Abstract
It is one of the fundamental questions in biology how proteins efficiently fold into their native conformations despite off-pathway events such as misfolding and aggregation in living cells. Although molecular chaperones have been known to assist the de novo folding of certain types of proteins, the role of a binding partner (or a ligand) in the folding and in-cell solubility of its interacting protein still remains poorly defined. RNase P is responsible for the maturation of tRNAs as adaptor molecules of amino acids in ribosomal protein synthesis. The RNase P from Escherichia coli, composed of M1 RNA and C5 protein, is a prototypical ribozyme in which the RNA subunit contains the catalytic activity. Using E. coli RNase P, we demonstrate that M1 RNA plays a pivotal role in the in-cell solubility of C5 protein both in vitro and in vivo. Mutations in either the C5 protein or M1 RNA that affect their interactions significantly abolished the folding of C5 protein. Moreover, we find that M1 RNA provides quality insurance of interacting C5 protein, either by promoting the degradation of C5 mutants in the presence of functional proteolytic machinery, or by abolishing their solubility if the machinery is non-functional. Our results describe a crucial role of M1 RNA in the folding, in-cell solubility, and, consequently, the proteostasis of the client C5 protein, giving new insight into the biological role of RNAs as chaperones and mediators that ensure the quality of interacting proteins.
Keywords: aggregation, C5 protein, M1 RNA, protein folding, RNase P, RNA-mediated chaperoning function
Abbreviations
- RNase P
Ribonuclease P
- wt
wild-type
- IDP
intrinsically disordered protein
- IDR
intrinsically disordered region.
Introduction
Ribonuclease P (RNase P) is an essential enzyme in all 3 kingdoms of life, Bacteria, Archaea, and Eukarya,1,2 and is responsible for tRNA maturation by cleaving the 5′ leader sequence of its precursor.3,4 The machinery that carries out this cleavage process is composed of 2 distinct subunits: a single RNA and one or more cofactor proteins.4 Bacterial RNase P consists of a large RNA subunit of 350 – 400 nucleotides and a small protein of approximately 14kDa.5 RNase P is unique from other RNases in that it is a ribozyme; the RNA subunit exerts the catalytic activity while the protein subunit(s) provides an auxiliary but crucial role.4
Historically, the term ‘molecular chaperones’ refers to a group of proteins that assist the folding of proteins by preventing their misfolding or aggregation.6,7 Despite extensive studies on various types of molecular chaperones, the effect of a binding partner (or a ligand) on the in-cell solubility of its cognate protein, including the concepts of protein folding, misfolding, aggregation, and homeostasis, still remains largely unexplored. And yet, the current concept of molecular chaperones neither asks nor answers the potential role of RNA as a folding modulator of its interacting protein.
RNAs are abundant in the cytoplasm, where they directly or indirectly interact with proteins over the course of their life cycle. All proteins are synthesized by ribosomal machinery, and as such, are linked to or in close contact with rRNAs (rRNAs) from the beginning of their synthesis.8-11 In addition, a great number of proteins interact with their RNA ligands and form RNA-protein complexes (RNP complexes). It has been continually reported that mutations or abnormalities in the RNA subunit of an RNP complexes causing its disruption can be detrimental to cells and even lead to neurodegeneration.12 Most of these conditions are closely associated with the aggregation of proteins.13-16 One very interesting feature of these responsible proteins is that they are intrinsically unstable and have a high tendency to form disordered structures on their own,17 and are induced to fold and form ordered structures only when interacting with their specific ligands such as nucleic acids (RNA or DNA), small molecules, other proteins, and membranes.18-22 In addition, it has been reported that RNAs—either in an RNP complex or naked, in the form of a ribosomal complex, its 50S subunit, or 23S rRNA—assist protein folding in vitro in a trans-acting manner.23-26 The chaperoning role of RNA on protein folding in a cis-acting manner, via its highly negative charges and gigantic size was also proposed.27-29 Taken together, it is worth examining the role of RNA on the folding and proteostasis of its interacting proteins.
Here we report that M1 RNA plays a pivotal role in the in-cell solubility of its interacting partner, C5 protein, both in vitro and in vivo. Hampering the interaction between M1 RNA and C5 protein crucially affected the maintenance of in-cell solubility and the proteostasis of C5 protein. These results shed light on an exciting possibility that a variety of RNAs which transiently or covalently interact with proteins could function as a protein folding modulator, either independently or in concert with a protein-based molecular chaperone, in the highly crowded cellular environment.
Results
M1 RNA facilitates in vitro refolding of C5 protein
To investigate the effect of M1 RNA on the folding of C5 protein, in vitro refolding of C5 protein was performed in the presence or absence of M1 RNA as shown in Figure 1. To monitor the refolding of C5 protein in real-time, EGFP, a folding reporter, was fused to the C-terminus of C5 protein (C5-EGFP), as it is challenging to directly monitor the proper folding of C5 protein. First, the C5 protein of RNase P (unlike M1 RNA) has no known enzymatic activity.4 Second, the enzymatic assay of RNase P is based on the cleavage of a precursor tRNA, which is incompatible with the time-scale of the fast folding of C5 protein and assembly into RNase P. The problem was circumvented by EGFP fusion, which is commonly used for monitoring of the folding of the passenger protein in vitro and in vivo.30,31 Although the folding of EGFP in itself is not affected by the presence of M1 RNA (Fig. 1D), the folding status of C5 protein, i.e. properly folded or misfolded, directly affect the folding of its physically linked partner EGFP. Thus, this system allowed us to monitor the C5 protein folding in real-time.
Figure 1.
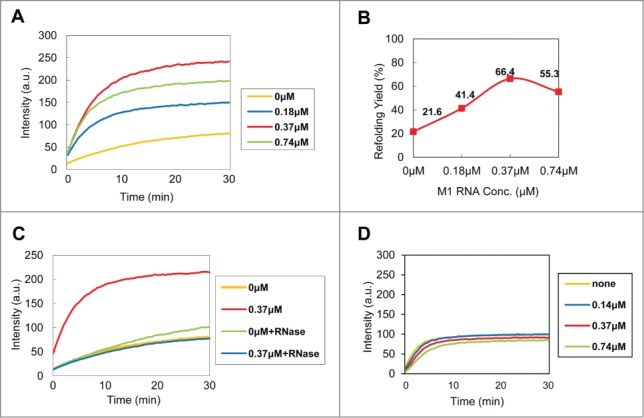
In vitro refolding of C5 protein in the presence of M1 RNA. (A) Refolding of C5-EGFP (0.37 µM) with varying concentrations of M1 RNA, i.e. 0, 0.5X, 1X and 2X of molar equivalents of C5-EGFP. The data from 3 independent experiments were summarized. (B) The refolding yield of C5-EGFP (at 30 min) relative to purified native C5-EGFP. (C) Effect of RNase treatment on the refolding yield. (D) Refolding of wt EGFP (0.37 µM) at various concentrations of M1 RNA.
The purified C5-EGFP was denatured and refolded in the presence of various concentrations of M1 RNA (0, 0.18, 0.37, and 0.74 µM) while the concentration of C5-EGFP was fixed at 0.37 µM (Fig. 1A). The refolding of C5-EGFP was continuously monitored by measuring EGFP fluorescence. C5-EGFP refolding was proportional to the concentration of M1 RNA, with the exception of a slight decline at the highest concentration (0.74 µM). The refolding was enhanced by M1 RNA more than 3-fold (21.6% and 66.4%, in the absence and the presence of M1 RNA, respectively) (Fig. 1B). As a control, the addition of the same concentrations of M1 RNA to native C5-EGFP yielded no detectable effect on the fluorescence intensity (data not shown). Consistent with this observation, the RNase treatment almost completely abolished the refolding yield of C5 protein (Fig. 1C, blue), and the refolding of wild-type (wt) EGFP (0.37 µM) was not affected by the presence of M1 RNA (Fig. 1D). Taken together, these results indicate that M1 RNA potently increases the folding of C5-EGFP in vitro.
Mutations in the regions involved in interactions between M1 RNA and C5 protein greatly affect the M1 RNA-mediated C5 refolding in vitro
The dependence of the folding of C5 protein on M1 RNA in vitro was further investigated by employing M1 RNA derivatives, mM1 RNA-1 (Δ62-74) and mM1 RNA-2 (Δ229-232), both of which have an impaired C5 protein-binding ability (Fig. 2A) based on a high-resolution structure of RNase P holoenzyme,32 interaction models between C5 protein and M1RNA,33 and in vitro binding assays.34-36
Figure 2.
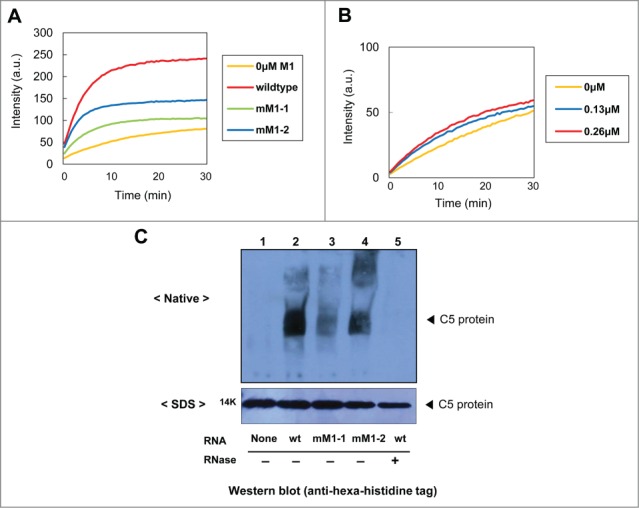
Effect of C5 protein and M1 RNA mutants on the folding of C5-EGFP in vitro. (A) Refolding of C5-EGFP (0.37 µM) in the presence of M1 RNA derivatives. (B) Refolding of the C5 (K66A/R67A)-EGFP (0.13 µM) at various M1 RNA concentrations. The data shown in (A) and (B) were obtained from 3 independent experiments. (C) Binding assays for C5 protein with M1 RNA derivatives. C5 protein was produced by in vitro translation, complexed with M1 RNA, separated on native-PAGE, and visualized with Western blot (upper panel). The total amounts of C5 protein for the binding assay were separated on SDS-PAGE and visualized with Western blot (lower panel). Wt C5 protein only, lane 1; wt C5 protein + wt M1 RNA, lane 2; wt C5 protein + mM1 RNA-1, lane 3; wt C5 protein + mM1 RNA-2, lane 4; wt C5 protein + wt M1 RNA + RNase A, lane 5.
The refolding yields of C5 protein by mM1 RNA-1 and mM1 RNA-2 were significantly lower than that by wt M1 RNA: 58% and 42% of wt M1 RNA, respectively (Fig. 2A). These results indicate that the M1 RNA derivatives provide, if any, only an impaired chaperoning ability compared to the wt M1 RNA.
To further support these results, we constructed a C5 protein mutant, C5 (K66A/R67A), whose mutations would affect the formation of a prokaryotic RNase P complex.37,38 Importantly, these residues are exposed on the outer surface of the C5 protein and thus the mutations are not expected to have a significant effect on the overall stability of its conformation.39 As shown in Figure 2B, the wt M1 RNA failed to provide a chaperoning effect on the C5 protein mutant, although the refolding yield was slightly increased at higher concentrations of M1 RNA.
In vitro binding assays (Fig. 2C) showed that the M1 RNA mutants had a significantly reduced affinity to C5 protein (lanes 3 and 4) compared to wt M1 RNA (lane 2). Unlike the RNase P complex, whose pI values is 5.5, the unbound C5 protein cannot be detected on the same native gel because of its extreme pI value, that is, 12.2.40 Consequently, the unbound C5 protein moves in the opposite direction under the electrophoretic conditions. Of note, the affinity of mM1 RNA-1 was relatively lower than mM1 RNA-2, which is consistent with its inferior refolding yield of C5 protein shown in Figure 2A (green vs. blue). RNA-protein complexes were not detected in either negative control: in the absence of M1 RNA (lane 1) or by pretreatment with RNase A (lane 5).
Taken together, these results show that specific interaction between M1 RNA and C5 protein is required for M1 RNA-mediated folding of C5 protein in vitro.
The solubility of C5 protein is dependent on M1 RNA in vivo
To investigate its effect on the in-cell solubility of C5 protein, M1 RNA was coexpressed with C5 protein in vivo, under IPTG and L-arabinose induction conditions. As its first-hand indicators, the parameter of the in-cell solubility including protein folding, stability, aggregation, and proteostasis in cells has been studied by separating the cell lysate into its soluble and insoluble fractions.28,29,66,67 As shown in Fig. 3A, the C5 protein remained partially soluble (62%), but its solubility was greatly increased by the coexpression of M1 RNA (92%). This result was further confirmed with Western blot analysis using an anti-hexa-histidine tag antibody (Fig. 3A, lower panel). Without the induction by L-arabinose, C5 protein was below detection threshold, but could be identified in an LC-MS analysis (data not shown). As negative controls, the mock vector (pLysE) or the coexpression of a non-cognate RNA, i.e. E. coli tRNALys, had little effect, if any, on the solubility of C5 protein (Fig. 3B). The overexpression of M1 RNA was confirmed by reverse transcription-PCR of total RNA from the E.coli extract (Fig. 3C). The endogenous M1 RNA without inducers was below the detection level, whereas the band intensity of M1 RNA was greatly increased upon IPTG induction. Interestingly, the M1 RNA band intensity was slightly increased by the overexpression of C5 protein only, confirming that the lifetime of M1 RNA highly depends on the existence of C5 protein.41
Figure 3.

Effect of the M1 RNA coexpression on the solubility of C5 protein in vivo. (A) Coexpression of C5 protein and M1 RNA. The expression of C5 protein and M1RNA was induced by L-Arabinose and IPTG. The wt C5 protein is indicated with an arrow. Relative solubility of proteins was estimated by densitometric scanning of the stained gel. M, T, S, and I represent the molecular weight marker, total, soluble, and insoluble fraction, respectively. The data from 3 independent experiments were summarized in the right panel. The expression patterns of C5 protein were further confirmed by Western blot, which is shown in the lower panel. (B) Coexpression of C5 protein with a mock vector (pLysE) or tRNALys as negative controls. (C) RT-PCR analysis of M1 RNA expression. The analysis was performed on cDNAs from the E. coli lysates shown in Fig. 3A.
To further examine the M1 RNA-dependent enhancement of C5 protein folding in vivo, we used inducers to temporally control the expression of M1 RNA and C5 protein and examined the solubility of C5 proteins. Thus, M1 RNA was induced at 0.5, 1, and 2 hours after the induction of C5 protein. As shown in Figure 4, the solubility of C5 protein gradually decreased with longer delays to the induction of M1 RNA. Probably due to the existence of endogenous M1 RNA and/or unknown cellular conditions, the effect of M1 RNA on the solubility of C5 protein in vivo appeared to be lower than that on the refolding yield of C5-EGFP in vitro. Overall, the results indicate that M1 RNA increases the solubility of C5 protein and prevents the aggregation of C5 protein both in vivo and in vitro.
Figure 4.
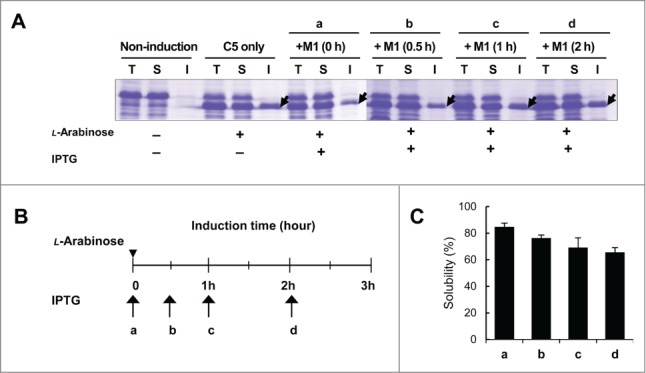
Effect of delayed induction of M1 RNA on the solubility of C5 protein. M1 RNA was induced by IPTG at 0 h (a), 0.5 h (b), 1 h (c), or 2 h (d) after induction of C5 protein. The induction points of C5 protein are indicated by an arrowhead and that of M1 RNA are indicated by arrows and small letters (B). The expression patterns were analyzed by SDS-PAGE (A) and the relative solubility of C5 protein was calculated by densitometric scanning (C).
Mutations in the regions involved in interactions between M1 RNA and C5 protein greatly affect the M1 RNA-mediated C5 protein solubility in vivo
We then tested if the M1 RNA derivatives (mM1 RNA-1 and mM1 RNA-2) were defective in providing chaperoning function and the solubility of C5 protein in vivo (Fig. 5A). The coexpression of C5 protein with each M1 RNA derivative was performed under the same condition used in Figure 3A. The coexpression of M1 RNA mutants failed to rescue the solubility of C5 protein (56.8% and 60.2% for mM1 RNA-1 and mM1 RNA-2, respectively, as compared to wt M1 RNA (91.1%)) (Fig. 5A). To examine the expression level of M1 RNA derivatives, we performed RT-PCR analysis (Fig. 5A, right panel). Because the primer set for the detection of M1 RNA covers nucleotides 20 through 190, mM1 RNA-1, which has deletions from nucleotides 62 to 74, was detected at slightly lower positions, indicating that M1 RNA derivatives was well-expressed in vivo. These results support that the interaction with M1 RNA leads to the folding of C5 protein in vivo, confirming and further extending our findings in vitro (Fig. 2A).
Figure 5.
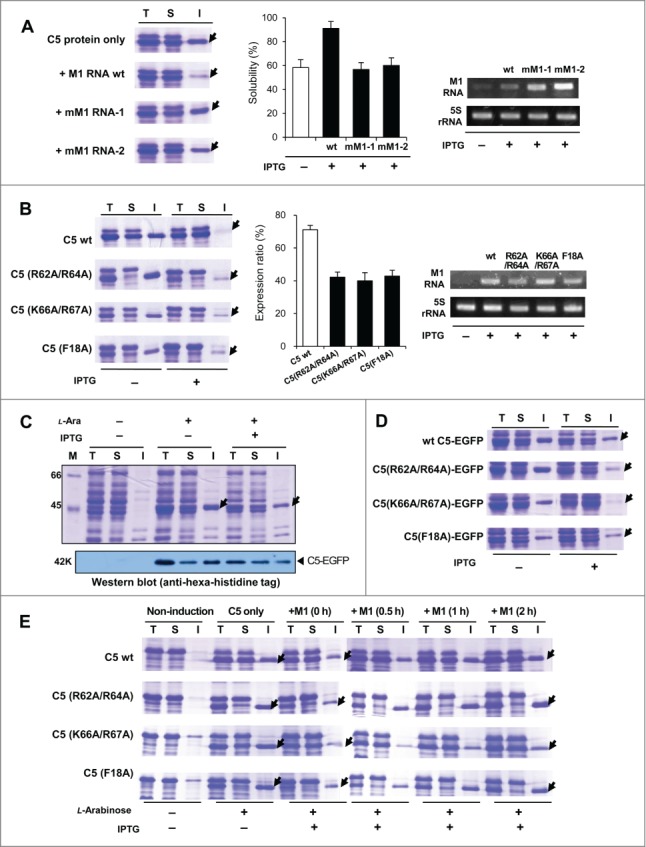
Effect of the mutations altering C5 protein-M1 RNA interaction on the solubility of C5 protein in vivo. (A) Coexpression of wt C5 protein with M1 RNA mutants (mM1 RNA-1 and mM1 RNA-2). Arrows in (A) and (B) indicate the expressed C5 protein. The solubility from 3 independent experiments was summarized in the middle panel. RT-PCR analysis of M1 RNA derivatives and 5S rRNA (control) was shown in the right panel. (B) Coexpression of C5 mutants with wt M1 RNA (left panel). The band intensity of the expressed C5 mutants in the total fractions was quantified by gel densitometer and the relative expression ratio was calculated by taking the ratio of the amount of C5 mutants with to without the induction of wt M1 RNA. Three independent experiments were performed and summarized in the middle panel. RT-PCR results of M1 RNA and 5S rRNA (right panel) (C) Coexpression of C5-EGFP in the presence or absence of M1 RNA. The expression patterns of C5-EGFP were further confirmed by Western blot analysis (lower panel). Arrows in (C) and (D) indicate expressed C5-EGFP. (D) Coexpression of C5-EGFP derivatives with M1 RNA in vivo. Three C5 protein mutants, C5 (R62A&R64A)-EGFP, C5 (K66A&R67A)-EGFP, and C5 (F18A)-EGFP, were coexpressed with M1 RNA. (E) Effects of delayed induction of wt M1 RNA on the solubility of C5 protein mutants under the same condition as in Fig. 4.
In addition to the C5 protein mutant (K66A/R67A) used in Figure 2B, 2 additional mutants (R62A/R64A, and F18A) that affect the interaction with RNA37,38 were constructed. In general, the solubility of the mutants was lower than wt C5 protein, suggesting that direct interaction with M1 RNA is indeed important for promoting the solubility of C5 protein (Fig. 5B). Overexpression of M1 RNA partially rescued the solubility of C5 protein mutants; however, the rescue was significantly lower than that of wt C5 protein. Interestingly, the coexpression with M1 RNA strongly decreased the overall expression levels of all C5 protein mutants (Fig. 5B, middle panel). Similar observations were made in the same sets of mutations in the C5-EGFP fusion constructs (Fig. 5C-D). This finding was further verified in a separate experiment in which the expression of M1 RNA and C5 protein was temporally controlled. As shown in Figure 5E, the expression level of C5 protein mutants was decreased in the presence of M1 RNA at the early phase of induction. However, as the induction of M1 RNA was delayed, the overall expression level of the mutants gradually increased, and after a 2-hour interval, the expression level was similar to that of C5 protein mutants in the absence of M1 RNA. These results suggest that C5 protein mutants, by interacting with M1 RNA weakly, could maintain the RNase P complex in a soluble state preventing their aggregation (Fig. 6A).
Figure 6.
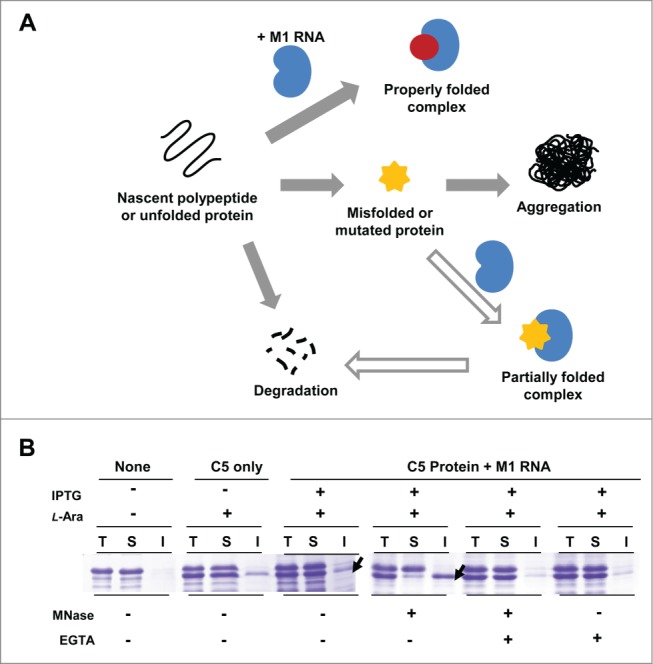
The role of M1 RNA in controlling the fate of C5 protein. (A) Proposed model for M1 RNA-mediated chaperoning function and proteostasis. The binding of the ligand (M1 RNA) to the unfolded C5 protein induces C5 protein into a stabilized conformation, which prevents aggregation, and leads to the assembly of RNase P. In the absence of M1 RNA, the unfolded protein is prone to aggregation. When misfolded or mutated, C5 protein still interacts with M1 RNA. Albeit defective, the interaction still confers partial solubility to C5 protein, facilitating its proteosomal degradation. (B) Effect of micrococcal nuclease treatment on the solubility of C5 protein. Two arrows indicate C5 protein with and without MNase, respectively. As a control, EGTA was added to inhibit MNase activity.
M1 RNA interaction is crucial for the stability of C5 protein after folding
As illustrated in Figure 6A, we demonstrated the possibility that the RNA subunit not only facilitates the folding of its cognate protein, but also dictates its overall in-cell solubility. A substantial fraction of proteins must bind to their partner(s) to reach and maintain their proper native structures. To elucidate the effect of M1 RNA on the stability of C5 protein in vivo, the lysates of C5 protein was treated with micrococcal nuclease (MNase) (Fig. 6B). The depletion of RNA greatly affected the solubility of C5 protein; C5 protein in the soluble fraction was greatly decreased enriching the insoluble fraction. As a control, MNase was treated with EGTA to inhibit its enzymatic activity, and under this condition, the solubility of C5 protein was not affected. These results indicate that M1 RNA not only promotes the folding but also has a pivotal role in maintaining the stability of C5 protein and forming an RNP complex.
Unbalanced proteostasis renders M1 RNA to hamper the folding of C5 protein
According to the “generic view” of protein aggregation, protein homeostasis is strongly associated with the maintenance of in-cell solubility of proteins, and thus, dysregulation in protein quality control mechanisms can eventually cause various diseases including neurodegenerative diseases.42-44 Recently accumulating data further support that mutation or misregulation of RNAs drives pathogenetic consequences, known as RNAopathies.12 It also should be noted that polyanions, especially RNAs, have been reported to stimulate PrP conversion and facilitate de novo prion formation.45-49 Taken together with our findings, it is tempting to speculate that RNAs, in spite of its positive chaperoning role in the normal environment, are also capable of aversive functioning leading to protein aggregation (and consequently amyloid formation) in certain abnormal conditions. The issue could be addressed by treating protease inhibitors and examining the pattern of C5 protein mutants in the presence of M1 RNA. This approach is difficult, however, because of the lack of commercially available protease inhibitors that can effectively function in vivo. Instead, we used an alternative E.coli cell line, BL21star(DE3), which has a deletion of the lon protease and a mutation in the outer membrane protein protease VII. Interestingly, the results were opposite to those from using a protease positive strain: the expression level of C5 protein mutants in the absence of M1 RNA appeared to have been decreased and the mutants were significantly aggregated in the presence of M1 RNA (Fig. 7A). To further examine whether M1 RNA has a role in these opposite aggregation patterns, we investigated the presence of M1 RNA in the insoluble fraction of C5 mutants using RT-PCR. E.coli lysates were separated into soluble and insoluble fractions by centrifugation and total RNA was isolated from the lysates. As shown in Figure 7B, M1 RNA was detected in the insoluble fraction and the intensity was greatly increased with the induction of M1 RNA. The result suggests that M1 RNA could promote the aggregation of mutant C5 protein in the proteosomally imbalanced condition. Taken together, the results support the hypothesis that the effect of RNA on proteostasis is delicately balanced and greatly influenced by the availability of proteosomal degradation machinery, though the exact mechanisms require further exploration.
Figure 7.
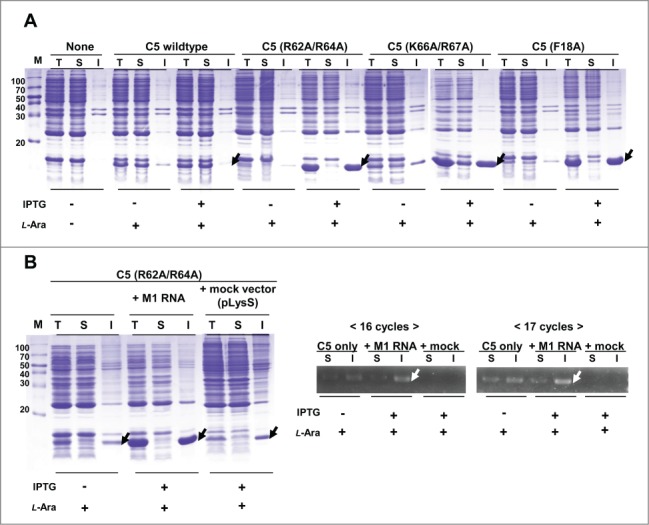
The deterrent role of M1 RNA in the folding of C5 protein. (A) Coexpression of C5 protein mutants in the presence or absence of wt M1 RNA in E.coli host BL21star(DE3). The arrows indicate C5 protein derivatives in the presence of M1 RNA. (B) Coexpression of C5 (R62A/R64A) with wt M1 RNA or mock vector (left panel) and RT-PCR analysis of M1 RNA expression (right panel). White arrows indicate detected M1 RNA band from the lysates of the insoluble fractions.
M1 RNA binding with intrinsically disordered regions (IDRs) in C5 protein
Nascent polypeptides can interact with their partners during their biogenesis and folding processes. Furthermore, a substantial fraction of proteins has been reported to be intrinsically disordered proteins (IDPs) or have intrinsically disordered regions (IDRs),50-52 which are able to form stable structures in a binding-coupled manner.53 Along with these reports, our results suggest that ligands could play important roles in mediating the folding of and providing chaperoning functions to their interacting partners. To verify whether there are IDRs present in the C5 protein and whether such regions are associated with M1 RNA binding, we predicted the IDRs of C5 protein by both sequence- and structure-based prediction algorithms.54,55 Our In silico analysis found the predominant disordered region of the C5 protein in its middle part, in addition to the extreme ends of its N- and C-termini (Fig. 8), confirming previous experimental observations.56 Notably, the major IDR overlapped with the known M1 RNA binding sites (red square, in Fig. 8A).37,38 Consistent with this, 2 mutants with mutations in the IDR region, C5 (R62A/R64A) and C5 (R67A/K67A), inhibited M1 RNA binding (Fig. 8B). Because the F18 residue is also known to bind to the 5′ leader sequence of pre-tRNA,39,56 the perturbed solubility and folding observed in the C5 (F18A) mutant could be ascribed to the failure of binding to the tRNA substrate as well as M1 RNA. Taken together, these results further suggest that M1 RNA binding shields the disordered region from exposure to water preventing its subsequent misfolding and induces a stable conformation of C5 protein, as various partners of IDPs do.57 Taken together, our results strongly suggest that RNAs, similar to protein-based molecular chaperones, can assist protein folding, either by operating independently or in concert with molecular chaperones.
Figure 8.
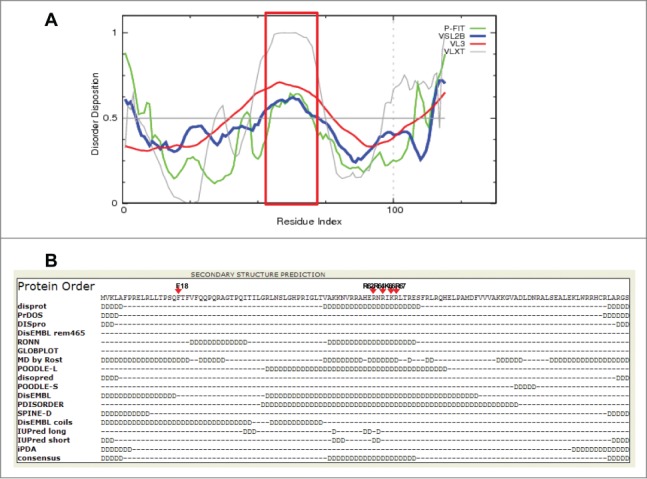
M1 RNA interaction regions in C5 protein were predicted as IDRs. (A) Prediction of IDR by FONDR-FITTM and other associated predictors including VSL2B, VL3, and VLXT. Peptide residues experimentally verified as M1 RNA binding region are shown in a red squre. Disorder Disposition > 0.5 indicates possible IDP regions. (B) Predicted results by Genesilico Metaserver. Various IDP predictors used in (B) were listed in the left column. Amino acids with high propensity of disorderedness are represented with letter 'D'. Consensus IDP regions among the predictors were aligned in the bottom line. Arrows in red indicate the amino acids used for our mutation experiments.
Discussion
This study clearly shows that M1 RNA, a ribozyme responsible for the maturation of tRNAs, also provides a chaperoning function to its cognate C5 protein. This chaperoning function is mediated by a direct interaction between the 2 mutations in either M1 RNA or C5 protein that affect their mutual interaction were detrimental to the chaperoning function, and a close correlation was observed between the affinity and chaperoning ability (Fig. 2 and 5). The C5 protein mutants were mostly aggregated in the absence of M1 RNA coexpression, whereas, under coexpression, their overall expression levels were substantially decreased as compared to wt C5 protein (Fig. 5). Ligands are generally thought to protect their partners from proteolytic degradation by inducing their folding or increasing their stability, as is the case of M1 RNA to wt C5 protein. With C5 protein mutants, however, their weak interaction with M1 RNA, while maintaining the RNP complex in a soluble state, may render the complex amenable to proteolytic degradation, consequently affecting the overall homeostasis of C5 protein (Fig. 6A). Thus, the role of M1 RNA in the stability of C5 protein is strikingly similar to molecular chaperones, which play a dual role in both the folding and proteostasis of their client proteins.58,59 Besides the chaperoning function, M1 RNA is also responsible for maintaining the conformational and chemical stability of C5 protein; mutations affecting their interaction resulted in the decrease of protein stability (Fig. 2 and 5), and the depletion of the RNA component by micrococcal nuclease treatment resulted in the precipitation of C5 protein (Fig. 6B). Similarly, thermal melting studies and CD spectra also show M1 RNA-dependent stabilization and folding of C5 protein in vitro, further supporting our findings.56 In proteolytically imbalanced conditions, M1 RNA also showed a negative function on the folding of C5 protein mutants (Fig. 7) compared to its positive role in normal condition. Thus, we suggest that M1 RNA also functions as a mediator that quality-controls C5 protein, on multiple aberrant conditions: either by promoting its degradation in the presence of functional proteolytic machinery, or by eliminating its solubility if the machinery is non-functional.
Through the IDR prediction of C5 protein (Fig. 8), we speculated that RNA, as a binding partner of IDPs, has a potential role in controlling and maintaining the folding and stability of IDPs in a ligand binding-dependent manner. Overall, the results signify that M1 RNA not only dictates the folding pathway of the nascent polypeptide into mature form but also maintains C5 protein in a stable conformation as an RNase P complex. Taken together, our results suggest that M1 RNA controls the fate of C5 protein, from birth to its eventual turnover.
C5 protein, as the sole client protein to its cognate M1 RNA, is strictly dependent on M1 RNA for its folding, stability, solubility, and turnover. However, considering that a substantial number of proteins transiently or covalently interact with RNAs and form RNP complexes,27 our finding, as a model system for validating a general principle, potentially has far-reaching implications on elucidating the role of RNA subunits on the folding and proteostasis of their cognate proteins. Besides RNA, proteins interact with various partners such as protein, DNA, and ions to reach their native state. Such partners promote protein folding by providing folding energies and maintain stable structure by shielding aggregation-prone regions.57 Consistent with our results, it has been reported that coexpression of binding partners effectively increase solubility and maintain stability of their proteins by preventing protein misfolding and aggregation.60-62 It could be speculated that binding partners, either protein or RNA, contribute to the folding and stability of their interacting partner complex.63,64 In accordance with the present data, it should be noted that an unbalanced proteome and protein-quality control mechanisms following aneuploidy lead to proteotoxic stresses and diseases.65 Taken together, our results provide new insight into the concept of protein folding and homeostasis mediated by binding partner(s).
With respect to in vitro refolding, it should be noted that the thermal stability and structure of C5 protein is strongly dependent on the presence of M1 RNA.56 Further extending this, here we demonstrated that M1 RNA not only promotes the folding but has a pivotal role in the overall in-cell solubility of C5 protein. Because of the complexity of a cellular environment, such as macromolecular crowding, cotranslational folding, molecular chaperones, and binding partners, protein folding in vivo has large differences compared to the simplified condition in vitro.64 It should be remembered that our in vivo data are based on an over-expression system combined with mutagenesis. Although this system has been commonly used in other protein folding studies,28,29,66-68 this may not accurately reflect the real cellular environment where endogenous C5 protein is present in low-abundance. Examining the folding of endogenous C5 protein would be technically difficult, however, especially because of its low abundance in E. coli (below detection level either by Western blot or LC-MS analysis, data not shown) and the absence of proper genetic tools for its analysis.
The protein subunit of the RNase P complex provides an auxiliary but crucial function to the RNA subunit, contributing to its substrate specificity and catalytic efficiency by stabilizing the RNA subunit in RNP complexes,35 binding to the 5′ leader sequence of immature tRNA,69 and assisting the release of mature tRNA.70 Our results further suggests the possibility that M1 RNA, by directly dictating the folding of its cognate protein, enhances the generation of the repertoire of tRNAs as adaptors to activated amino acids for ribosome-assisted polypeptide synthesis, further extending the RNA world concept.67,71-73 Considering the variety of non-coding RNAs of yet unknown functions in the human genome,74 the potential role of RNA in the folding and proteostasis of its interacting proteins in cellular environment, in concert with protein-based molecular chaperones, remains to be further explored.
Materials and Methods
Expression vector construction for C5 protein, M1 RNA, and their derivatives
The vector modified from pGEMEX-1 (Promega) was used for the construction of the expression vectors for the E. coli wt C5 protein. C5 protein, its mutants, and reporter-fused forms, C5 (R62A/R64A), C5 (K66A/R67A), C5 (F18A), C5-EGFP, and C5 (K66/R67A)-EGFP, were constructed under the control of pBAD promoter. C5 gene was generated by PCR using E.coli lysate as a template. The genes of 3 mutants, C5 (R62A/R64A), C5 (K66A/R67A) and C5 (F18A), were prepared from PCR-overlapping mutagenesis of the C5 template. The linker (Gly-Ser-Gly-Glu-Gly-Asp-Gly) was introduced between C5 protein and EGFP, and the hexa-histidine tag was added to the C-terminus of all proteins for Ni-affinity purification.
The M1 RNA and the 2 deletion mutants, mM1 RNA-1 (with deletions of nucleotides 62 to 74) and mM1 RNA-2 (with deletion of nucleotides 229 to 232), were also constructed. M1 RNA gene was obtained from E.coli cell lysate, following similar procedures as described above. The two M1 RNA derivatives were generated by PCR-overlapping mutagenesis of the M1 RNA gene. The genes were cloned into the plasmid pE-tRNALys, 28, derived from pLysE (Novagen), by replacing the E. coli tRNALys gene under the control of T7 promoter and Lac operator.
In vitro synthesis of M1 RNA and its derivatives
The DNA fragments of M1 RNA and its derivatives obtained by PCR amplification were cut with restriction enzymes. RNAs were synthesized in vitro using the purified linear DNA fragments as a template with RiboMAXTM large scale RNA production system-T7 (Promega). After treating with phenol-chloroform-isoamyl alcohol, RNAs were precipitated by isopropanol (Sigma) and washed by ethyl alcohol (Sigma). The precipitate was diluted with nuclease-free water and unincorporated rNTPs were removed by spin columns, illustraTM MicroSpin G-25 (GE healthcare). Purified RNAs were quantified by measuring the absorbance at 260 nm using a spectrophotometer (Thermo) and stored at −80°C until use.
In vitro refolding
C5-EGFP and C5 (K66A/R67A)-EGFP were transformed into the E. coli host HMS174(DE3) and grown overnight at 37°C in LB media containing 50 µg/ml of ampicillin and 34 µg/ml of chloramphenicol. Each single colony of the transformants was inoculated into 3 ml of LB medium containing the same antibiotics and transfer into 50 ml of LB medium for scale-up. 50 ml of the overnight cultured broth was diluted into 500 ml of fresh LB medium and cultured to the optical density of 0.8 at 600 nm at 37°C. The expression of the tested proteins was induced with 0.02% L-arabinose (Sigma) at 20°C for 5 hours. After induction, the cultures were harvested by centrifugation at 8,000 rpm for 10 min (4°C) and the cells were lysed in A buffer (50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 10 mM imidazole, 10% glycerol, 2 mM 2-mercaptoethanol, and 0.5 % NP-40) by sonication. The proteins were purified on a Ni-affinity column with a 0.2-300 mM linear gradient of imidazole by ÄTKA prime (Amersham) and concentrated with Centriprep (Amicon). The purified proteins in the buffer (50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 1 mM DTT and 50% glycerol) were stored at −80°C until use. The purified proteins were denatured in 6 M guanidine-HCl and 1 mM DTT for 20 min at 40 °C. Then the denatured proteins were diluted 25-fold into the refolding buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2 and each RNA derivative. 500 µg of RNase A was added to the mixture to eliminate M1 RNA for 30 min at room temperature. The fluorescence emission at 509 nm after excitation at 490 nm was monitored for 30 min at 25°C by Cary Eclipse fluorescence spectrophotometer (Varian).
Coexpression of C5 protein and M1 RNA
The E. coli host HMS174(DE3) was cotransformed by 2 independent plasmids coding for C5 protein and M1 RNA, respectively, and was grown at 37°C in 3 ml of LB medium containing the same antibiotics as above. Expression of the protein and RNA was induced with 0.02% L-arabinose (Sigma) and 0.01% IPTG (Sigma) and cultured for 1 hour at 42°C and for 2 hours at 37°C. The cells harvested from 10 ml of the cultured broth were lysed in PBS by sonication and separated into soluble and insoluble fractions by centrifugation for 10 min at 4°C. The cell lysates were used for the analysis of protein expression patterns on an SDS-PAGE and for Western blot analysis. The expression of M1 RNA and C5 protein was temporally regulated by adjusting the time of induction (see Results). The coexpression using the E. coli host BL21star(DE3) was performed under the same condition as HMS174(DE3), except the induction conditions (for 3 hours at 37°C).
Reverse transcription-PCR
After induction as described above, the cells from 1 ml of LB broth were harvested for the RNA isolation. Total RNAs were extracted from E. coli lysates using the TRIZOL reagent (Invitrogen) or Hybrid-R total RNA purification kit (GeneAll) and quantified by the spectrophotometer. Then, cDNA was generated using 100 ng-2 µg of total RNA as a template by Omniscript® Reverse Transcriptation kit (Qiagen). To analyze the level of expression of M1 RNA, PCR was performed using a primer set for the M1 RNA gene, which covers nucleotides 20 to 190, generating about 170 bp products. As a positive control, RT-PCR was performed on the same RNA preparation with 5S rRNA-specific primers.
In vitro translation of C5 protein and RNA-Protein binding assay
The C5 protein for the RNA-protein binding assay was produced using the cell-free translation system of EasyXpress Protein synthesis kit (Qiagen) following the manufacturer's instructions. RNase inhibitors were supplemented to the reaction tube to inhibit RNA degradation. Following in vitro translation for 1 hour at 37°C, 2 µM of M1 RNA derivatives were added into the reaction mixtures and then incubated for the binding assay at 37°C for 15 min. The C5 proteins complexed with M1 RNA derivatives were separated by 5% native-PAGE and visualized by Western blotting using an anti-histidine tag antibody for the detection of the C-terminally histidine-tagged C5 protein. As a negative control, 500 µg of RNase A was added into the mixture to eliminate M1 RNA.
Micrococcal Nuclease treatment
The cells from 10 ml of LB broth were harvested and lysed in PBS by sonication. Then each E. coli lysate was incubated with 10 units of micrococcal nuclease (NEB) for 30 min at 37°C. The samples were then separated into total fraction, soluble fraction, and insoluble fraction by centrifugation and analyzed on SDS-PAGE gel. As a control, 3 mM EGTA was used to inhibit the micrococcal nuclease reaction. EGTA-only treatment was also included as a control for the effect of EGTA on the solubility of C5 protein.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank H. Jeong and YH. Jang for helpful discussions.
Funding
This work was supported by Global Ph.D. Fellowship Program Grant NRF-2012H1A2A1049133 from the Ministry of Education (A Son) and Grant HI13C0826 by the Ministry of Health and Welfare of the Korean Government (BL Seong).
References
- 1.Robertson HD, Altman S, Smith JD. Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid presursor. J Biol Chem 1972; 247:5243-51; PMID:4560501 [PubMed] [Google Scholar]
- 2.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol 2006; 41:77-102; PMID:16595295; http://dx.doi.org/ 10.1080/10409230600602634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vioque A, Arnez J, Altman S. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J Mol Biol 1988; 202:835-48; PMID:2459398; http://dx.doi.org/ 10.1016/0022-2836(88)90562-1 [DOI] [PubMed] [Google Scholar]
- 4.Frank DN, Pace NR. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem 1998; 67:153-80; PMID:9759486; http://dx.doi.org/ 10.1146/annurev.biochem.67.1.153 [DOI] [PubMed] [Google Scholar]
- 5.Srisawat C, Houser-Scott F, Bertrand E, Xiao SH, Singer RH, Engelke DR. An active precursor in assembly of yeast nuclear ribonuclease P. RNA 2002; 8:1348-60; PMID:12403471; http://dx.doi.org/ 10.1017/S1355838202027048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell 1998; 92:351-66; PMID:9476895; http://dx.doi.org/ 10.1016/S0092-8674(00)80928-9 [DOI] [PubMed] [Google Scholar]
- 7.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 2002; 295:1852-8; PMID:11884745; http://dx.doi.org/ 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- 8.Steitz TA. From the structure and function of the ribosome to new antibiotics (Nobel Lecture). Angew Chem Int Ed Engl 2010; 49:4381-98; PMID:20509130; http://dx.doi.org/ 10.1002/anie.201000708 [DOI] [PubMed] [Google Scholar]
- 9.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic Eubacterium. Cell 2001; 107:679-88; PMID:11733066; http://dx.doi.org/ 10.1016/S0092-8674(01)00546-3 [DOI] [PubMed] [Google Scholar]
- 10.Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature 2000; 407:327-39; PMID:11014182; http://dx.doi.org/ 10.1038/35030006 [DOI] [PubMed] [Google Scholar]
- 11.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science 1999; 285:2095-104; PMID:10497122; http://dx.doi.org/ 10.1126/science.285.5436.2095 [DOI] [PubMed] [Google Scholar]
- 12.Belzil VV, Gendron TF, Petrucelli L. RNA-mediated toxicity in neurodegenerative disease. Mol Cell Neurosci 2012; 56:406-19; PMID:23280309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prusiner SB. Shattuck lecture - Neurodegenerative diseases and prions. New Engl J Med 2001; 344:1516-26; PMID:11357156; http://dx.doi.org/ 10.1056/NEJM200105173442006 [DOI] [PubMed] [Google Scholar]
- 14.Ueda K, Fukushima H, Masliah E, Xia Y, Iwai A, Yoshimoto M, Otero DA, Kondo J, Ihara Y, Saitoh T. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci USA 1993; 90:11282-6; PMID:8248242; http://dx.doi.org/ 10.1073/pnas.90.23.11282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glenner GG, Wong CW, Quaranta V, Eanes ED. The amyloid deposits in Alzheimer's disease: their nature and pathogenesis. Appl Pathol 1984; 2:357-69; PMID:6242724 [PubMed] [Google Scholar]
- 16.Zoghbi HY, Orr HT. Polyglutamine diseases: protein cleavage and aggregation. Curr Opin Neurobiol 1999; 9:566-70; PMID:10508741; http://dx.doi.org/ 10.1016/S0959-4388(99)00013-6 [DOI] [PubMed] [Google Scholar]
- 17.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci 2009; 14:5188-238; http://dx.doi.org/ 10.2741/3594 [DOI] [PubMed] [Google Scholar]
- 18.Uversky VN, Narizhneva NV. Effect of natural ligands on the structural properties and conformational stability of proteins. Biochemistry (Mosc) 1998; 63:420-33; PMID:9556525 [PubMed] [Google Scholar]
- 19.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J Mol Graph Model 2001; 19:26-59; PMID:11381529; http://dx.doi.org/ 10.1016/S1093-3263(00)00138-8 [DOI] [PubMed] [Google Scholar]
- 20.Uversky VN, Gillespie JR, Fink AL. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins 2000; 41:415-27; PMID:11025552; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 21.Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J Mol Biol 1999; 293:321-31; PMID:10550212; http://dx.doi.org/ 10.1006/jmbi.1999.3110 [DOI] [PubMed] [Google Scholar]
- 22.Plaxco KW, Gross M. Cell biology - The importance of being unfolded. Nature 1997; 386:657-9; PMID:9109481; http://dx.doi.org/ 10.1038/386657a0 [DOI] [PubMed] [Google Scholar]
- 23.Das B, Chattopadhyay S, Bera AK, Dasgupta C. In vitro protein folding by ribosomes from Escherichia coli, wheat germ and rat liver: the role of the 50S particle and its 23S rRNA. Eur J Biochem 1996; 235:613-21; PMID:8654409; http://dx.doi.org/ 10.1111/j.1432-1033.1996.00613.x [DOI] [PubMed] [Google Scholar]
- 24.Chattopadhyay S, Das B, Dasgupta C. Reactivation of denatured proteins by 23S ribosomal RNA: role of domain V. Proc Natl Acad Sci USA 1996; 93:8284-7; PMID:8710862; http://dx.doi.org/ 10.1073/pnas.93.16.8284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudlicki W, Coffman A, Kramer G, Hardesty B. Ribosomes and ribosomal RNA as chaperones for folding of proteins. Fold Des 1997; 2:101-8; PMID:9135982; http://dx.doi.org/ 10.1016/S1359-0278(97)00014-X [DOI] [PubMed] [Google Scholar]
- 26.Samanta D, Mukhopadhyay D, Chowdhury S, Ghosh J, Pal S, Basu A, Bhattacharya A, Das A, Das D, DasGupta C. Protein folding by domain V of Escherichia coli 23S rRNA: specificity of RNA-protein interactions. J Bacteriol 2008; 190:3344-52; PMID:18310328; http://dx.doi.org/ 10.1128/JB.01800-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi SI, Ryu K, Seong BL. RNA-mediated chaperone type for de novo protein folding. RNA Biol 2009; 6:21-4; PMID:19106620; http://dx.doi.org/ 10.4161/rna.6.1.7441 [DOI] [PubMed] [Google Scholar]
- 28.Choi SI, Han KS, Kim CW, Ryu KS, Kim BH, Kim KH, Kim SI, Kang TH, Shin HC, Lim KH, et al. Protein solubility and folding enhancement by interaction with RNA. PLoS One 2008; 3:e2677; http://dx.doi.org/ 10.1371/journal.pone.0002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi SI, Lim KH, Seong BL. Chaperoning roles of macromolecules interacting with proteins in vivo. Int J Mol Sci 2011; 12:1979-90; PMID:21673934; http://dx.doi.org/ 10.3390/ijms12031979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol 1999; 17:691-5; PMID:10404163; http://dx.doi.org/ 10.1038/10904 [DOI] [PubMed] [Google Scholar]
- 31.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci 2007; 120:4247-60; PMID:18057027; http://dx.doi.org/ 10.1242/jcs.005801 [DOI] [PubMed] [Google Scholar]
- 32.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 2010; 468:784-9; PMID:21076397; http://dx.doi.org/ 10.1038/nature09516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai HY, Masquida B, Biswas R, Westhof E, Gopalan V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J Mol Biol 2003; 325:661-75; PMID:12507471; http://dx.doi.org/ 10.1016/S0022-2836(02)01267-6 [DOI] [PubMed] [Google Scholar]
- 34.Niranjanakumari S, Day-Storms JJ, Ahmed M, Hsieh J, Zahler NH, Venters RA, Fierke CA. Probing the architecture of the B. subtilis RNase P holoenzyme active site by cross-linking and affinity cleavage. RNA 2007; 13:521-35; PMID:17299131; http://dx.doi.org/ 10.1261/rna.308707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buck AH, Kazantsev AV, Dalby AB, Pace NR. Structural perspective on the activation of RNAse P RNA by protein. Nat Struct Mol Biol 2005; 12:958-64; PMID:16228004; http://dx.doi.org/ 10.1038/nsmb1004 [DOI] [PubMed] [Google Scholar]
- 36.Sharkady SM, Nolan JM. Bacterial ribonuclease P holoenzyme crosslinking analysis reveals protein interaction sites on the RNA subunit. Nucleic Acids Res 2001; 29:3848-56; PMID:11557817; http://dx.doi.org/ 10.1093/nar/29.18.3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gopalan V, Baxevanis AD, Landsman D, Altman S. Analysis of the functional role of conserved residues in the protein subunit of ribonuclease P from Escherichia coli. J Mol Biol 1997; 267:818-29; PMID:9135114; http://dx.doi.org/ 10.1006/jmbi.1997.0906 [DOI] [PubMed] [Google Scholar]
- 38.Jovanovic M, Sanchez R, Altman S, Gopalan V. Elucidation of structure-function relationships in the protein subunit of bacterial RNase P using a genetic complementation approach. Nucleic Acids Res 2002; 30:5065-73; PMID:12466529; http://dx.doi.org/ 10.1093/nar/gkf670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin JS, Kim KS, Ryu KS, Han K, Lee Y, Choi BS. Structural analysis of Escherichia coli C5 protein. Proteins 2011; 80(3):963-7 . [DOI] [PubMed] [Google Scholar]
- 40.Talbot SJ, Altman S. Kinetic and thermodynamic analysis of RNA-protein interactions in the RNase P holoenzyme from Escherichia coli. Biochemistry 1994; 33:1406-11; PMID:8312259; http://dx.doi.org/ 10.1021/bi00172a017 [DOI] [PubMed] [Google Scholar]
- 41.Kim Y, Lee Y. Novel function of C5 protein as a metabolic stabilizer of M1 RNA. FEBS Lett 2009; 583:419-24; PMID:19114042; http://dx.doi.org/ 10.1016/j.febslet.2008.12.040 [DOI] [PubMed] [Google Scholar]
- 42.Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci 1999; 24:329-32; PMID:10470028; http://dx.doi.org/ 10.1016/S0968-0004(99)01445-0 [DOI] [PubMed] [Google Scholar]
- 43.Chiti F, Calamai M, Taddei N, Stefani M, Ramponi G, Dobson CM. Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases. Proc Natl Acad Sci USA 2002; 99:16419-26; PMID:12374855; http://dx.doi.org/ 10.1073/pnas.212527999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vendruscolo M, Knowles TP, Dobson CM. Protein solubility and protein homeostasis: a generic view of protein misfolding disorders. Cold Spring Harb Perspect Biol 2011; 3:a010454; PMID:21825020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USA 2007; 104:9741-6; PMID:17535913; http://dx.doi.org/ 10.1073/pnas.0702662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geoghegan JC, Valdes PA, Orem NR, Deleault NR, Williamson RA, Harris BT, Supattapone S. Selective incorporation of polyanionic molecules into hamster prions. J Biol Chem 2007; 282:36341-53; PMID:17940287; http://dx.doi.org/ 10.1074/jbc.M704447200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva JL, Lima LMTR, Foguel D, Cordeiro Y. Intriguing nucleic-acid-binding features of mammalian prion protein. Trends Biochem Sci 2008; 33:132-40; PMID:18243708; http://dx.doi.org/ 10.1016/j.tibs.2007.11.003 [DOI] [PubMed] [Google Scholar]
- 48.Tribouillard-Tanvier D, Dos Reis S, Gug F, Voisset C, Beringue V, Sabate R, Kikovska E, Talarek N, Bach S, Huang C, et al. Protein folding activity of ribosomal RNA is a selective target of two unrelated antiprion drugs. PLos One 2008; 3:e2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pang YH, Kurella S, Voisset C, Samanta D, Banerjee D, Schabe A, Das Gupta C, Galons H, Blondel M, Sanyal S. The antiprion compound 6-aminophenanthridine inhibits the protein folding activity of the ribosome by direct competition. J Biol Chem 2013; 288:19081-9; PMID:23673663; http://dx.doi.org/ 10.1074/jbc.M113.466748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta 2010; 1804:1231-64; PMID:20117254; http://dx.doi.org/ 10.1016/j.bbapap.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunker AK, Gough J. Sequences and topology: intrinsic disorder in the evolving universe of protein structure. Curr Opin Struct Biol 2011; 21:379-81; PMID:21530236; http://dx.doi.org/ 10.1016/j.sbi.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 52.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Bio 2005; 6:197-208; http://dx.doi.org/ 10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- 53.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struc Biol 2009; 19:31-8; http://dx.doi.org/ 10.1016/j.sbi.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 2010; 1804:996-1010; PMID:20100603; http://dx.doi.org/ 10.1016/j.bbapap.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozlowski LP, Bujnicki JM. MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics 2012; 13(1):111; PMID:22624656; http://dx.doi.org/ 10.1186/1471-2105-13-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo X, Campbell FE, Sun L, Christian EL, Anderson VE, Harris ME. RNA-dependent folding and stabilization of C5 protein during assembly of the E. coli RNase P holoenzyme. J Mol Biol 2006; 360:190-203; PMID:16750220; http://dx.doi.org/ 10.1016/j.jmb.2006.05.001 [DOI] [PubMed] [Google Scholar]
- 57.Fernandez A, Crespo A. Protein wrapping: a molecular marker for association, aggregation and drug design. Chem Soc Rev 2008; 37:2373-82; PMID:18949110; http://dx.doi.org/ 10.1039/b804150b [DOI] [PubMed] [Google Scholar]
- 58.Kandror O, Busconi L, Sherman M, Goldberg AL. Rapid degradation of an abnormal protein in Escherichia coli involves the chaperones GroEL and GroES. J Biol Chem 1994; 269:23575-82; PMID:7916344 [PubMed] [Google Scholar]
- 59.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem Vol 82 2013; 82:323-55; PMID:23746257; http://dx.doi.org/ 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- 60.Li C, Schwabe JWR, Banayo E, Evans RM. Coexpression of nuclear receptor partners increases their solubility and biological activities. Proc Natl Acad Sci USA 1997; 94:2278-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fribourg S, Romier C, Werten S, Gangloff YG, Poterszman A, Moras D. Dissecting the interaction network of multiprotein complexes by pairwise coexpression of subunits in E.coli. J Mol Biol 2001; 306:363-73; PMID:11237605; http://dx.doi.org/ 10.1006/jmbi.2000.4376 [DOI] [PubMed] [Google Scholar]
- 62.Mossakowska DE. Expression of nuclear hormone receptors in Escherichia coli. Curr Opin Biotech 1998; 9:502-5; PMID:9821279; http://dx.doi.org/ 10.1016/S0958-1669(98)80036-0 [DOI] [PubMed] [Google Scholar]
- 63.Uversky VN, Fink A. Protein misfolding, aggregation and conformational diseases: Part B: Molecular mechanisms of conformational diseases. Springer Science & Business Media, 2007; 6. [Google Scholar]
- 64.Choi SI, Kwon S, Son A, Jeong H, Kim KH, Seong BL. Protein folding in vivo revisited. Curr Protein Pept Sci 2013; 14:721-33; PMID:24384034; http://dx.doi.org/ 10.2174/138920371408131227170544 [DOI] [PubMed] [Google Scholar]
- 65.Oromendia AB, Amon A. Aneuploidy: implications for protein homeostasis and disease. Dis Model Mech 2014; 7:15-20; PMID:24396150; http://dx.doi.org/ 10.1242/dmm.013391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 2005; 122:209-20; PMID:16051146; http://dx.doi.org/ 10.1016/j.cell.2005.05.028 [DOI] [PubMed] [Google Scholar]
- 67.Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, et al. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci USA 2006; 103:15800-5; PMID:17043235; http://dx.doi.org/ 10.1073/pnas.0607534103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ullers RS, Luirink J, Harms N, Schwager F, Georgopoulos C, Genevaux P. SecB is a bona fide generalized chaperone in Escherichia coli. Proc Natl Acad Sci USA 2004; 101:7583-8; PMID:15128935; http://dx.doi.org/ 10.1073/pnas.0402398101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niranjanakumari S, Stams T, Crary SM, Christianson DW, Fierke CA. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc Natl Acad Sci USA 1998; 95:15212-7; PMID:9860948; http://dx.doi.org/ 10.1073/pnas.95.26.15212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reich C, Olsen GJ, Pace B, Pace NR. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science 1988; 239:178-81; PMID:3122322; http://dx.doi.org/ 10.1126/science.3122322 [DOI] [PubMed] [Google Scholar]
- 71.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983; 35:849-57; PMID:6197186; http://dx.doi.org/ 10.1016/0092-8674(83)90117-4 [DOI] [PubMed] [Google Scholar]
- 72.Lazcano A, Miller SL. The origin and early evolution of life: prebiotic chemistry, the pre-RNA world, and time. Cell 1996; 85:793-8; PMID:8681375; http://dx.doi.org/ 10.1016/S0092-8674(00)81263-5 [DOI] [PubMed] [Google Scholar]
- 73.Gesteland RF, Atkins JF. The RNA world : the nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Lab; 1993; 271-302 [Google Scholar]
- 74.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet 2001; 2:919-29; PMID:11733745; http://dx.doi.org/ 10.1038/35103511 [DOI] [PubMed] [Google Scholar]


