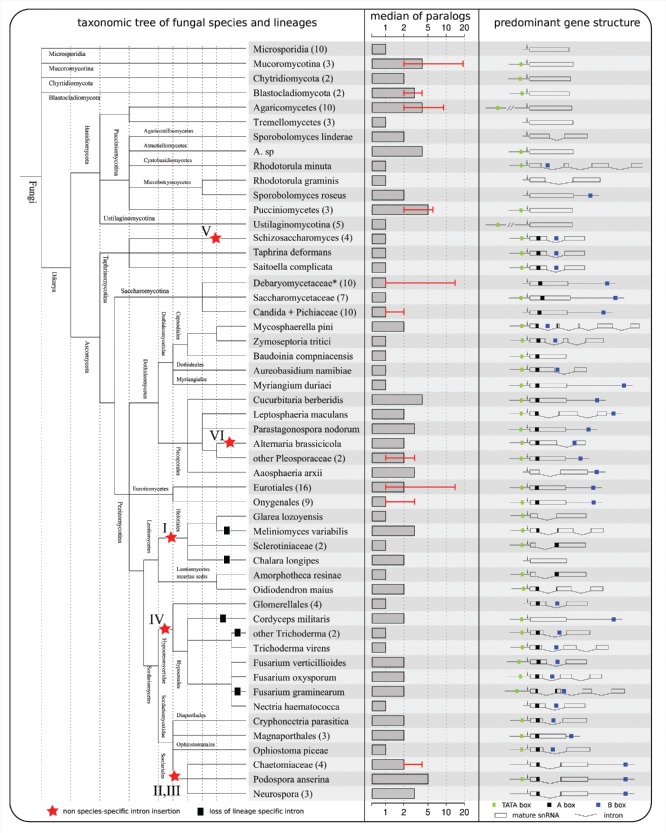
Figure 1.
Condensed taxonomic tree of all analyzed fungal organisms is shown on the left. Organisms showing similar gene structure with respect to intron insertions and Pol III promoter motifs are summarized into their respective lineages. The amount of different organisms contained in one lineage is given in parentheses. A red star indicates potential intron insertion events that are based on recognizable intron sequence homology and on precise homologous insertion positions within the mature snRNA. For detailed information see the intron homology section. The median amount of paralogous snRNA genes that were found in each organism is shown in the middle. The minimal and maximal values, in case they differ from the median, are given in red error bars. On the right side, the predominant gene structure is shown, i.e., this structure was found in at least one paralog of (nearly) all organisms that were grouped in the specific lineage. In case a single species is described, the structure containing the most Pol III motifs is shown.
In most species that encode more than one U6 gene, gene structures differ with respect to a potential intron insertion, the presence and the precise location of several Pol III associated promoter elements.
Various sequence alignments of precursor and mature U6 snRNAs can be found in the supplement. We further provide pictures showing the gene structure of each detected U6 transcript, including introns and promoter elements.
Intron interrupted U6 snRNA genes
Among the 145 fungi that encode 334 U6 snRNA transcripts in this study we detected 46 snRNAs distributed over 42 different organisms, that are interrupted by at least one intron-like fragment. In total, we discovered 59 intron candidates. Most of the intron harboring U6 genes are interrupted by precisely one intron, however, six, two, and one transcripts are split by 2, 3, and 4 introns, respectively, cf. Fig. 1.
U6 introns are canonical
In their survey covering 11.000 fungal mRNA-introns of 5 species Kupfer et al.22 extracted general properties of fungal pre-mRNA introns including intron length distributions as well as splice and branch site sequence motifs.
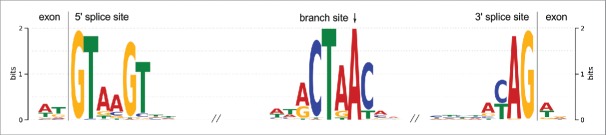
The dominant peak in the intron length distribution was found to be located between 50 ant 70nt.22 Some fungal species, such as the Microsporidia Encephalitozoon cuniculi and the Mucoromycotina Rhizopus oryzae, have even smaller introns with a median length of 32 and 57nt, respectively.23 The length distribution of our detected U6 introns is in good agreement with these values: The median length is 56nt and both central quartiles of the 59 introns are between 51 and 59nt long, see Fig. 2.
Figure 2.
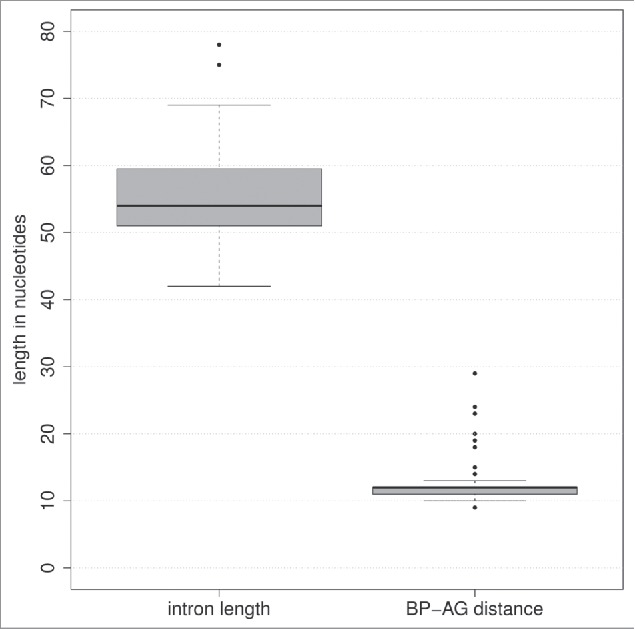
Boxplot of intron length and distances between the branch point adenosine and the 3′ splice site (BP-AG distance) gathered from the 59 putative U6 snRNA introns.
Kupfer et al.22 and Rep et al.24 defined the canonical intron splice sites as one of 5′GT-AG3′, 5′GC-AG3′, and 5′AT-AC3′. More than 98% of the introns in their data use the first motif. Of our U6 snRNA intron-like fragments, 57 out of 59 show the predominant 5′GT-AG3′ motif (96,6%), one encodes 5′GC-AG3′ and the intron in one of the both U6 paralogs in C.parasitica uses a non-canonical 5′GT-GG3′ junction.
The consensus sequence for the 5′ splice site derived from 5 fungal organisms was found to be 5′GTRWGT.22 The overall consensus sequence calculated by MEME on our U6 intron candidates was 5′GTAAGT and thus matches very well with the fungi consensus and even the metazoan consensus 5′ splice site motif. The fungi consensus acceptor sites are very similar to the higher eukaryotes sharing the a YAG3′ consensus pattern.22,23 More than 96% of our putative U6 introns encode this motif. The MEME-derived sequence logos for both splice sites are shown in Fig. 3.
Figure 3.
Sequence logos derived by MEME from the 59 potential U6 snRNA introns. From left to right: sequence logos for the last dinucleotides of the upstream exon, 5′ donor site, branch site, 3′ acceptor site, and the first dinucleotides of the downstream exon. The precise branch point is indicated by an arrow. The y axes displays the frequency of occurrences of a nucleotide in bits. The relative height of a letter is proportional to the relative frequencies of the nucleotide in the respective multiple alignment column.
Branch site consensus sequence
The branch site is the key element for lariat formation during the splicing process.25 In fungi, the general branch site motif was determined to be RCTRAY where the A in pos. +5 is the precise branch point whose 2′OH group performs the nucleophilic attack on the first nucleotide of the intron at the 5′ splice site.22,23 Within our dataset, 85% of the potential U6 introns provide a perfect match to the consensus branch site. Remarkably, the branch point adenosine is conserved in each putative intron sequence. The average distance between the branch point A and the 3′ splice site differs significantly between species. A general distance in fungi was denoted to be 8 to 36nt.22,26 The median distance between the branch point A and the 3′ splice site in our intron set is 12nt, see Fig. 2(b).
The U6 intron in S.pombe is not exceptional
The finders of the U6 intron in S.pombe14 suggested it to be “the only known example of a split snRNA gene from any organism – animal, plant, or yeast.” In this study we found that the closely related species Taphrina deformans and Saitoella complicata comprise U6 genes that are likewise interrupted by an intron. Interestingly, these are located at different positions and show no indisputable sequence homology. The latter organisms and the fungi in the Schizosaccharomyces genus are all part of the Taphrinomycotina lineage and encode exactly one U6 snRNA with the intron located in the most conserved region that is thought to be important in U4:U6 interaction.27
Later, experimentally detected U6 snRNA genes in the Basidiomycota species Rhodosporidium dacryoidum and Rhodotorula hasegawae were found harboring one and four introns, respectively.15 These introns, however, show no significant sequence similarity. All four introns were experimentally shown in R. hasegawae to be excised using the pre-mRNA splicing machinery.15 We found additional introns interrupting the U6 genes of the closely related species of Sporobolomyces linderae, Rhodotorula graminis, and Rhodotorula minuta. Again, the introns show neither an obvious sequence similarity to one another nor to the previously detected U6 introns in Basidiomycota. Since all other Basidiomycota contain intronless U6 snRNA genes, the phenomenon of intron interrupted U6 snRNAs can be narrowed to Pucciniomycota, a subgroup of the multifarious Basidiomycota, confer Fig. 1.
We further screened all fungi U6 snRNAs for potential introns and were able to detect additional intron interrupted U6 genes in the subgroups Leotiomycetes, Sordariomycetes and Dothidiomycetes of Pezizomycotina. In Eurotiomycetes, the last subgroup of Pezizomycotina, the U6 snRNA genes are not interrupted by introns. The overall gene structure (number and positions of introns in U6) and number of paralogous U6 snRNA genes in each of these species varies significantly. Most U6 genes encode a single intron, albeit we found U6 genes that encode 3 (e.g. Fusarium graminearum) or even 4 introns (Mycosphaerella pini) in the precursor of the 100nt short snRNA. The Fusarium genus is a perfect example for the rapid changes within the U6 gene structure: each of the 3 Fusarium species harbors exactly 2 U6 genes. However, these differ extraordinarily in their intron count. F.verticillioides on the one hand encodes 2 genes with 1 intron each, while F.oxysporum encodes 1 U6 snRNA with 1 intron and a second U6 snRNA with 2 introns. The previously mentioned F.graminearum encodes 1 U6 gene that is not interrupted by an intron and a second U6 gene harboring 3 introns, while the closely related Nectria haematococca encodes solely 1 U6 gene with 1 intron.
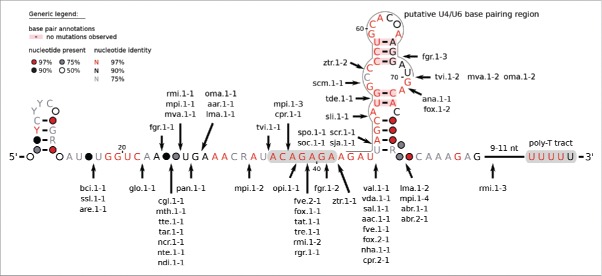
Since U6 genes are highly conserved even among distantly related species, the intron positions can precisely be assigned and are comparable within the snRNA transcript. Contradicting previous remarks that known U6 introns are predominantly located in restricted regions,12,15 all 59 introns presented here are quite uniformly distributed within the snRNA sequence, see Fig. 4. It is apparent, however, that closely related species frequently share introns located at the same positions. This may indicate a common origin of these introns.
Figure 4.
Consensus sequence of all fungal U6 snRNA genes containing at least one intron in their precursor. Intron positions are rather randomly distributed within the U6 gene. Each intron position is precisely indicated by an arrow, introns are denoted by the species 3-letter-abbreviation, the transcript number, and the intron number. The potential base pairing of U4 and U6 snRNAs is indicated by a black line. The marked ACAGAGA region is highly conserved across Fungi and Metazoa and provides the binding site for the 5′ splice site of the intron.28
Intron encoded ncRNAs
In eukaryotes, introns are known hosts for short non-coding RNAs. We tested our introns for similarity to any Rfam annotated RNA family using the GotohScan approach. However, we did not identify such potential short RNA molecules that might be hidden within the U6 introns.
Intron homology
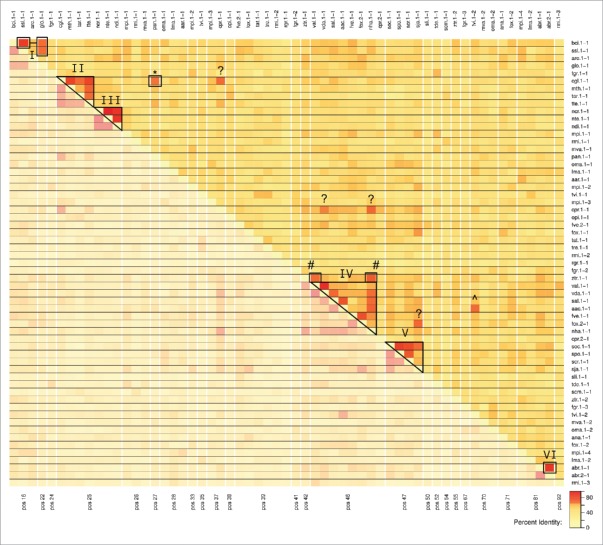
To determine whether introns of different U6 transcripts are related we calculated the pairwise sequence identity of all intron pairs and checked for similar intron positions. Operationally, a set of introns is defined as homologous if each of its members shares a sequence identity of at least 65% to at least 2 other cluster members. We classified 6 such intron clusters, containing 23 introns and 3 loosely linked introns, that might share a common ancestor (Fig. 5). The intron positions and the overall transcript structure is also highly similar within each cluster. Naturally, the evolution of introns is not constrained very much, such that a signal of common origin may be lost already within a few million years. Thus, we cannot interpret lower similarities as proof that sequences are not related by common descent.
Figure 5.
Heatmap representation of the pairwise sequence identities of all U6 introns. The introns are ordered with respect to their absolute position in the U6 snRNA sequence. Clusters of introns showing more than 65% pairwise sequence identity are boxed. Clustered sequences might indicate a common origin. It is apparent that introns that are located at the same position often show a significant sequence similarity.
A subgroup of Sordariales (C.globosum, M.thermophila, T.terrestris, and T.arenaria) shares introns with a mean pairwise identity of 70% (cluster II at position 25 within the snRNA). The single intron of P.anserina U6 (marked with asterisk in Fig. 5), shows 65% sequence identity to the C.globosum intron, but its position is shifted 2 nucleotides downstream (position 27). Introns of the closely related Neurospora species have a mean pairwise similarity of 92% (III) and the exact intron insertion site as the second cluster. Nevertheless, the identities between those 2 clusters range from 43% to 56%, hence we cannot strictly rule out that they either arose from independent intron insertion events that are coincidentally located at the same position or that they in fact descend from a common ancestor that emerged at the root of Sordariales.
The four species of the Glomerellales lineage, V.alfalfae, V.dahliae, S.alkalinus, and A.alcalophilum, show a high sequence similarity (cluster IV at position 46) and share the same intron insertion point indicating a common origin. With the inclusion of the N.haematococca intron, which shares over 65% sequence identity to all 4 Glomerellales, the point of origin might even be shifted to the root of Hypocreomycetidae (incl. Fusarium and Trichoderma, see Fig. 1). This becomes even more plausible with the 2 Fusarium introns of F.verticillioides transcript 1 and F.oxysporum transcript 2, which share a pairwise identity of 78% and, again, are located in the same position. The first intron of Z.tritici on the other hand (ztr.1-1, marked with a ‘#’) has a convincing sequence identity to the nha.1-1 and val.1-1 intron (72% and 67%, respectively), although it is located 4 nucleotides farther upstream. The intron tvi.1-2 of T.virens (denoted with ‘^’) shares 67% sequence identity with its closely related species A.alcalophilum but the insertion point is at position 70, 24 nucleotides further downstream.
Another cluster (V at position 47) comprises the introns of the Schizosaccharomyces U6 snRNA genes with a mean pairwise identity of 70%. Closely related species of T.deformans and S.complicata show neither convincing sequence similarity to this cluster (47% and 34%, respectively) nor to one another (42%). In addition, the introns of these 2 species are shifted 5 and 7 nucleotides downstream, respectively. These facts suggest that there might have been independent intron insertions in the Taphrinomycotina lineage.
A common origin is very plausible for the introns of both A.brassicicola transcripts (VI at position 81), since they share nearly 90% identity. The striking conservation of the mature snRNA but the missing similarity in the flanking regions suggest a gene duplication after the intron insertion.
There are several additional high similarity connections between 2 introns of different species (denoted with a question mark in Fig. 5). The identities of cgl.1-1 with cpr.1-1 (67%), cpr.1-1 with nha.1-1 (68%), and cpr.1-1 with vda.1-1 (65%) potentially indicate a link between the cluster II and IV, although it might not appear to be highly parsimonious. Another high similarity was detected between the single intron of S.japonicus and the intron of F.oxysporum transcript 1 (68%). However, this is probably no true homology, since these 2 species are very distantly related and no other supporting connection in more closely related species was found. Also, note that a large fraction of the intron (approx. 35% of the sequence) holds the promoter specific motifs, hence it is likely to find some similar introns by coincidence.
The remaining 32 introns share only marginal sequence similarity beyond the splice site motifs. They are further located at various different positions within the snRNA gene, even among closely related species. This points at multiple species-specific intron insertions rather than a common ancestral state for these cases.
Pol III promoter elements
We screened all 334 U6 transcripts and their 300nt up- and downstream flanking regions for the characteristic Pol III promoter elements.
TATA box
A TATA box conforming to the consensus motif TATAWW is present in 201 (60,2%) of U6 loci. The median distance between the TATA box and the transcription start is 29nt, with an interquartile range between 27 and 86nt. 59 of these motifs were found in early branching fungi such as Microsporidia, Blastocladiomycota, or Basidiomycota (out of 123 transcripts detected in 43 organisms, 48,0%), while 142 elements were discovered among 211 Ascomycota U6 genes (encoded by 104 organisms, 67,3%).
A box element
An A box promotor element has been identified previously within the mature transcript of the U6 snRNA.11 Our motif search in each mature transcript and both the 5′ and 3′ flanking regions (300nt) returned 163 potential A box sequences in the snRNA genes of Ascomycota. No A box motifs were found in the flanking regions; and none were found outside of Ascomycota in early branching fungi. The consensus sequence is TGGTCAAWTTR, with the invariant bases G, T, C, and A in position 2, 4, 5, and 7 (underlined). See Fig. 6 for the respective sequence logo.
Figure 6.

Sequence logos of different Pol III associated promoter elements derived by MEME. (a) Intronic B box elements were detected 26 times among the 59 U6 snRNA intron sequences. (b) B box motifs in the flanking regions were exclusively found in the 300nt downstream region of 111 U6 transcripts. (c) A box motifs were detected in the mature snRNA of 163 genes. (d) TATA box elements were found in 201 of the 334 300nt long upstream flanking region.
B box element
Intrigued by the finding that the Pol III associated B box promoter element is translocated into the intron sequence,14 we analyzed the 59 potential snRNA introns with respect to a present consensus B box motif. We detected B box motifs in 26 distinct introns of 26 distinct U6 transcripts with the consensus sequence GTTCGAWWC (Fig. 6). While these transcripts harbor 36 introns, interestingly, 25 of the potential B boxes are located in the first intron and only a single B box is found in the second intron.
The independent search for potential B box elements in the 300nt downstream region of all 334 U6 genes returned 111 candidates with the consensus GTTCGARWC (Fig. 6). Each B box belongs to the flanking region of a different transcript. An intronic B box was found in only a single U6 gene, that of T.reesei, which also has a B box motif in its downstream region. Thus, in total we found 136 U6 snRNAs in fungi with a B box either in the first or second intron or within the first 300nt downstream region of the gene. Within early branching fungi, only 2 of 123 transcripts are associated with a B box motif (1,6%). In Ascomycota, on the other hand, over 63% of all detected U6 genes (134 of 211 transcripts) have a B box motif.
Overall, the promoter structure appears to be quite flexible in Ascomycota. Even paralogous transcripts or genes of closely related species combine the 3 promoter motifs in various different ways.
Discussion
We systematically analyzed the U6 snRNA gene family in fungi. With 2 exceptions, U6 snRNAs were found in all fungal genomes. We found 59 introns inserted into 46 distinct snRNA genes. The previously described intron interrupted U6 genes are thus not exceptional but rather frequent in fungal U6 genes. A single U6 gene may harbor up to 4 introns. All introns clearly conform to the usual spliceosomal introns in fungi w.r.t. donor, acceptor, and branch point sequences and their length is concerned.
Only closely related species show conservation in intron sequence, count, and position within the snRNA U6 gene. Those introns can clearly be traced back to a shared ancestral state. In contrast, we cannot use the absence of high levels of sequence conservation to conclude that introns have originated independently. As introns evolve very rapidly, evolution may have had enough time to eradicate ancestral sequence similarities. Another possible view on these cases is that the insertion of intron(s) may have happened multiple times during evolution.
U6 genes in fungi also show high diversity in the presence and location of Pol III promotor elements. Some genes are transcribed due to a TATA box, some exhibit a B box while others might be transcribed because of a cooperated promoter consisting of a TATA box, A box, and/or B box. This raises interesting biological questions about the meaning of these differences and the specific transcription levels of the distinct paralogous U6 genes.
Randomly distributed intron insertion points within the mature U6 snRNA, overall low sequence conservation – except of course for the donor, acceptor, and branch point motifs – and the absence of introns in many U6 genes rather suggest that fungal U6 genes acquired introns in multiple independent events. Introns of closely related species, on the other hand, are frequently located at homologous positions and share recognizable levels of sequence similarity. These introns thus form homologous groups. Overall, the (re)organization of the U6 transcript structure seems to be subjected to short time scales since even organisms of the same genus encode several but completely individually organized transcripts (confer the Fusarium or Trichoderma species).
The precise mechanism of intron insertion remains unclear. The randomly distributed introns appear to be at odds with the theory that U6 introns are a product of reverse splicing, i.e., the excised mRNA introns are incorporated in close proximity to the catalytic domain of U6.15 Instead, this might point at a more general and non-spliceosomal insertion mechanism as it was suggested for the mRNA-type intron found in the U3 snoRNA in S.cerevisiae.29 The lineage- and species-specific intron insertion events as they were discovered for fungal U3 snoRNAs30 features significant similarities to the insertion patterns that were observed in this study.
Introns in other spliceosomal RNAs than U6 are found exclusively in the fungi R.hasegawae. In addition to the 4 introns in the U6 gene, there is also one intron each in the U1 and U2 snRNAs and 2 introns in its U5 snRNA.31,32 Whether these results are truly species specific or solely the tip of the iceberg remains to be investigated.
The analysis presented here is entirely based on computational evidence. Therefore, we cannot completely rule out false positives. In those cases where we detected only a single U6 snRNA gene in the genome, this is most unlikely due to the high levels of sequence similarity with the most similar unspliced U6 snRNA sequences and the presence of secondary structure features characteristic for U6 snRNAs. As the U6 snRNA is essential for pre-mRNA splicing in Eukarya, it is also very unlikely that the detected sequence is a pseudogene. In contrast, in genomes where multiple paralogous U6 snRNA sequences were identified by the computational screen, it is indeed possible that only some of the sequences are functional. This is particularly likely in cases where the U6 candidates feature different sequence motifs in their putative promoter regions. Where available, we cross-checked our annotations with available RNA-seq data and found that these are consistent with our homology based gene models. Especially in F.graminearum it is clearly confirmed that all 3 introns are spliced at the predicted canonical splice sites, see Fig. 7. Additional figures can be found in the supplement.
Figure 7.
Mapping between RNA-seq reads and the intron interrupted U6 gene in F.graminearum. Both upper tracks contain the computationally identified exonic and intronic regions of this transcript while the mapped reads are shown below.19
Footnotes
www.bioinf.uni-leipzig.de/publications/supplements/15-046
http://meme-suite.org/doc/fimo.html
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported in part by the Deutsche Forschungs Gemeinschaft (Project STA 850/15-1).
References
- 1.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol February 2014; 15(2):108-21; PMID:24452469; http://dx.doi.org/ 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bringmann P, Appel B, Rinke J, Reuter R, Theissen H, Lührmann R. Evidence for the existence of snrnas u4 and u6 in a single ribonucleoprotein complex and for their association by intermolecular base pairing. EMBO J June 1984; 3(6):1357-63; PMID:6204860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hashimoto C, Steitz JA. U4 and u6 rnas coexist in a single small nuclear ribonucleoprotein particle. Nucleic Acids Res April 1984; 12(7):3283-93; PMID:6201826; http://dx.doi.org/ 10.1093/nar/12.7.3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brow DA, Guthrie C. Spliceosomal rna u6 is remarkably conserved from yeast to mammals. Nature July 1988; 334(6179):213-8; PMID:3041282; http://dx.doi.org/ 10.1038/334213a0 [DOI] [PubMed] [Google Scholar]
- 5.Singh R, Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal u6 small nuclear rna. Proc Natl Acad Sci U S A November 1989; 86(21):8280-3; PMID:2813391; http://dx.doi.org/ 10.1073/pnas.86.21.8280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunkel GR, Maser RL, Calvet JP, Pederson T. U6 small nuclear rna is transcribed by rna polymerase iii. Proc Natl Acad Sci U S A November 1986; 83(22):8575-9; PMID:3464970; http://dx.doi.org/ 10.1073/pnas.83.22.8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy R, Henning D, Das G, Harless M, Wright D. The capped u6 small nuclear rna is transcribed by rna polymerase iii. J Biol Chem January 1987; 262(1):75-81; PMID:3793736 [PubMed] [Google Scholar]
- 8.Das G, Henning D, Reddy R. Structure, organization, and transcription of drosophila u6 small nuclear rna genes. J Biol Chem January 1987; 262(3):1187-93; PMID:3027083 [PubMed] [Google Scholar]
- 9.Moenne A, Camier S, Anderson G, Margottin F, Beggs J, Sentenac A. The u6 gene of saccharomyces cerevisiae is transcribed by rna polymerase c (iii) in vivo and in vitro. EMBO J January 1990; 9(1):271-7; PMID:2403927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brow DA, Guthrie C. Transcription of a yeast u6 snrna gene requires a polymerase iii promoter element in a novel position. Genes Dev August 1990; 4(8):1345-56; PMID:2227412; http://dx.doi.org/ 10.1101/gad.4.8.1345 [DOI] [PubMed] [Google Scholar]
- 11.Marck C, Kachouri-Lafond R, Lafontaine I, Westhof E, Dujon B, Grosjean H. The rna polymerase iii-dependent family of genes in hemiascomycetes: comparative rnomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res 2006; 34(6):1816-35; PMID:16600899; http://dx.doi.org/ 10.1093/nar/gkl085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tani T, Ohshima Y. The gene for the u6 small nuclear rna in fission yeast has an intron. Nature January 1989; 337(6202):87-90; PMID:2909894; http://dx.doi.org/ 10.1038/337087a0 [DOI] [PubMed] [Google Scholar]
- 13.Potashkin J, Frendewey D. Splicing of the u6 rna precursor is impaired in fission yeast pre-mrna splicing mutants. Nucleic Acids Res October 1989; 17(19):7821-31; PMID:2798130; http://dx.doi.org/ 10.1093/nar/17.19.7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frendewey D, Barta I, Gillespie M, Potashkin J. Schizosaccharomyces u6 genes have a sequence within their introns that matches the b box consensus of trna internal promoters. Nucleic Acids Res April 1990; 18(8):2025-32; PMID:2336389; http://dx.doi.org/ 10.1093/nar/18.8.2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tani T, Ohshima Y. mrna-type introns in u6 small nuclear rna genes: implications for the catalysis in pre-mrna splicing. Genes Dev June 1991; 5(6):1022-31; PMID:2044950; http://dx.doi.org/ 10.1101/gad.5.6.1022 [DOI] [PubMed] [Google Scholar]
- 16.Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, et al.. Rfam 12.0: updates to the rna families database. Nucleic Acids Res January 2015; 43(Database issue):D130-7; PMID:25392425; http://dx.doi.org/ 10.1093/nar/gku1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertel J, de Jong D, Marz M, Rose D, Tafer H, Tanzer A, Schierwater B, Stadler PF. Non-coding rna annotation of the genome of trichoplax adhaerens. Nucleic Acids Res April 2009; 37(5):1602-15; PMID:19151082; http://dx.doi.org/ 10.1093/nar/gkn1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 1994; 2:28-36; PMID:7584402 [PubMed] [Google Scholar]
- 19.Wong P, Walter M, Lee W, Mannhaupt G, Münsterkötter M, Mewes HW, Adam G, Güldener U. Fgdb: revisiting the genome annotation of the plant pathogen fusarium graminearum. Nucleic Acids Res January 2011; 39(Database issue):D637-9; PMID:21051345; http://dx.doi.org/ 10.1093/nar/gkq1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bähler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell October 2012; 151(3):671-83; PMID:23101633; http://dx.doi.org/ 10.1016/j.cell.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ries L, Pullan ST, Delmas S, Malla S, Blythe MJ, Archer DB. Genome-wide transcriptional response of trichoderma reesei to lignocellulose using rna sequencing and comparison with aspergillus niger. BMC Genomics 2013; 14:541; PMID:24060058; http://dx.doi.org/ 10.1186/1471-2164-14-541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kupfer DM, Drabenstot SD, Buchanan KL, Lai H, Zhu H, Dyer D W, Roe BA, Murphy JW. Introns and splicing elements of five diverse fungi. Eukaryot Cell October 2004; 3(5):1088-100; PMID:15470237; http://dx.doi.org/ 10.1128/EC.3.5.1088-1100.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irimia M, Roy SW. Evolutionary convergence on highly-conserved 3′ intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet 2008; 4(8):e1000148; PMID:18688272; http://dx.doi.org/ 10.1371/journal.pgen.1000148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rep M, Duyvesteijn RG, Gale L, Usgaard T, Cornelissen BJ, Ma LJ, Ward TJ. The presence of gc-ag introns in neurospora crassa and other euascomycetes determined from analyses of complete genomes: implications for automated gene prediction. Genomics March 2006; 87(3):338-47; PMID:16406724; http://dx.doi.org/ 10.1016/j.ygeno.2005.11.014 [DOI] [PubMed] [Google Scholar]
- 25.Reed R, Maniatis T. The role of the mammalian branchpoint sequence in pre-mrna splicing. Genes Dev October 1988; 2(10):1268-76; PMID:3060403; http://dx.doi.org/ 10.1101/gad.2.10.1268 [DOI] [PubMed] [Google Scholar]
- 26.Mertins P, Gallwitz D. Nuclear pre-mrna splicing in the fission yeast schizosaccharomyces pombe strictly requires an intron-contained, conserved sequence element. EMBO J June 1987; 6(6):1757-63; PMID:3649292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinke J, Appel B, Digweed M, Lührmann R. Localization of a base-paired interaction between small nuclear rnas u4 and u6 in intact u4/u6 ribonucleoprotein particles by psoralen cross-linking. J Mol Biol October 1985; 185(4):721-31; PMID:2932555; http://dx.doi.org/ 10.1016/0022-2836(85)90057-9 [DOI] [PubMed] [Google Scholar]
- 28.Kandels-Lewis S, Séraphin B. Involvement of u6 snrna in 5′ splice site selection. Science December 1993; 262(5142):2035-9; PMID:8266100; http://dx.doi.org/ 10.1126/science.8266100 [DOI] [PubMed] [Google Scholar]
- 29.Myslinski E, Ségault V, Branlant C. An intron in the genes for u3 small nucleolar rnas of the yeast saccharomyces cerevisiae. Science March 1990; 247(4947):1213-6; PMID:1690452; http://dx.doi.org/ 10.1126/science.1690452 [DOI] [PubMed] [Google Scholar]
- 30.Marz M, Stadler PF. Comparative analysis of eukaryotic u3 snorna. RNA Biol 2009; 6(5):503-7; PMID:19875933; http://dx.doi.org/ 10.4161/rna.6.5.9607 [DOI] [PubMed] [Google Scholar]
- 31.Takahashi Y, Urushiyama S, Tani T, Ohshima Y. An mrna-type intron is present in the rhodotorula hasegawae u2 small nuclear rna gene. Mol Cell Biol September 1993; 13(9):5613-9; PMID:8355704; http://dx.doi.org/ 10.1128/MCB.13.9.5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi Y, Tani T, Ohshima Y. Spliceosomal introns in conserved sequences of u1 and u5 small nuclear rna genes in yeast rhodotorula hasegawae. J Biochem September 1996; 120(3):677-83; PMID:8902636; http://dx.doi.org/ 10.1093/oxfordjournals.jbchem.a021465 [DOI] [PubMed] [Google Scholar]