ABSTRACT
XIST is a long non-coding RNA, which expressed exclusively from the inactive X chromosome. Although it has been revealed that the A-repeat contributes to the X chromosome inactivation (X-inactivation), the role of the longest D-repeat has not yet been investigated. Here, a sgRNA directed CRISPR/Cas9 system which have multiple target sites within repeat D of XIST, were used to generate D-repeat deletion and studied its roles on X-inactivation. The results showed that the deletion of D-repeat caused a significantly decreased expression of XIST, and up regulated expression of X-linked genes, suggesting that the D-repeat may play an important role in the regulation of XIST expression and silencing of the X-linked genes, which could provide a new idea in the molecular mechanisms of X-inactivation.
KEYWORDS: CRISPR/Cas9, D-repeat, long non-coding RNA, XIST; X-inactivation
Introduction
XIST is one of the long non-coding RNA, which is located in X-inactivation center (Xic) and is required for the X-inactivation.1 Human XIST encodes approximately 17 kb of transcripts, and several tandem repeats named A-F repeat are conserved among mammals.2,3 It has been shown that the A-repeat, located in the proximal part of the XIST RNA, contributes to XIST RNA localization and X chromosomal silencing, while other repeats may play an alternative role in transcriptional localization.4,5
Recently, the bacterial clustered regularly interspaced short palindromic repeats (CRISPR) - associated endonuclease 9 (CRISPR/Cas9) system has been utilized as a genome-editing tool for generating mutations specifically and efficiently in varieties of organisms.6,7 While the undesirable off-target effect has been frequently reported in culture cells8-10 and mouse zygotes 11 caused by the mismatches sequence of the single guide RNA (sgRNA). However, the off-target mutation mediated by CRISPR/Cas9 could be used for the disruption of multi-homeologous and paralogous genes members in rice.12
Although it has been well-documented that the A-repeat is responsible for X chromosomal silencing, the role of the longest D-repeat within exon 1 of XIST has not yet been investigated. To investigate the role of the D-repeat in X-inactivation, we developed an efficient strategy to generate a D-repeat knockout by a sgRNA directed CRISPR/Cas9 system. In this study, we reported the novel findings regarding the D-repeat as one of the elements required for X-inactivation.
Materials and methods
Vector construction
The PX459 plasmid (Addgene ID 48139) described previously was used for the cloning of the sgRNA in this study.13 Briefly, the PX459 vector was digested with BbsI and gel purified using the Gel Extraction Kit (Tiangen, Beijing, China). A pair of complementary 20-nt oligonucleotides of guide sequences was annealed at 95°C for 5 min and ramp down to 25°C to generate the dsDNA fragment, and then the sgRNA for the targeting site was cloned into the linearized PX459 vector and sequence confirmed.
Cell culture and DNA transfection
Human embryonic kidney (HEK) 293FT and Hela cell line (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (HyClone), 2mM GlutaMAX (Life Technologies), 100 U/ml penicillin and 100 mg/ml streptomycin and incubated at 37°C in an atmosphere of 5% CO2. The cells were seeded into 6-well plates at a density of 120,000 per-well and transfected using Turbofect™in vitro Transfection Rengent (Fermentas) according to the manufacturer's instructions. Transfected cells were diluted 1:32 and cultured in selective medium using 2.0 mg/mL puromycin (Sigma-Aldrich), then the cell clones were picked up and collected for further study
Mutation detection by PCR
The genomic DNA was extracted from sgRNA and empty vector transfected (Con) cell using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) according to the manufacturer's instructions. The pair of primers used for the amplification of D-repeat as follows: XIST-F, 5′ TGACAATTACATCGTATCCTTCCT 3′, XIST-R, 5′ TGTCCAAGGGACATTGTTGT 3′. Then the PCR products were gel purified using the TIANgel Midi Purification Kit (TIANGEN, Beijing, China) and subjected to pGM-T vector cloning (Tiangen, Beijing, China). Ten positive plasmid clones were sequenced, and the Vector NTI (Invitrogen) and DNAman were used for sequence analysis.
Quantitative real-time PCR (q-PCR)
Total RNA was extracted from sgRNA transfected and control cells using TRNzol-A+ reagent (TIANGEN, Beijing, China) according to the manufacturer's instructions. The isolated RNA was treated with DNase I (Fermentas) to eliminate the residual DNA. The cDNA was then synthesized using the BioRT cDNA First Strand Synthesis Kit (Bioer Technology, Hangzhou, China).
q-PCR was carried out using the BioEasy SYBR Green I Real Time PCR Kit (Bioer Technology, Hangzhou, China) according to the manufacturer's instructions. The GAPDH was used as endogenous gene. The relative gene expression was presented as means ± standard deviation (SD) from 3 cell clones, and analyzed by 2−ΔΔCT formula. Data was statistically analyzed by Graphpad prism software (T test) and a p value < 0.05 was considered statistically significant. The primers used for q-PCR are shown in supplementary table S1.
Off-target analysis
Potential off-target sites (POTS) of the sgRNAs were predicted using the CRISPR/Cas9 online design tool (http://tools.genomeengineering.org). The top 5 POTSs which were most likely to produce off-target mutations were selected and subjected to PCR and sequence analysis. The primers are listed in supplementary table S2.
Karyotype analysis
Metaphase slide preparation and G-banding were performed according to standard procedures. 100 well-spread metaphases cells were captured with Nikon microscope (ts100) and analyzed by Lucia computer analysis system for karyotyping.
Results and discussion
Selection of target sequence and the vector construction
X-inactivation is a chromosome-wide gene silencing regulated by long non-coding XIST RNA, which inactivates one of the 2 X chromosomes in female mammals.14 XIST transcript is composed of 6 tandem repeats defined A to F which are considered to be crucial factors in X-inactivation.5 Although previous study has reported that A-repeat is an essential motif responsible for chromosomal silencing, research on the role of the longest D-repeat within exon 1 is lacking.15 The D-repeat locates in +6086 to +8312, and contains 7.6 copies of 290 bp tandem repeat units in XIST RNA (Fig. 1A). To investigate the role of the D-repeat in X-inactivation, we chose 20bp in the consensus of D-repeat as a sgRNA sequence, which was cloned into PX459 backbone using BbsI site and confirmed by sequencing. The corresponding POTSs in each tandem repeat unit is shown in Fig. 1B. The schematic representation of the vector using in this study is shown in Fig. 1C.
Figure 1.
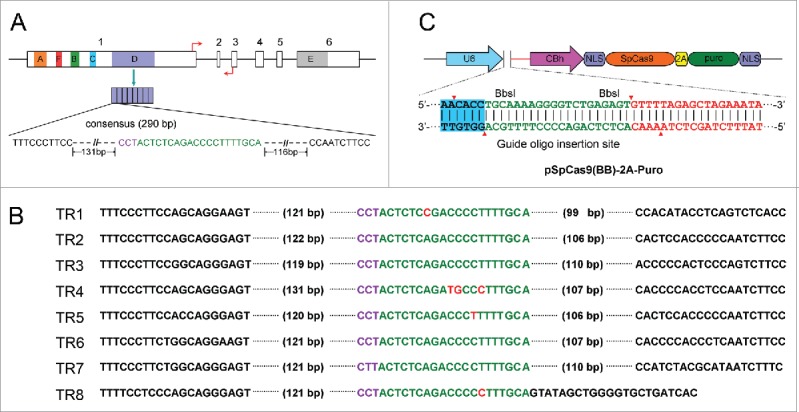
Schematic of CRISPR/Cas9-mediated D-repeat knockout and vector construction. (A) Target site of CRISPR/Cas9-mediated mutagenesis in D-repeat of XIST gene. Gene structure of the human XIST with 6 exons represented as blank boxes and the tandem repeat of A-F are shown with boxes of different colors. The sequence of the sgRNA is shown according to the consensus of 7.6 D tandem repeat (TR) units. The PAM sequence (NGG) is shown in purple and the target sequence is shown in green. The q-PCR primers for the XIST are represented with a pair of red arrows. (B) The alignment of the sgRNA in TR1-TR8 is shown; the red means the mismatches sequence compared to the sgRNA. (C) Vector used in this study. The vector contains sgRNA insertion sites (pink) and the human codon optimized S. pyogenes Cas9 (hSpCas9) (orange).
Knockout of D-repeat using one sgRNA
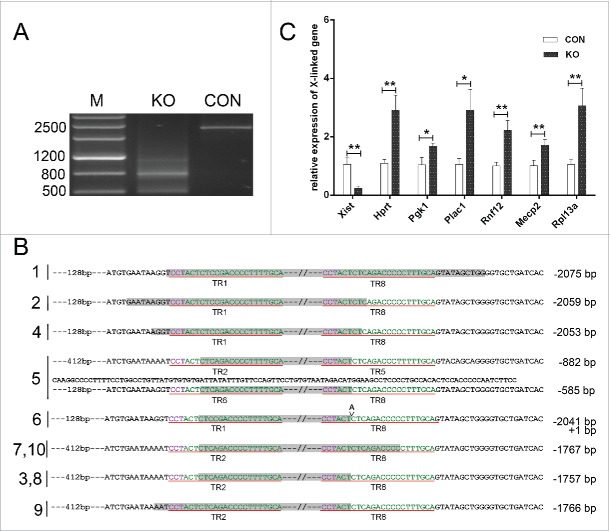
To determine the efficiency of long tandem repeat knockout of D-repeat using one sgRNA, the sgRNA and empty plasmid was transfected into 293FT cells, and then the PCR was performed to amplify the D-repeat using the isolated genomic DNA from sgRNA transfected and control cells. The agarose gel analysis showed a specificity and intact band of 2537 bp in control cells, while there were obscure bands and no PCR product of 2537 bp was detected in sgRNA targeting D-repeat samples, indicating that various types of mutations were occurred after the sgRNA transfection. The band of ˜500 bp suggested that the length of the fragment deletions mediated by the sgRNA was up to 2000 bp (Fig. 2A). To verify this finding, 10 T-vector subclones were sequenced for sgRNA transfected cell group. As shown in Fig. 2B, the deletion of D-repeat mediated by a sgRNA was varied from 1500 bp to 2000 bp, 5 to 7 repeat units, respectively.
Figure 2.
The expression of XIST and X-linked genes after the knockout of D-repeat. (A) The sgRNA- mediated D-repeat mutation were analyzed by PCR and agarose gel. Genomic DNA was extracted from sgRNA transfected and control cells, subjected to PCR and 2% agarose gel analysis. M, marker III. KO, knockout of D-repeat. Con, empty vector. (B) The mutation efficient of were analyzed by T-vector sequences analysis. WT represent the sequence of 7.6 tandem repeat units in D-repeat. The PAM sequence is marked in purple and the off-target sites marked in green. The sequence of region deletions is under gray background and the length of deletions is noted to the right of each sequence (- deletion). The sgRNA targeted TRs were demonstrated in red underlined.(C) The expression of XIST and X-linked genes were investigated by q-PCR. Data are shown as mean ±SEM (n = 3). Asterisks denote significant differences between Con and KO samples; *p < 0.05,**p < 0.001.
Off-target mutation is a critical issue in the application of CRISPR/Cas9 system. To test whether undesired off-target mutation occurred in the human genome in sgRNA transfected cells, 5 POTSs were predicted using the CRISPR design tool and chose to perform off-target analysis. The sequence analysis data revealed that none of the sequencing reads exhibited mutations, suggesting that no off-target occurred in the 5 POTSs (Fig. S1). In addition, to determine if the XIST locus is structurally intact, the primers specificity RT-PCR and Sanger sequence were performed on sgRNA transfected and control cells, which indicated there is no additional inversion or rearrangement in XIST (data not shown).
It has been suggested that the 20 bp sequence in the sgRNA and the 5′-NGG and 5′-NAG protospacer adjacent motif (PAM) play an important role in the specificity of CRISPR/Cas9-mediated DNA cleavage. The ‘seed region’ of sgRNA is approximately 12 bases proximal to the PAM, which is crucial for sgRNA pairing and DNA cleavage.9,10 While in our study, off-target mutation also occurred at the POTS in TR1 which has a mismatch at 7 bp proximal to the PAM. Thus, we suspect that some of these cleavages are not the product of DSB activity but rather an unpredictable non-homologous end joining (NHEJ) ocurring in neighboring tandem repeats. In fact, off-target effects are not easily predicted, as they depend not only on the location but also on mismatch distribution and chromatin structure.16,17 In addition, off-target mutations occurred in D-repeat while not in POTSs in the human genome, which is not surprising because there are more mismatches in these 5 top POTSs than in the sgRNA targeted D-repeat sequence. Moreover, the efficiency of long tandem repeats deletion can be improved by 2 or more sgRNAs according to the specific sequence of each repeat unit.
The gene expression analysis of XIST and X-linked genes after knockout of D-repeat
To investigate the role of the D-repeat in X-inactivation, the expression of XIST and X-linked genes were compared between sgRNA transfected and control cells by q-PCR. The result demonstrated that the XIST was down regulated in D-repeat knockout cell group, while the expression of 6 X-linked genes, HPRT, PGK1, PLAC1, RNF12, MECP2 and RPL13A were all increased significantly compared to the control cells (Fig. 2C). In all, the q-PCR results revealed that deletion of D-repeat induced the down-regulated expression of XIST and X-activation.
Previously studies have demonstrated that 5′ A-repeat in XIST gene contributes to both the localization of the RNA and the establishment of X-inactivation, while other repeats play a role in the RNA localization.5 In our study, the failure of X-inactivation resulted from the D-repeat deletion could be ascribed to a reduction either in the stability or the expression of XIST RNA. Therefore, we raised a hypothesis that the D-repeat in XIST is an essential element required for X-inactivation (Fig. 3). Although our study provided a new insight into the mechanisms of X chromosomal silencing, the further attempts to identify the mechanism of D-repeat regulation in XIST expression and X-inactivation are needed.
Figure 3.
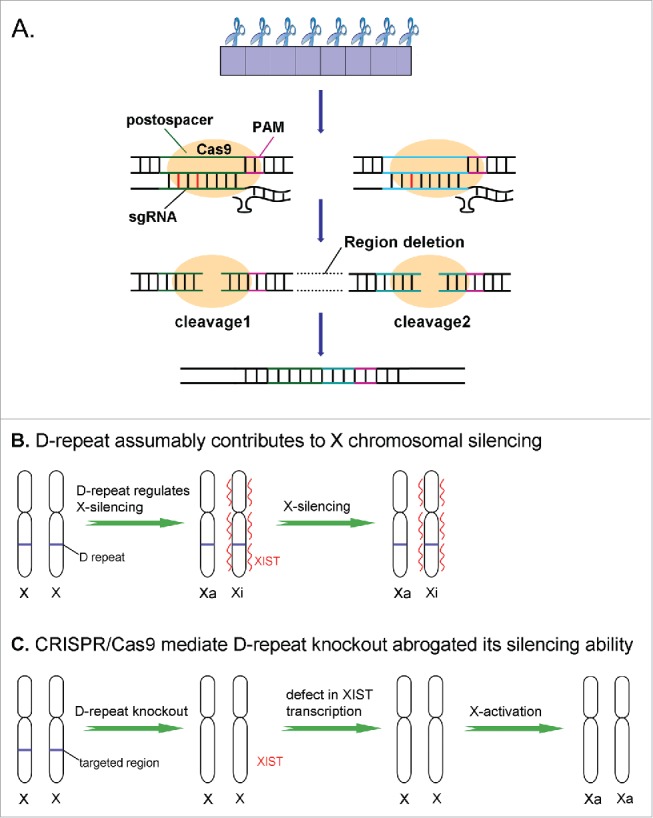
Summary of the CRISPR/Cas9 mediated D-repeat knockout and its roles on X-inactivation (A) Schematic representation of D-repeat knockout in XIST using CRISPR/Cas9 system. The Cas9 protein binds to the sgRNA scaffold and in the presence of the postorspacer sequence and the PAM sequence it generates a cleavage. The cleavage triggers the endogenous cellular DNA repair machinery that catalyzes non-homologous end joining (NHEJ). The red segment indicated the misparing between sgRNA and genome site. The green part represents the OTS1, the blue represents the OTS2. (B) D-repeat regulated the expression of XIST (red) on the inactive X chromosome (Xi). Subsequently, the XIST RNA coats the whole X chromosome induced the X-inactivation.(C) CRISPR/Cas9 mediated D-repeat knockout resulted in the downregulated expression of XIST, which lead to the compromise of X-inactivation.
We chose 293FT cell in this study because of its high transfection efficiency, but the unstable karyotype and diversity of the mutation genotypes in transiently transfected cells is unavoidable. In order to strengthen the gene expression results, the stable 293FT cell lines with the homozygous genotype were generated by puromycin selection. As shown in Fig S2, significantly decreased expression of XIST while increased X-linked genes were observed in the D-repeat knockout cells, which confirmed that D-repeat is required for X chromosome inactivation (Fig S2). In addition, the unstable karyotype in 293FT cells could also affect the gene expression. So here we performed karyotype analysis in mutant and control cells, the data indicated the chromosome number of 293FT cells is mainly in 63–78, while there is no significantly different between the mutant and control cells (Fig S3). Based on the previously study have shown the 293FT cell could be used for the study of XCI (X-chromosome inactivity)18-19 and the same result observed in Hela cells (Fig S2), we believe the downregulated expression of XIST and X-chromosome activation is not related to different chromosome numbers while induced by the D-repeat deletion in this study.
In conclusion, our study established an efficient way to generate D-repeat knockout in human XIST gene using a sgRNA directed CRISPR/Cas9 system, which provides a novel method for disruption of tandem repeat. Moreover, we proposed and preliminary demonstrated that D-repeat is one of the key elements in the process of X chromosomal silencing, which will provide further insight into the mechanism of X-inactivation.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This project was gratefully funded from the National Natural Science Foundation of China (Grant No. 31201080 and 31272394). Agricultural science and technology research of Guangdong province (2011A020102003).
Reference
- 1.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991; 349:38–44; PMID:1985261; http://dx.doi.org/ 10.1038/349038a0 [DOI] [PubMed] [Google Scholar]
- 2.Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992; 71:527–42; PMID:1423611; http://dx.doi.org/ 10.1016/0092-8674(92)90520-M [DOI] [PubMed] [Google Scholar]
- 3.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse XIST gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992; 71:515–26; PMID:1423610; http://dx.doi.org/ 10.1016/0092-8674(92)90519-I [DOI] [PubMed] [Google Scholar]
- 4.Minks J, Baldry SE, Yang C, Cotton AM, Brown CJ. XIST-induced silencing of flanking genes is achieved by additive action of repeat a monomers in human somatic cells. Epigenetics Chromatin 2013; 6:23; PMID:23915978; http://dx.doi.org/ 10.1186/1756-8935-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of XIST RNA. Nat Genet 2002; 30:167–74; PMID:11780141; http://dx.doi.org/ 10.1038/ng820 [DOI] [PubMed] [Google Scholar]
- 6.Pellagatti A, Dolatshad H, Valletta S, Boultwood J. Application of CRISPR/Cas9 genome editing to the study and treatment of disease. Archives Toxicol 2015; 89:1023–34; PMID:25827103; http://dx.doi.org/ 10.1007/s00204-015-1504-y [DOI] [PubMed] [Google Scholar]
- 7.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife 2013; 2:e00471; PMID:23386978; http://dx.doi.org/ 10.7554/eLife.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 2013; 31:839–43; PMID:23934178; http://dx.doi.org/ 10.1038/nbt.2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al.. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013; 31:827–32; PMID:23873081; http://dx.doi.org/ 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013; 31:822–6; PMID:23792628; http://dx.doi.org/ 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res 2013; 41:e187; PMID:23997119; http://dx.doi.org/ 10.1093/nar/gkt772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endo M, Mikami M, Toki S. Multi-gene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol 2014; 56:41–7; PMID:25392068; pcu154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protocols 2013; 8:2281–308; PMID:24157548; http://dx.doi.org/ 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 2008; 42:733–72; PMID:18729722; http://dx.doi.org/ 10.1146/annurev.genet.42.110807.091711 [DOI] [PubMed] [Google Scholar]
- 15.Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, Sasaki H, Sado T. A proximal conserved repeat in the XIST gene is essential as a genomic element for X-inactivation in mouse. Development 2009; 136:139–46; PMID:19036803; http://dx.doi.org/ 10.1242/dev.026427 [DOI] [PubMed] [Google Scholar]
- 16.Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 2014; 24:1020–7; PMID:24696462; http://dx.doi.org/ 10.1101/gr.171264.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 2013; 31:833–8; PMID:23907171; http://dx.doi.org/ 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow JC, Hall LL, Baldry SE, Thorogood NP, Lawrence JB, Brown CJ. Inducible XIST-dependent X-chromosome inactivation in human somatic cells is reversible. Proc Natl Acad Sci 2007; 104; 10104–9; PMID:17537922; http://dx.doi.org/ 10.1073/pnas.0610946104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernández-Muñoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci U S A 2005; 102; 7635–40; PMID:15897469; http://dx.doi.org/ 10.1073/pnas.0408918102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.