ABSTRACT
Incorporating miRNA-like features into vector-based hairpin scaffolds has been shown to augment small RNA processing and RNAi efficiency. Therefore, defining an optimal, native hairpin context may obviate a need for hairpin-specific targeting design schemes, which confound the movement of functional siRNAs into shRNA/artificial miRNA backbones, or large-scale screens to identify efficacious sequences. Thus, we used quantitative cell-based assays to compare separate third generation artificial miRNA systems, miR-E (based on miR-30a) and miR-3G (based on miR-16-2 and first described in this study) to widely-adopted, first and second generation formats in both Pol-II and Pol-III expression vector contexts. Despite their unique structures and strandedness, and in contrast to first and second-generation RNAi triggers, the third generation formats operated with remarkable similarity to one another, and strong silencing was observed with a significant fraction of the evaluated target sequences within either promoter context. By pairing an established siRNA design algorithm with the third generation vectors we could readily identify targeting sequences that matched or exceeded the potency of those discovered through large-scale sensor-based assays. We find that third generation hairpin systems enable the maximal level of siRNA function, likely through enhanced processing and accumulation of precisely-defined guide RNAs. Therefore, we predict future gains in RNAi potency will come from improved hairpin expression and identification of optimal siRNA-intrinsic silencing properties rather than further modification of these scaffolds. Consequently, third generation systems should be the primary format for vector-based RNAi studies; miR-3G is advantageous due to its small expression cassette and simplified, cost-efficient cloning scheme.
KEYWORDS: amiRNA, miRNA, RNAi, shRNA, siRNA
Introduction
In mammalian cells, sequence-specific gene silencing by way of RNA interference (RNAi) is most often triggered by the introduction of synthetic small interfering RNAs (siRNAs) or short hairpin containing RNAs, which are subsequently processed into siRNAs.1,2 While RNAi has become a transformative molecular genetic tool for loss-of-function studies, underlying this technology are complex, small-RNA-driven biological phenomena devoted to properly maintaining cellular homeostasis, development, and protection from genome invasion.3 Early attempts at generating vector-based RNAi systems made use of artificial, short, fully base-paired hairpin structures, which superficially resemble pre-microRNAs (pre-miRNAs) with minimized stem and loop segments.4-8 While capable of inducing a potent RNAi effect in some instances, such first generation short hairpin triggers (hereafter noted as shRNAs) were found to be poor substrates for small RNA biogenesis factors, are processed into a heterogeneous mix of small RNAs, and accumulation of their precursor transcripts has been shown to induce sequence-independent, non-specific effects in vivo.9-13
Subsequent attempts to improve upon the efficacy and specificity of vector-based RNAi formats employed endogenous miRNA like scaffolds (termed sh-miRs or artificial miRNAs, and noted hereafter as amiRNAs), including single-stranded, stem-flanking sequence elements, which can influence the accuracy and efficiency of small RNA processing.14-20 Indeed, expression of amiRNAs exhibited reduced sequence-independent, non-specific effects relative to shRNAs, but various reports suggested this may be at the expense of RNAi potency.21-23 To counteract the inconsistent performance of vector-based RNAi systems, a promising strategy using large-scale, sensor-based assays was developed, whereby thousands of amiRNAs against a given target can be evaluated in parallel, and this approach has successfully identified potent, hairpin-derived guide RNA sequences.24 However, the platform used to carry out such screens is specialized and not accessible for most labs, and scaling up for multi-gene or genome-wide target site identification would be time and cost intensive.
More recently, the overall efficiency of vector-based RNAi has been improved as a consequence of an evolving and deeper understanding of small RNA biogenesis pathways. A direct example of this was the optimization of human miR-30a-based amiRNAs, leading to creation of the “miR-E” format through the unmasking of a conserved CNNC motif, which was first identified as a putative miRNA-processing enhancer within the miRNA stem 3p flanking sequences, and was inadvertently destroyed when creating earlier variants of miR-30-based amiRNAs.25,26
Although miR-E was shown to be a more effective form of the miR-30a-like amiRNA context, endogenous miR-30a displays relatively symmetric processing of both 5p and 3p strands of this stem into functional small RNAs, suggesting it may not be an optimal native stem loop format for maximizing siRNA guide relative to passenger strand delivery.17,27,28 Subtle modification of the endogenous miR-30a stem region appears to minimize this shortcoming.14,26 However, we reasoned that separate miRNA contexts, which express high levels of precise, asymmetrically processed ˜21-nucleotide (nt) RNAs may provide enhanced silencing relative to miR-E. In addition, cloning into miR-E, via >100 nt-long DNA oligonucleotides (oligos) or some combination of long oligos plus PCR, is both costly and time consuming. Therefore, an amiRNA context that enables a simplified, shortened oligo-based cloning scheme would provide significant benefit.
Following bioinformatic and experimental assessment, we have identified human miR-16-2 to be a distinct, highly functional amiRNA context (referred to as miR-3G herein), which we have modified so that unique targeting sequences can be cloned through a single-step, <100 nt oligo based approach. Despite unique sequences, structures, strandedness, and engineering schemes, miR-3G and miR-E operated with remarkable consistency to one another across all evaluated guide RNAs and with broadly improved efficacy relative to first and second generation hairpin formats. Deep sequencing revealed that the third generation hairpin contexts provided the highest aggregate expression of precisely defined guide RNAs, consistent with their superior performance in our assays. Moreover, targeting sequences with potency equal to or greater than those obtained in sensor-based assays could be readily identified by combining a standard siRNA design algorithm with the miR-3G or miR-E-based contexts.
Together, our data supports the conclusion that the third generation amiRNAs provide favorable contexts for siRNA processing and efficacy, and subsequent gains in vector based RNAi function will derive from improvements in expression of the hairpin-containing transcripts themselves, through combinatorial hairpin expression schemes, or through further definition of sequence-specific features that can be used to predict those siRNAs that are most likely to be processed asymmetrically, precisely, and at comparatively high volume. The use of miR-3G over miR-E has net advantages due to miR-3G's compact expression cassette and low-cost, single step cloning process.
Results
In an effort to develop potent, expression system-independent amiRNA vectors that are also cost-effective to clone, we first surveyed human and mouse miRNA datasets within miRBASE (www.mirbase.org) and cross-checked tissue-specific expression via small RNA sequencing data.28,29 Specifically, we hypothesized that a given miRNA context would make for an optimized amiRNA platform if it were 1) expressed across a broad tissue range and not reported to undergo post-transcriptional regulation, so that potency is expected to be maintained in most cell types, 2) naturally asymmetric with the mature, single stranded ∼21-nt miRNA derived exclusively from either the 5p or 3p arm of the stem and processed such that the 5′ nt of the mature ∼21-nt miRNA is invariant in order to minimize off-targeting, and 3) the stem and loop segment of the miRNA was relatively small and rigid so as to simplify and reduce costs associated with amiRNA vector cloning. Both human and mouse miR-16-2 met these criteria, and we utilized the human locus as the template for our third generation amiRNA vector development.
In order to construct and evaluate miR-16-2-based systems as RNAi triggers, we cloned an ˜175 bp fragment containing the native miR-16-2 stem and loop, as well as its flanking regions (˜35 bps on each side of the stem) into the 3′ UTR of a turbo RFP-reporter gene whose expression was driven by a CMV (Pol-II) promoter. Within this context we exchanged the first 21 nts of the reported miR-16-2 mature 5p (targeting/guide/antisense) and corresponding 3p (passenger/sense) sequences for 6 unique ff-luc targeting siRNAs. Notably, the mature miR-16-2 sequence is reported to be 22 nts in length, and a 3′ terminal G may be processed along with each unique 21-mer cloned into a miR-16-2 context (Fig. 1A). Three of these targeting sequences had previously been identified as functional within a Drosophila miR-1 amiRNA context (Hal), a miR-30a sensor-based assay (Han, also known as Luci.1309), and as an siRNA (Tus).24,30,31 Sequence “Han” was characterized as being particularly potent, and this was used as a baseline positive control for optimal silencing efficiency in our assays. Three additional siRNAs were designed using the DSIR algorithm with the “21-nt siRNA” settings.32 During incorporation of the ff-luc targeting sequences, the native miR-16-2 stem structure was maintained by mismatching nts 1, 11, 12, and 21 relative to the guide RNA. In parallel, we cloned the identical targeting sequences into a modified variant of the miR-16-2 context (termed miR-3G), which contained novel MluI and EcoRI cloning sites in the 5p and 3p arm-flanking sequences, respectively, and a fully base paired guide-passenger strand stem configuration, except for a mismatch at position 1 relative to the guide strand. The engineered restriction sites in miR-3G facilitate the generation of new targeting constructs via 88-mer duplexed DNA oligonucleotides without compromising the predicted secondary structure of the miR-16-2 hairpin and flanking elements (Fig. S1 and S2). Importantly, the native miR-16-2 3p flanking sequence contains 2 CNNC motifs as well as a putative and newly-identified GHG motif, which may act as small RNA processing enhancers.25,33 One of the CNNC motifs, which is outside of the optimal RNA processing enhancer region, as well as the potential GHG motif were modified in miR-3G in order to facilitate oligo-based cloning and to maintain the predicted, native miR-16-2 secondary structure (Fig 1A, red text). While the siRNAs designed for and used in this study possess inherent thermodynamic asymmetry, which is expected to favor loading of the guide strand into RISC, the mismatch at position 1 of the target strand, within the miR-3G context, may reinforce this imbalance by facilitating the generation of a “frayed” siRNA duplex post-DICER cleavage.34,35
Figure 1.
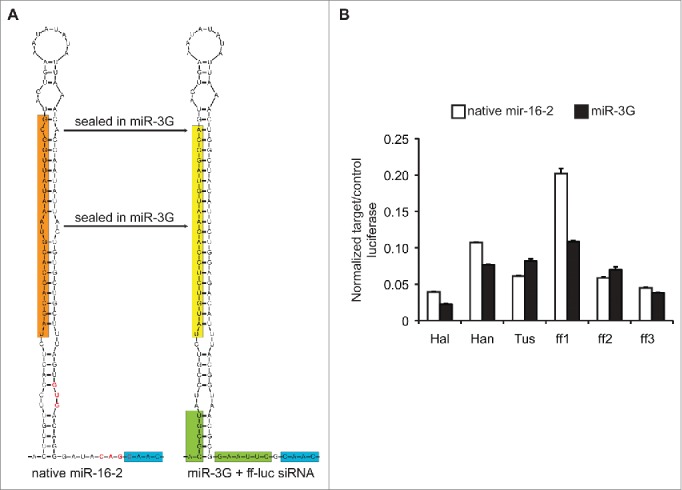
Structure and function of miR-16-2-based RNAi triggers. (A) Comparison of the native miR-16-2 hairpin context relative to the miR-3G format. The mature, primary, 5p miR-16-2 sequence is highlighted in orange, whereas the targeting (antisense) sequence derived from miR-3G is highlighted in yellow. The novel MluI (ACGCGU) and EcoRI (GAAUUC) sites for cloning unique hairpins into miR-3G are highlighted in green. Note, the native miR-16-2 context contains a GHG motif and 2 putative CNNC miRNA processing enhancers, red text and blue highlight. The sequences in red text, including the GHG motif and one of the putative CNNC motifs, has been modified in miR-3G so as to incorporate the EcoRI cloning site. Structures were predicted using the default settings from the mFold web server (http://unafold.rna.albany.edu/?q = mfold/rna-folding-form).50 These are partial sequence views, and structures of the full ∼175 nt contexts are available in Supplemental Figure 2. (B) Knockdown efficiency of distinct targeting sequences within the native miR-16-2 (black bars) or miR-3G (white bars) hairpin contexts. ff-luc (target) was normalized to Rr-luc (control) luciferase expression, and all amiRNAs were co-transfected in a 1:30 w/w ratio to the target plasmid. Knockdown was measured ∼40 hour post transfection into HEK293T cells. Shown is a representative experiment (N = 2 ) performed in technical triplicate. Error bars represent one standard deviation of uncertainty, based on the sampled triplicates, and bars represent the mean for individual data sets.
To assess the ability of the endogenous or “miR-3G” modified variants of miR-16-2 to induce gene silencing, we compared these formats, with the luciferase-targeting sequences described above, side-by-side in a transient, quantitative dual luciferase assay under target excess conditions (1:30 amiRNA:target plasmid concentration) in HEK293T cells (Fig. 1B). Here, the target (ff-luc) knockdown was measured relative to a co-transfected internal control, Rr-luc, in the presence of each unique amiRNA expression plasmid ∼40 hours post-transfection. While the native and modified variants induced ˜80-95% silencing under these conditions, the miR-3G format provided equally potent silencing with less variation sequence-by-sequence. Together, these results suggest the miR-16-2 context can be used effectively to induce RNAi, and that the miR-3G modifications, including the disruption of one of the 2 CNNC motifs and the GHG motif, do not hinder silencing relative to the native context.
To assess the broader functionality of miR-3G, we compared 10 ff-luc and 8 Rr-luc targeting sequences expressed from miR-3G to the same sequences expressed from distinct first, second, and third generation hairpin contexts in the dual luciferase assay format described above (Fig. 2). Specifically, for first generation shRNA systems, we chose to evaluate the widely-adopted TRC format as well as a hybrid shRNA/miR-30a system (noted here as BD).21,36 For second generation amiRNAs, we used the pSM2/miR-30a-based context.14 The third generation miR-E format used for comparison was discussed above.26 The six ff-luc targeting sequences described in Fig. 1B were used in these experiments, as well as 4 additional ff-luc-specific DSIR-designed guides. All Rr-luc targeting sequences were generated using the DSIR algorithm. See Fig. S3 for an alignment of the minimal expression cassette for each of the evaluated hairpin formats, and Table S1 for the oligonucleotide sequences used to assemble individual targeting vectors.
Figure 2.
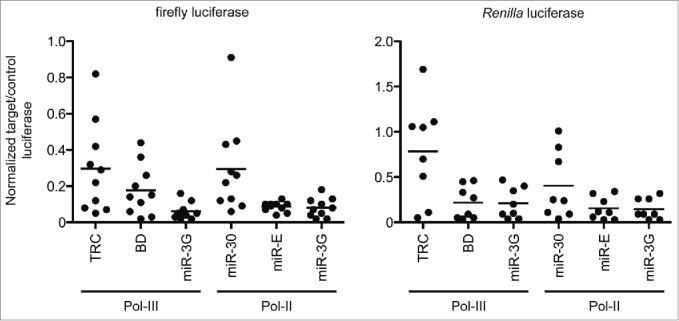
Evaluation of knockdown efficiency between first, second, and third generation vector-based RNAi triggers. Left, knockdown efficiency of ff-luc relative to Rr-luc within distinct hairpin expression contexts. 10 distinct targeting sequences were evaluated with hairpins expressed from a human U6 promoter (Pol-III) or CMV (Pol-II) promoter, all of which were co-transfected in a 1:30 w/w ratio to the target expression plasmid. Evaluated were the first generation shRNA system, TRC, the hybrid first and second generation system, BD, the second generation amiRNA, miR-30, and distinct third generation systems, miR-E and miR-3G. Right, as left, with 8 unique sequences targeting Rr-luc. Knockdown was normalized to firefly luciferase. All knockdown was measured ∼40 hours post-transfection into HEK293T cells. Shown is a representative experiment (N = 2 ) performed in technical triplicate. Each point represents an average of the technical triplicates, and bars represent the mean for individual data sets.
So that efficacy was monitored under the optimal expression context for each hairpin format, first generation systems and the complete miR-3G ∼175 mer fragment were expressed from the identical Pol-III promoter element, and the second or third generation amiRNA formats (both miR-E and miR-3G) were expressed from the Pol-II-driven, turbo RFP-3´ UTR configuration described above. Accordingly, we were able to confirm the BD hairpin functioned best among first and second generation formats (Fig. 2 and Table S2). However, in contrast to all tested first and second generation systems, the third generation amiRNA contexts enabled more robust and consistent silencing with the bulk of targeting sequences against either luciferase variant. Interestingly, while it has been reported that amiRNAs function poorly when expressed from a Pol-III promoter, in aggregate, miR-3G performed as well or better than the first and second generation systems within this context, based on the overall potency and consistency of knockdown.15
During these evaluations we noted near identical silencing efficiency, sequence-by-sequence, between the miR-E or miR-3G formats. To ascertain a true difference in promoting knockdown, if any, between the 2 contexts, we further sensitized the dual-luciferase assay conditions by increasing the amiRNA:target plasmid concentration differential (1:60) and expanding our assessment from 10 to 24 total ff-luc targeting sequences (Fig. 3). In sum, no separation in performance was observed for these amiRNA formats despite modest disparities between a few individual sequences between systems. When looking across both formats, ˜40% of sequences performed as well or better than Han under these assay conditions (Fig. 3B, 9/23 sequences). Conversely, targeting sequences with similar potency were found at rates of ˜2.5% on average within the second-generation miR-30a format, as determined by large-scale sensor based assays and without pre-selection of sequences by way of a target design algorithm.24 Altogether, we conclude that the miR-E and miR-3G formats, in spite of their dissimilar loops, stem sequences, flanking nucleotide sequences and length, as well as strandedness, are functional equivalents, pair effectively with modern siRNA design algorithms, and provide optimized silencing over earlier generation hairpin contexts.
Figure 3.
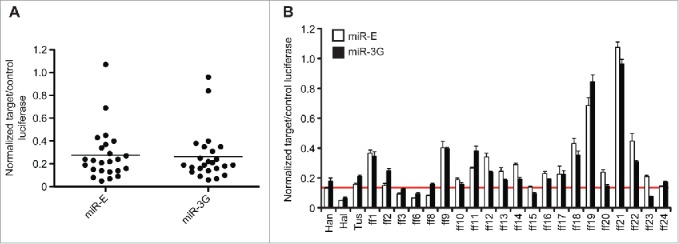
Expanded and sensitized comparison of miR-3G vs. miR-E-based knockdown. (A) 24 unique firefly luciferase targeting sequences were expressed from either the miR-E or miR-3G hairpin contexts. All amiRNA plasmids were co-transfected into HEK293T cells in a 1:60 ratio with the target (ff-luc) expression vector, and knockdown was evaluated relative to control (Rr-luc) ∼40 hours post-transfection. Each point represents and average of 4 technical triplicates, derived from a representative experiment, and bars represent the mean for individual data sets. (B) All conditions described in (A) shown as pairwise comparisons between each individual targeting sequence delivered via miR-3G (black bars) or miR-E (white bars). Shown is a representative experiment (N=2 ) performed in technical quadruplicate. Error bars represent one standard deviation of uncertainty, based on the sampled quadruplicates. A red bar marks the efficiency of the Han targeting sequence in the miR-E format.
Previous studies have suggested that the relative proportion of guide RNAs expressed from an shRNA or amiRNA is a primary driver of vector-based RNAi efficiency.26,37 To explore the relationship between guide RNA production and knockdown potency or consistency across the hairpin formats tested in this study, we employed deep sequencing of small RNAs from cells transfected with pools of 6 common targeting sequences expressed from each context. This approach also allowed us to perform unbiased analyses of small RNA processing features from each context, as hairpin-derived sequence heterogeneity is thought to impact the on and off-target silencing potential for vector-based RNAi. The aligned sequencing reads for each hairpin context, as well as endogenous miR-21 and miR-25 small RNA alignments used for sequencing quality control and expression normalization, can be found in Table S3.
Examination of the observed and predicted small RNA species for each hairpin context revealed that the TRC and BD formats produced relatively few guide RNAs that initiated from the designated 5′ nt (Fig. 4A). Specifically, most guide RNAs derived from the TRC hairpin were shifted ∼4 nt 3′ of the expected 5′ start site, and the BD-derived guide RNAs were shifted ˜1 nt 3´, suggesting the targeting sequence may be out of optimal alignment within each of these contexts. The single nt shift of siRNAs processed from the BD format could contribute to altered silencing potential and/or guide versus passenger strand loading into RISC, as 2 of the 6 guides tested show inverted strand accumulation patterns relative to the second and third generation formats, which display consistent patterns of processing asymmetry for all evaluated targeting sequences (Table S4).3,38 Together, these results suggest that small RNAs derived from more complete miRNA-like RNAi triggers, which include single-stranded stem loop segments, are processed with higher fidelity, relative to the artificial, pre-miRNA-like structures that comprise the TRC and BD contexts. This result also suggests that imprecise small RNA processing, which would deliver a heterogeneous mixture of guide and passenger strand RNAs, could be a contributing factor to the previously observed shRNA-dependent toxicity.9,12
Figure 4.
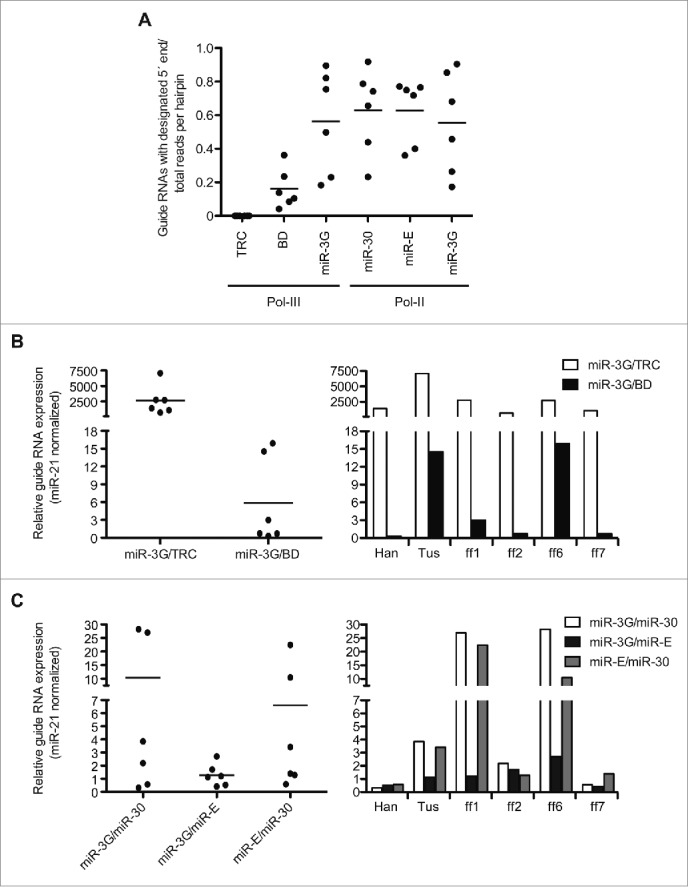
Differences in guide RNA processing fidelity and expression across first, second, and third generation hairpin formats. (A) Displayed are the fraction of guide RNA reads initiating from the designated 5′ end relative to the total small RNA reads for the respective targeting sequences and hairpin contexts (B) Left, relative expression of guide RNA reads derived from the TRC, BD, or miR-3G (Pol-III promoter) contexts that initiated from the designated 5′ end. The counts for each targeting sequence and context were normalized to endogenous miR-21 reads detected for each of the independently sequenced libraries. Right, pairwise comparisons of the individual, normalized guide RNA sequences are displayed. (C) As in (B) for the Pol-II expressed miR-3G, miR-30, and miR-E vector contexts. Bars on each dot plot represents the mean for individual data sets.
Accounting for only those guide RNAs with the designated 5′ nt, we find that, on average, the third generation systems express similar guide RNA levels to one another and modestly (BD) to substantially (TRC) higher guide levels relative to the first and second generation systems. (Figs. 4B and 4C). This result is consistent with the superior performance of the third generation systems in our silencing assays, and is in agreement with previous results suggesting that relative guide RNA expression level positively correlates with knockdown potency and consistency.26,37
Detailed assessment of the guide and passenger strand reads from miR-3G and miR-E suggests strengths and liabilities, with respect to targeting specificity, for each system that may be improved through refined target sequence prediction schemes and alterations of the stem loop sequence/structure. In particular, we noted that while >80% of the sequences for each miR-E derived guide RNA initiated from the designated 5′ nt, 2/6 miR-3G derived guides did not reach this threshold (Fig. 5A). This is consistent with the finding that the 5′ terminal nt of miRNAs derived from the 5p arm of their stem loops, like miR-3G derived guides, can be less precisely defined than the 5′ ends of 3p arm-derived small RNAs, such as those from miR-E26. Contrary to those observations, however, and not unlike observations made in the Drosophila system, 4/6 miR-3G-derived guide RNAs were processed with equal precision to those from miR-E, suggesting that siRNA-intrinsic sequences may impact small RNA processing fidelity, independent of the miR-16-2 scaffold, and definition of these sequence features could be exploited to improve the design and expression of miR-3G adapted siRNAs.39 Separately, a comparison of guide vs. passenger strand accumulation patterns (inclusive of all small RNAs containing +/− 1 5´nt of the predicted guide RNA and +/− 1 5 nt of the dominant passenger sequence) revealed that miR-3G generates small RNAs with a ∼2-fold average greater ratio of asymmetry, relative to those derived from miR-E (Fig. 5B). The observed skewing of guide strand accumulation for miR-3G compared to miR-E occurs despite similar trends in sequence-by-sequence asymmetry for the 2 contexts. This could reflect the intrinsically higher asymmetric processing of endogenous miR-16-2, with respect to miR-30a, and suggests that further manipulation of the miR-E stem loop may lead to more favorable guide versus passenger strand ratios and decreased off-target silencing potential.
Figure 5.
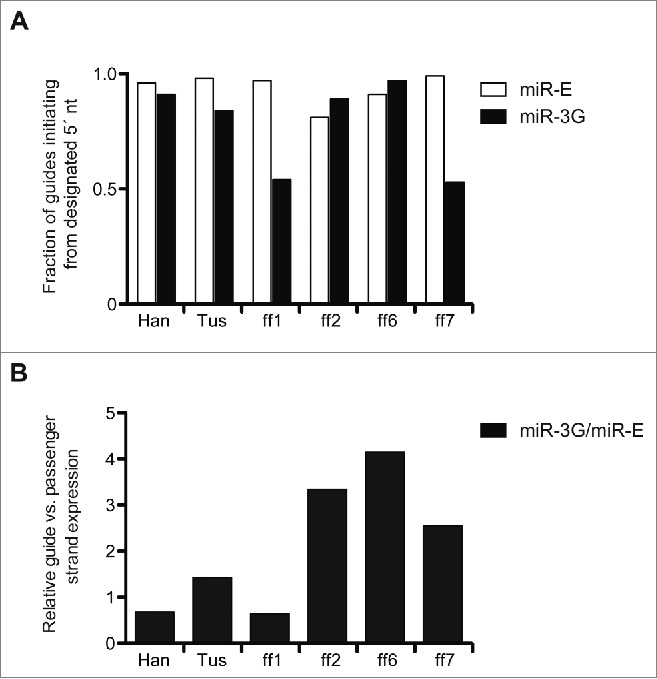
Potential liabilities identified for miR-3G and miR-E-based RNAi. (A) Total guide RNA values were determined by adding all reads with the designated guide RNA 5′ nt, as well as those +/− 1 nt 5′ shifted from the appropriate start site. Displayed are the ratios, for each targeting sequence and hairpin context, of guides with the appropriate 5′ nt relative to the total guide RNA read count. (B) Total guide RNA read counts (computed as in A) and passenger strand reads, which included the dominant passenger strand read and all reads with a +/− 1 nt 5′ shift are compared for miR-3G relative to miR-E.
Discussion
RNAi has proven to be a revolutionary approach for generating loss-of-function phenotypes in a host of mammalian systems. Vector-based RNAi technology, in particular, provides the opportunity and flexibility to evaluate gene function in vitro or in vivo under stable, inducible, or reversible knockdown conditions.40 Over the past decade, vector-based RNAi systems, formatted as either shRNAs or amiRNAs, have also been adapted for genome-scale screening applications, either in arrayed or pooled settings.40-43 Collectively, however, one conclusion reached from the numerous studies utilizing vector-based RNAi is that effective hairpin-derived targeting sequences are rare and difficult to predict, with up to 80% of hairpins being described as non-functional in some shRNA libraries.44
We hypothesized that the paucity of effective sequences delivered by way of vector-based RNA triggers was not the result of gross deficiencies in the target site identification schemes per se, but rather was due to sub-optimal functionality of the first or second generation hairpin contexts themselves. Therefore, we and others have evaluated endogenous hairpin structures, modified to enable simplified cloning, that promote idealized small RNA processing and expression. This led to a pair of distinct amiRNA concepts, miR-E, based on miR-30a, and as described here, miR-3G, based on miR-16-2, both of which we define as third generation amiRNA systems.26
Both miR-3G and miR-E provide substantial and consistent improvement in gene silencing vs. first and second generation systems and, as tested for miR-3G, can be used effectively in both Pol-II and Pol-III expression contexts. Importantly, existing siRNA design algorithms can be paired with third generation hairpin formats to streamline identification of potent targeting sequences. Although we utilized the DSIR algorithm in our study, new algorithms, like shERWOOD, which was derived from hundreds of thousands of experimentally generated data points, and are based on vector-mediated, not synthetic siRNA-triggered RNAi, have the potential to further increase our ability to predict efficacious vector-derived targeting sequences.37 However, while the shERWOOD algorithm was shown to have increased predictive value over DSIR when using the second generation miR-30a amiRNA format, a direct comparison of DSIR or shERWOOD-derived sequences was not performed in the optimal hairpin formats, and it is unclear if the hairpin format itself may balance out the algorithm-dependent effects.
The rationales for choosing miR-30a (miR-E) or miR-16-2 (miR-3G) as amiRNA formats were unique. Yet, in spite of their distinct origins and engineering schemes, we found little or no difference in guide RNA expression levels nor functionality between these 2 contexts across all evaluated targeting sequences. We conclude that this reflects the peak delivery and efficacy of each unique targeting sequence from a vector-based platform, and that, based on our current understanding of small RNA processing and expression, the third generation contexts represent the optimal formats for single-hairpin, vector-based RNAi studies. Nevertheless, of the 2 third generation systems, miR-3G provides a simpler, more cost-effective cloning scheme, where relatively short oligonucleotides are used to generate each unique amiRNA expression vector. In addition, the miR-3G expression cassette is less-than half the size of miR-E (˜175 vs. ∼375 nts), which may be of value for packaging-size limited viral delivery of amiRNA containing transgenes.
The benefits of adopting these new hairpin formats are expected to be numerous. For example, increasing the number of effective amiRNAs per gene, by using miR-3G or miR-E and advanced target design algorithms, would allow for reduction in the overall complexity of deep coverage, pooled hairpin libraries, simplifying downstream experimental setup and hit calling. Being able to express optimized amiRNAs via Pol-II and Pol-III promoter types opens the door for more flexible delivery options, within both basic research and therapeutic settings, the latter of which may require maximal delivery and silencing from allele-specific targeting sequences that do not necessarily conform to optimized siRNA design schema.45-47 While the third generation vectors provide enhanced small RNA expression and knockdown, relative to first and second generation systems, we have observed potential liabilities, with respect to guide RNA processing fidelity (miR-3G) and passenger strand accumulation into RISC (miR-E), which could impact on and off-target silencing in sensitive settings, such as human gene therapy. At least some of these effects can be attributed to siRNA-sequence intrinsic factors, and further deep sequencing analyses of an expanded cohort of small RNAs derived from third and future generation vector systems may provide insight into the small RNA sequence-dependent, hairpin-scaffold-independent features that comprise optimal vector adapted siRNAs. Looking forward, the removal of significant functional constraints thought to be inherent to vector-based RNAi systems is predicted to enhance all varieties of knockdown studies, and in lieu of future breakthroughs in small RNA biogenesis mechanisms, it is likely that subsequent gains in vector based RNAi potency will come from continued refinement in the expression vectors themselves (promoters, etc.), combinatorial or tandem hairpin expression concepts, and enhanced target design algorithms or large-scale sensor-like target identification schemes.23,24,31,37,48
Materials and methods
shRNA and amiRNA vector construction
Previously unreported siRNAs against firefly (Photinus pyralis) luciferase (ff-luc, pGL3, Promega) and Renilla reniformis luciferase (Rr-luc, pRLTK, Promega) were designed via the DSIR algorithm, using the 21 nt siRNA settings. First generation shRNAs were engineered as described, and were cloned as AgeI-EcoRI or KpnI-EcoRI-overhanged duplexed oligos into a TRC2-pLKO-puro vector (Sigma-Aldrich, product #SHC201).21,36 The miR-3G context was cloned into a variant of TRC2-pLKO-puro as follows. A turboRFP (Evrogen) expression cassette was exchanged for the puromycin resistance gene within TRC2-pLKO-puro. We term the resulting vector TRC2-pLKO-tRFP. Subsequently, we cloned an ˜160 bp sequence, as duplexed oligonucleotides containing the 5p and 3p miR-3G flanking sequence and the MluI-EcoRI unique amiRNA cloning sites, just downstream of the U6 promoter. Individual miR-3G hairpins were then cloned into this vector as duplexed 88-mer oligonucleotides (Fig. S1). Second and third generation amiRNAs were cloned into a modified variant of pLENTI6.3 (LifeTechnologies), which was modified by way of gene synthesis to include a turboRFP expression cassette downstream of the CMV promoter, as well as the appropriate miR-3G, pSM2/miR-30a, or miR-E expression contexts within the turboRFP 3′ UTR. We term this vector pLENTI6-tRFP. Native miR-3G targeting sequences were cloned into pLENTI6-tRFP as ˜175 bp SpeI-XhoI overhanged oligos. miR-3G sequences were cloned into pLENTI6-tRFP as 88 mer, duplexed MluI-EcoRI overhanged oligos, pSM2/miR-30a as 110 mer XhoI-EcoRI overhanged oligos, and miR-E as 125 mer XhoI-EcoRI overhanged oligos. Propagation of vectors was performed using Stbl3 cells (LifeTechnologies) to avoid recombination. DNA was prepared for each vector using the HiSpeed Plasmid Maxi system (Qiagen). The quantity and quality of each preparation was evaluated with a Nanodrop 8000 (Thermo Scientific). See Table S1 for all shRNA/amiRNA insert sequences. Full expression vector sequences will be made available upon request.
Cell culture and reporter assay
All dual luciferase assays were performed using HEK293T cells in 96 well format, using white, flat, clear bottom tissue culture plates (Corning). One day prior to transfection, 10,000 cells were seeded (100 ul of media and cells/well). pGL3 and pRLTK were co-transfected with each shRNA/amiRNA, as described below. For each transfection at 1:30 shRNA/amiRNA to target, a 100 ul mastermix containing OPTIMEM (LifeTechnologies), 4 ul Fugene6 (Promega), 970 ng of the target vector, 180 ng of the control vector, 30 ng of the shRNA/amiRNA expression vector was prepared. Six ul of the mastermix was added to each replicate well. The amount of each amiRNA expression vector was reduced to 15 ng for 1:60 amiRNA/target transfection conditions. ∼40 hours post transfection the cells were lysed and evaluated for ff or Rr-luc expression via the DualGlo luciferase assay (Promega) using an Envision system (PerkinElmer). Knockdown was normalized relative to the control luciferase. All assays were performed in biological duplicate and either technical triplicate or quadruplicate, as noted in the figure legends.
Deep sequencing and analysis of shRNA and amiRNA-derived small RNAs
Independent pools for each hairpin context, including miR-3G in the Pol-II and Pol-III expression formats, were created by combining, 1:1, ff-luc targeting sequences Han, Tus, ff1, ff2, ff6, and ff7, with a GFP expression vector. For each pool, a transfection mix containing 100 ul OPTIMEM, 4 ul Fugene6, and a total ∼1 ug of DNA [900 ng GFP expression vector and 75 ng shRNA/amiRNA pool (12.5 ng/ul for each targeting sequence)] was added to 200,000 HEK293T cells, per well, in quadruplicate 6 well plates (Corning). ∼72 hours post-transfection, total RNA was collected from each well using the miRNeasy mini kit (Qiagen) following the manufacturer's instructions. Total RNA from each well was quantified using a Nanodrop 8000 (Thermo Scientific), and material from the replicate wells was concentration normalized with RNAase-free water and pooled. Small RNAs derived from each shRNA/amiRNA pool were adapted for deep sequencing with the TruSeq Small RNA preparation kit (Illumina). The libraries were multiplexed and sequenced using an Illumina HiSeq2500 (50 basepair, single-end reads), and 5′ and 3′ adaptor sequences were trimmed from the resultant reads. Library preparation and sequencing was performed at Elim Biopharmaceuticals (Hayward, CA). Reads lacking the 3′ adaptor or having a post-trimming sequence of <18 basepairs were eliminated from further analysis. BLAST was used to align the trimmed reads to the shRNA/amiRNA reference sequences or control miRNA loci.49 A valid alignment was required to be exact, have no gaps or mismatches, and have at least 18 identities.
Supplementary Material
Disclosure of potential conflicts of interest
C.W., T.L.C., and B.H. were employees of Genentech, a member of the Roche Group, at the time this work was performed, and they declare competing financial interests.
Acknowledgments
I would like to thank the Genentech oligo synthesis and DNA sequencing groups for their exceptional help and support throughout this project. I am indebted to Christian Siebel, David Davis, Yuxin Liang, Honglin Chen, and Mark McCleland for the suggestions and reagents each provided during the course of these experiments.
References
- 1.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov 2004; 3:318-29; PMID:15060527; http://dx.doi.org/ 10.1038/nrd1345 [DOI] [PubMed] [Google Scholar]
- 2.Singer O, Verma IM. Applications of lentiviral vectors for shRNA delivery and transgenesis. Curr Gene Ther 2008; 8:483-8; PMID:19075631; http://dx.doi.org/ 10.2174/156652308786848067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002; 296:550-3; PMID:11910072; http://dx.doi.org/ 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- 5.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 2002; 16:948-58; PMID:11959843; http://dx.doi.org/ 10.1101/gad.981002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci U S A 2002; 99:1443-8; PMID:11818553; http://dx.doi.org/ 10.1073/pnas.032652399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol 2002; 20:505-8; PMID:11981566; http://dx.doi.org/ 10.1038/nbt0502-505 [DOI] [PubMed] [Google Scholar]
- 8.Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A 2002; 99:6047-52; PMID:11972060; http://dx.doi.org/ 10.1073/pnas.092143499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006; 441:537-41; PMID:16724069; http://dx.doi.org/ 10.1038/nature04791 [DOI] [PubMed] [Google Scholar]
- 10.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al.. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A 2008; 105:5868-73; PMID:18398004; http://dx.doi.org/ 10.1073/pnas.0801775105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denise H, Moschos SA, Sidders B, Burden F, Perkins H, Carter N, Stroud T, Kennedy M, Fancy SA, Lapthorn C, et al.. Deep Sequencing Insights in Therapeutic shRNA Processing and siRNA Target Cleavage Precision. Mol Ther Nucleic Acids 2014; 3:e145; PMID:24496437; http://dx.doi.org/ 10.1038/mtna.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell 2012; 151:900-11; PMID:23141545; http://dx.doi.org/ 10.1016/j.cell.2012.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntyre GJ, Yu YH, Lomas M, Fanning GC. The effects of stem length and core placement on shRNA activity. BMC Mol Biol 2011; 12:34; PMID:21819628; http://dx.doi.org/ 10.1186/1471-2199-12-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al.. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet 2005; 37:1281-8; PMID:16200065 [DOI] [PubMed] [Google Scholar]
- 15.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A 2005; 102:13212-7; PMID:16141338; http://dx.doi.org/ 10.1073/pnas.0506306102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 2005; 37:1289-95; PMID:16200064; http://dx.doi.org/ 10.1038/ng1651 [DOI] [PubMed] [Google Scholar]
- 17.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell 2002; 9:1327-33; PMID:12086629; http://dx.doi.org/ 10.1016/S1097-2765(02)00541-5 [DOI] [PubMed] [Google Scholar]
- 18.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem 2005; 280:27595-603; PMID:15932881; http://dx.doi.org/ 10.1074/jbc.M504714200 [DOI] [PubMed] [Google Scholar]
- 19.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 2005; 24:138-48; PMID:15565168; http://dx.doi.org/ 10.1038/sj.emboj.7600491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006; 125:887-901; PMID:16751099; http://dx.doi.org/ 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- 21.Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA 2008; 14:1834-44; PMID:18697922; http://dx.doi.org/ 10.1261/rna.1062908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Lin X, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA 2007; 13:1765-74; PMID:17698642; http://dx.doi.org/ 10.1261/rna.599107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebbink RJ, Lowe M, Chan T, Khine H, Wang X, McManus MT. Polymerase II promoter strength determines efficacy of microRNA adapted shRNAs. PLoS One 2011; 6:e26213; PMID:22031824; http://dx.doi.org/ 10.1371/journal.pone.0026213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al.. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell 2011; 41:733-46; PMID:21353615; http://dx.doi.org/ 10.1016/j.molcel.2011.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell 2013; 152:844-58; PMID:23415231; http://dx.doi.org/ 10.1016/j.cell.2013.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, et al.. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 2013; 5:1704-13; PMID:24332856; http://dx.doi.org/ 10.1016/j.celrep.2013.11.020 [DOI] [PubMed] [Google Scholar]
- 27.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002; 12:735-9; PMID:12007417; http://dx.doi.org/ 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- 28.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014; 42:D68-73; PMID:24275495; http://dx.doi.org/ 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al.. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129:1401-14; PMID:17604727; http://dx.doi.org/ 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411:494-8; PMID:11373684; http://dx.doi.org/ 10.1038/35078107 [DOI] [PubMed] [Google Scholar]
- 31.Haley B, Hendrix D, Trang V, Levine M. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev Biol 2008; 321:482-90; PMID:18598689; http://dx.doi.org/ 10.1016/j.ydbio.2008.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics 2006; 7:520; PMID:17137497; http://dx.doi.org/ 10.1186/1471-2105-7-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang W, Bartel DP. The Menu of Features that Define Primary MicroRNAs and Enable De Novo Design of MicroRNA Genes. Mol Cell 2015; 60:131-45; PMID:26412306; http://dx.doi.org/ 10.1016/j.molcel.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003; 115:199-208; PMID:14567917; http://dx.doi.org/ 10.1016/S0092-8674(03)00759-1 [DOI] [PubMed] [Google Scholar]
- 35.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003; 115:209-16; PMID:14567918; http://dx.doi.org/ 10.1016/S0092-8674(03)00801-8 [DOI] [PubMed] [Google Scholar]
- 36.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al.. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 2006; 124:1283-98; PMID:16564017; http://dx.doi.org/ 10.1016/j.cell.2006.01.040 [DOI] [PubMed] [Google Scholar]
- 37.Knott SR, Maceli AR, Erard N, Chang K, Marran K, Zhou X, Gordon A, El Demerdash O, Wagenblast E, Kim S, et al.. A Computational Algorithm to Predict shRNA Potency. Mol Cell 2014; 56(6):796-807; PMID:25435137; http://dx.doi.org/ 10.1016/j.molcel.2014.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan GC, Chan E, Molnar A, Sarkar R, Alexieva D, Isa IM, Robinson S, Zhang S, Ellis P, Langford CF, et al.. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res 2014; 42:9424-35; PMID:25056318; http://dx.doi.org/ 10.1093/nar/gku656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA* strands in flies. Curr Biol 2008; 18:147-51; PMID:18207740; http://dx.doi.org/ 10.1016/j.cub.2007.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fellmann C, Lowe SW. Stable RNA interference rules for silencing. Nat Cell Biol 2014; 16:10-8; PMID:24366030; http://dx.doi.org/ 10.1038/ncb2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods 2006; 3:707-14; PMID:16929316; http://dx.doi.org/ 10.1038/nmeth923 [DOI] [PubMed] [Google Scholar]
- 42.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods 2006; 3:715-9; PMID:16929317; http://dx.doi.org/ 10.1038/nmeth924 [DOI] [PubMed] [Google Scholar]
- 43.Mohr SE, Smith JA, Shamu CE, Neumuller RA, Perrimon N. RNAi screening comes of age: improved techniques and complementary approaches. Nat Rev Mol Cell Biol 2014; 15:591-600; PMID:25145850; http://dx.doi.org/ 10.1038/nrm3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bassik MC, Lebbink RJ, Churchman LS, Ingolia NT, Patena W, LeProust EM, Schuldiner M, Weissman JS, McManus MT. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat Methods 2009; 6:443-5; PMID:19448642; http://dx.doi.org/ 10.1038/nmeth.1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet 2006; 2:e140; PMID:16965178; http://dx.doi.org/ 10.1371/journal.pgen.0020140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding H, Schwarz DS, Keene A, Affar B, Fenton L, Xia X, Shi Y, Zamore PD, Xu Z. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell 2003; 2:209-17; PMID:12934714; http://dx.doi.org/ 10.1046/j.1474-9728.2003.00054.x [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez-Lebron E, Paulson HL. Allele-specific RNA interference for neurological disease. Gene Ther 2006; 13:576-81; PMID:16355113; http://dx.doi.org/ 10.1038/sj.gt.3302702 [DOI] [PubMed] [Google Scholar]
- 48.Sun D, Melegari M, Sridhar S, Rogler CE, Zhu L. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques 2006; 41:59-63; PMID:16869514; http://dx.doi.org/ 10.2144/000112203 [DOI] [PubMed] [Google Scholar]
- 49.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-10; PMID:2231712; http://dx.doi.org/ 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 50.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/ 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


