Abstract
The 40S ribosomal subunit cannot directly recognize the start codon of eukaryotic mRNAs. Instead, it recognizes the start codon after its association with the 5′-cap structure via translation initiation factors. Base-by-base inspection of the 5′UTR by a scanning ribosome is the generally accepted hypothesis of start codon selection. As part of an effort to confirm the underlying mechanism of start codon selection by the 40S ribosome, we investigated the role of eIF4G, which participates in the recruitment of 40S ribosomes to various translation enhancers, such as 5′-cap structure, poly(A) tail, and several internal ribosome entry sites. We found that an artificial translation factor composed of recombinant eIF4G fused with MS2 greatly enhanced translation of an upstream reporter gene when it was tethered to the 3′UTR. These data suggest that the 40S ribosome recruited to a translation enhancer can find the start codon by looping of the intervening RNA segment. The ‘RNA-looping’ hypothesis of translation start codon recognition was further supported by an analysis of the effect of 5′UTR length on translation efficiency and the mathematically predicted probability of RNA-loop–mediated interactions between the start codon and the 40S ribosome associated at the 5′-end.
Keywords: Eukaryotic mRNA, ribosome scanning, RNA looping, translation initiation
Does the 40S ribosome need to scan the 5′ untranslated region (5′UTR) base-by-base to find the start codon? Biased random collision of the 40S ribosomal subunit with various locations on an mRNA molecule could be an alternative molecular event underlying start codon identification
The probability that 2 objects (proteins, ribosomes, or various parts of polynucleotides) residing at different loci of a DNA (or RNA) molecule will interact through looping of the intervening segment depends on the length, flexibility, and conformation of the polynucleotides involved.1 Theoretically, the probability of an interaction between objects bound to a string can be expressed as the local concentration, jM (in moles per liter), of one binding site in proximity to the other. According to the theoretical formula (Fig. 1A), the interaction probability is very low if the 2 objects reside very close to each other because the stiffness of polynucleotide restrains the interaction. However, the probability of interaction is much higher than that of interaction through simple diffusion of the objects when the objects reside at an optimal distance on the polynucleotide (Fig. 1A). Many biological reactions occur through DNA or RNA looping, including enhancement of transcription by the interaction between transcription-activator proteins bound to enhancer sequences and other transcription factors bound to promoters, and enhancement of splicing by interactions between SR-proteins bound to a splicing enhancer and the splicing machinery bound to the 3′ splice site (ref. 1 and refs. therein).
Figure 1.
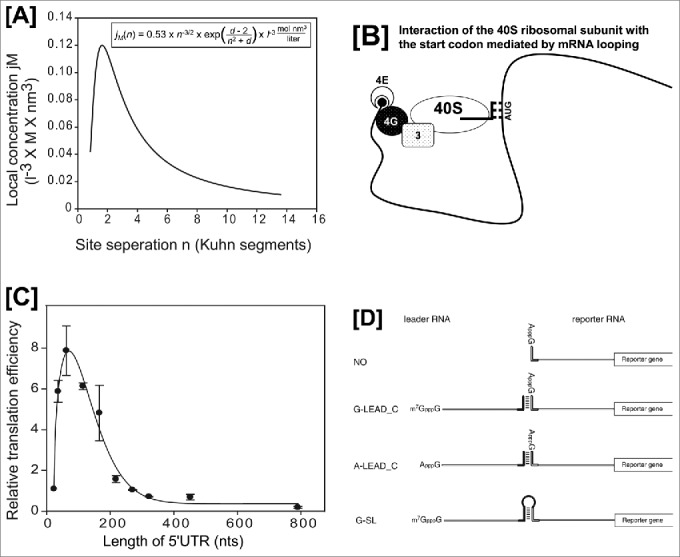
Evidence for cap-dependent translation mediated by RNA-looping. Panels [A] and [C] are modified versions of Fig. 5B and 5A from reference 2. [A] Graph depicting the relationship between the local concentration, jM, and the distance, n (number of Kuhn segments), predicted by the equation in the box. Because the experiments required to obtain the parameter d cannot be performed, we used d = 0 in the calculation.1 The peak of the graph occurred at n = 1.62. [B] Schematic diagram of the interaction between the AUG start codon and a 40S ribosome associated with the 5′-cap structure. It should be noted that the intervening RNA segment is looped out from the complex composed of the 43S ribosome and the start codon. Abbreviations: 4E, eIF4E; 4G, eIF4G; 3, eIF3; and 40S, 40S ribosomal subunit. The initiator tRNAi associated with the 40S ribosome is depicted as lines. [C] Relative translation efficiencies of mRNAs harboring 5′UTRs of various lengths. Reporter DNAs (kindly provided by Dr. Vincent Mauro, The Scripps Research Institute, La Jolla, CA) were transfected into HEK293T cells, and luciferase activity was measured 24 h after transfection. [D] A modified version of Fig. 1 from reference 3. Schematic diagram depicting the leader and reporter RNAs. The reporter RNA contains a reporter gene (firefly luciferase). The leader RNAs are either m7GpppG-capped (G-LEAD_C) or ApppG-capped (A-LEAD_C). The leader RNAs (G-LEAD_C and A-LEAD_C) contain a 3′ nucleotide sequence that is complementary (solid bar) to the 5′ sequence in the reporter RNAs. An RNA complex containing both leader and reporter RNAs covalently connected by a stem-loop structure (i.e., within a single molecule) and with an m7GpppG-cap (G-SL) was constructed to serve as a control mRNA.
Could translation initiation be directed by augmented interaction between the 40S ribosome associated at the 5´-end of an mRNA molecule and a start codon lying downstream of the ribosome-recruitment site via looping of an intervening RNA segment?
To answer this question, we analyzed the effect of the length of the 5′ untranslated region (5′UTR) on translation of eukaryotic mRNAs from the theoretical perspective of RNA looping.2 Translation efficiencies of mRNAs varied depending on the length of the 5′UTR, as shown in Fig. 1B. The optimal length was about 70 nucleotides, which is much longer than the length covered by a ribosome, and translation efficiency decreased precipitously beyond the optimal length (Fig. 1C). The relationship between mRNA translation efficiency and 5′UTR length was very similar to that of the mathematically predicted interaction probability between the 40S ribosome at the 5′UTR and start codons at various distances (Fig. 1, compare 1B with 1A). The experimental data and the theoretical analysis strongly suggest that most, if not all, cap-dependent translation events are executed by recognition of the start codon by 40S ribosome through ‘RNA looping’ rather than ‘ribosome scanning’.
The hypothesized cap-dependent translation initiation without base-by-base scanning was also supported by experiments using artificial mRNAs. As shown in Fig. 1D3, the translation efficiency of an uncapped reporter mRNA increased greatly when it was associated with an m7G-capped leader RNA (G-LEAD_C in Fig. 1D) through a double-stranded RNA (dsRNA) bridge. On the other hand, annealing an uncapped leader RNA (A-LEAD_C in Fig. 1D) to the reporter RNA did not affect translation of the reporter mRNA. Moreover, addition of a cap analog to the translation mixture dramatically reduced translation efficiency of a reporter RNA associated with the m7G-capped leader RNA, indicating that the m7G-capped leader RNA augments translation of the reporter RNA in a 5′-cap–dependent manner. Considering that a ribosome-scanning process would disrupt the dsRNA bridge, the cap-dependent translation mediated by the m7G-capped leader RNA must occur without base-by-base scanning by the ribosome. By comparing the translation efficiency of the reporter annealed to the m7G-capped leader with that of the control mRNA, which is composed of a single reporter RNA containing the leader sequence, a stem-loop structure and the 5′UTR of the reporter RNA (G-SL in Fig. 1D), we conclude that even under these adverse conditions of finding the initiating AUG codon translation occurs albeit only at 20-30% of the expected value with no break in the mRNA.3 If we consider the unstable nature of the dsRNA bridge in the presence of various RNA helicases in the translation mixture, this 20–30% value could be a considerable underestimate. It is reasonable to speculate that RNA looping mediates the enhanced translation caused by the m7G-capped leader because base-by-base scanning is not a plausible mechanism to explain the data.
If RNA looping is the underlying mechanism for recognition of the start codon by a ribosome associated with an mRNA, a logical prediction is that translation would occur through 40S ribosomal subunits recruited not only to the 5′-end, but also to various loci in the mRNA molecule
In other words, 40S ribosomal subunits recruited to downstream loci in a reporter gene could also mediate translation. We tested the possibility of orientation-independent translational activation by 40S ribosomal subunits recruited downstream of a reporter gene using artificial mRNAs (FLuc-MS2x6 and FLuc-MS2x24 in Fig. 2A) and an artificial translation activator (MS2-4GMC in Fig. 2B) composed of the RNA-binding domain of MS2 fused to the middle and C-terminal regions of eIF4G1 containing eIF3- and eIF4A-binding sites.2
Figure 2.
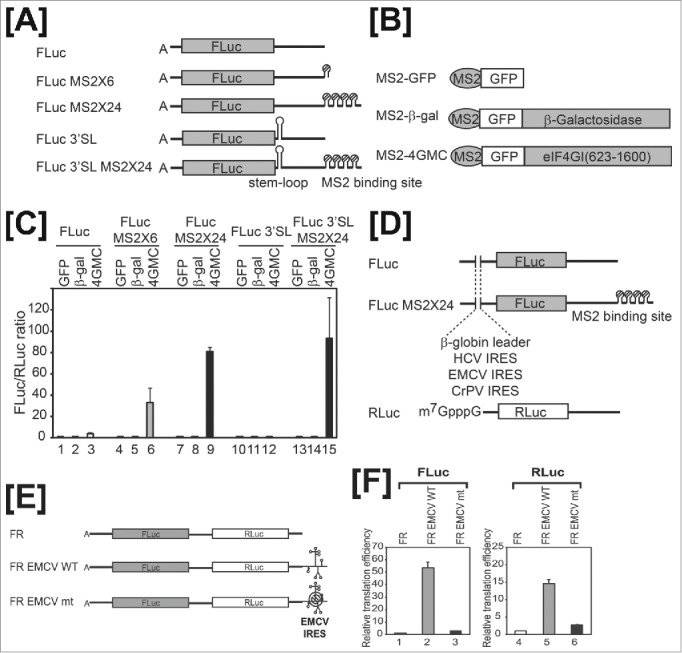
A 40S ribosomal subunit recruited to the 3′UTR of mRNA augments translation of upstream reporter genes. Panels [A] to [D] are modified versions of Fig. 1 from reference 2. [A] FLuc represents a reporter RNA containing the firefly luciferase gene as a reporter. MS2-binding sites (6 or 24 copies) were inserted into the reporter RNA to generate FLuc MS2 × 6 and FLuc MS2 × 24, respectively. A stable stem-loop was inserted downstream of the stop codon of reporter RNAs, FLuc and FLuc MS2 × 24, to generate FLuc 3′SL and FLuc 3′SL MS2 × 24, respectively. [B] Schematic diagram of MS2 fusion proteins. [C] The translation efficiencies of FLuc (lanes 1–3), FLuc MS2 × 6 (lanes 4–6), and FLuc MS2 × 24 (lanes 7–9) were determined by measuring firefly luciferase activity in cells expressing MS2-GFP (lanes 1, 4, and 7), MS2-GFP-β-galactosidase (lanes 2, 5, and 8), or MS2-GFP-eIF4G (lanes 3, 6, and 9). Firefly luciferase activity was normalized to that of Renilla luciferase. [D] Schematic diagram of reporter mRNAs. Each reporter contains a G-capped β-globin leader, HCV IRES, EMCV IRES, or CrPV IRES at the 5′UTR. G-capped reporter RNA containing the Renilla luciferase gene (Rluc) served as a control for mRNA transfection efficiency. Panels [E] and [F] are modified versions of Fig. 4 from reference 2. [E] Dual reporters contain the firefly luciferase gene (FLuc) followed by the Renilla luciferase gene (RLuc). Wild-type (WT) or mutant (mt) EMCV IRES resides at the 3′UTR of reporters. [F] ApppG-capped reporters were in vitro-translated using nuclease-untreated rabbit reticulate lysates in the presence of 150 mM KCl. Graphs depict 5′-end–dependent (FLuc, lanes 1–3) and -independent (Rluc, lanes 4–6) translation. Activity was normalized to firefly and Renilla luciferase activities of the FR reporter without EMCV IRES (defined as 1).
Co-transfection of mammalian cells with the reporter RNA and effector-expressing DNA results in association of the effector protein MS2-4GMC with the MS2-binding sites at the 3′ untranslated region (3′UTR) of the reporter mRNA through a specific protein-RNA interaction between the MS2 region of MS2-4GMC and the MS2-binding sites on the reporter RNA. MS2-4GMC is capable of recruiting the 40S ribosomal subunit to MS2-binding sites through consecutive interactions (↔) of the 4GMC region of MS2-4GMC with eIF3 and the 40S ribosomal subunit (MS2-4GMC↔eIF3↔40S).4 However, MS2-4GMC is not able to recruit the 40S ribosomal subunit to the cap structure at the 5′-end or the poly(A) tail at the 3′-end, because the region encoding the N-terminal domain required for the interactions with both eIF4E- and poly(A)-binding protein (PABP) is deleted from the eIF4G gene. Translation of the reporter RNA FLuc-MS2x24 was augmented by more than 80-fold by co-expression of the effector protein MS2-4GMC (Fig. 2C, lane 9). Moreover, translational activation by the downstream-recruited 40S ribosome occurred even when processive migration of 40S ribosomes by putative ‘backward scanning’ was blocked in the reporter RNA (FLuc-3′SL-MS2x24) by inserting a strong stem-loop structure at the 3′UTR (Fig. 2C, lane 15). Notably, translation by the 40S ribosomal subunit recruited to a site downstream of the reporter gene occurred even when the A-capped 5′UTR of the reporter gene was replaced with various elements, such as a G-capped 5′UTR, the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES), the hepatitis c virus (HCV) IRES, or the cricket paralysis virus (CrPV) IRES (Fig. 2D). These results indicate that putative transference of a translation initiation factor(s) from downstream to upstream of the reporter gene is not needed for translation by a 40S ribosome recruited downstream of the reporter gene, because translation enhancement occurred even when the 5′UTR of the reporter was replaced with the CrPV IRES, which does not require any translation initiation factors to initiate translation. eIF4G tethered downstream of a reporter gene stimulated translation, even that of dicistronic mRNAs. In the latter instance, translation of both the first and second cistrons was stimulated by eIF4G tethered to the 3′UTR of a dicistronic mRNA. Translation of the second cistron by the 40S ribosome recruited to the 3′UTR of a dicistronic mRNA was maintained even when translation of the first cistron was inhibited by insertion of a strong stem-loop structure at the 5′UTR. This indicates that translation by downstream-recruited 40S ribosomes can occur in a 5′-end–independent manner.
We further investigated translation initiation mediated by the 40S ribosome recruited to the 3′UTR using a natural mRNA element (EMCV IRES) that is known to recruit 40S ribosomes through interactions with eIF4G.5 We generated artificial mono- and dicistronic mRNAs containing the EMCV IRES at the 3′UTRs (Fig. 2E). As a negative control, we also generated mRNAs containing a mutant EMCV IRES with an adenine-to-uracil substitution at nucleotide 724 (A724U), which impairs EMCV IRES function by reducing affinity for eIF4G. The EMCV IRES at the 3′UTR of monocistronic mRNAs greatly enhanced translation of the upstream reporter genes (data not shown). The EMCV IRES at the 3′UTR of a dicistronic mRNA strongly augmented translation of both genes, consistent with the translational enhancement of both genes by eIF4G proteins tethered to MS2-binding sites (Fig. 2F). On the other hand, the mutant IRES only marginally augmented translation of upstream genes in mono- or dicistronic mRNAs (Fig. 2F).
Translational enhancement of upstream genes by translation-enhancing elements residing at the 3′UTR is not only observed in artificial mRNAs such as those shown above. Many translation-enhancing elements residing at the 3′UTR in natural mRNAs have been reported. The best-known examples are cap-independent translation elements (CITEs) in naturally uncapped plant viral mRNAs (ref. 6 and refs. therein). Most plant RNA viruses without a 5´ cap and 3´ poly(A) tail contain at least one CITE in their 3′UTR. The translation factors eIF4E or eIF4G, or an eIF4E/eIF4G complex, strongly interact with various CITEs depending on their particular structures. Alternatively, the 40S, 60S and/or 80S ribosome directly interacts with some CITEs.6 Nearly all 3´ CITEs are predicted to have long-distance ‘kissing-loop’ interactions with a 5´ hairpin located in the 5′UTR or in the nearby coding region. The kissing-loop interaction has been shown to augment CITE-dependent translation, even though it is not absolutely required for translation enhancement.6 It should be noted that the poly(A) tail present in most eukaryotic mRNAs is well known to augment translation. Preiss and Hentze showed that the poly(A) tail can direct translation of the upstream gene even in the absence of a 5´ cap structure.7 The underlying mechanism of action of these translation enhancers [CITE and poly(A) tail] is likely to be the same as that of the artificial ribosome-recruiting sites located in the 3′UTRs, described above. Taken together, the experimental data and theoretical analyses described above indicate that most, if not all, of the translation directed by translation enhancers [5´ cap, IRES, CITE, and poly(A) tail] occurs by RNA looping.
How is the authentic AUG start codon selected among all other AUG codons during translation initiation by RNA looping?
If RNA looping is the only underlying mechanism for finding the start codon, we speculate that the first AUGs are the most frequently used as start codon for the following reasons:
-
1.
Position effect (collision probability): According to the RNA-looping model, the probability of interaction depends on the length of the nucleotide between the cap and the AUGs (Figs. 1A and 1C). The optimal length between the 5′-cap structure (a ribosome recruiting site) and the start codon of individual mRNAs is also affected by the structure the 5′UTR and composition of the segment, reflecting the fact that the effective distance and stiffness of the RNA are determined by these factors. The average length of the 5′UTR of human mRNAs is about 150 nucleotides8,9, which is much longer than the minimal length required for 40S ribosome binding for putative ribosome scanning. Thus, the average length of a 5′UTR may reflect the optimal length for effective RNA looping. Notably, the first AUG is positioned close to the 40S ribosome recruited either to the 5´-end of an mRNA via consecutive 5´ cap↔eIF4E↔eIF4G↔eIF3↔40S ribosomal subunit interactions4 or to the 3´-end, because eukaryotic mRNAs form circular structures through consecutive 5´ cap↔eIF4E↔eIF4G↔PABP↔poly(A) tail interactions.4 In other words, the effective distance between the first AUG and the 40S ribosome recruited to the poly(A) tail is likely closer than the effective distance between the internal AUGs and the 40S ribosome associated with the poly(A) tail in a circularized mRNA.
-
2.
Context effect: The context surrounding an AUG plays a key role in its selection as a start codon.10 Most of the first AUGs of natural mRNAs that function as the start codon contain a good Kozak context.8 That is, purines (A and G) are preferred at the -3 and +4 positions relative to the ‘A’ of the AUG codon, designated +110. Changing the context of the 5′ proximal 2 AUGs from a weak to a strong Kozak sequence, or vice versa, alters the ratio of translation initiation from the 2 AUGs.10,11,12 It has been suggested that the mechanism underlying the context effect on start codon selection involves the participation of additional interactions13, including interactions between the purine at the -3 position of mRNA and the α subunit of eIF2 carrying the initiator tRNAi, and between the purine at the +4 position of mRNA and the nucleotides AA1818-1819 in helix 44 of 18S rRNA that form part of the A-site.13 It is conceivable that a collision between the 40S ribosome associated near the cap-structure and an AUG with a good Kozak context mediated by RNA looping would result in better translation initiation owing to the augmented affinity attributed to additional interaction(s) compared with a collision between the 40S ribosome and an AUG with a poor Kozak context. This is because the interaction between the AUG and the initiator tRNAi on the 40S ribosomal subunit is further stabilized by the additional interaction(s) with eIF2α and/or helix 44 of 18S rRNA if the AUG has a good context.
-
3.
Clearing effect: There is a chance that a putative translation-initiation complex transiently formed at a downstream AUG in the coding region may be removed by elongating 80S ribosomes migrating from an upstream AUG before translation is fully engaged. As a consequence, a significant portion of translational preinitiation complexes formed at downstream AUGs are likely to be cleared without production of a protein. An interesting phenomenon related to the clearing effect is the situation in which 2 AUGs are positioned very close to each other.11,12 For example, translation of proteins NB and NA of influenza B virus occurs from a single dicistronic mRNA (RNA segment 6). The start codons of NB and NA are located 4 nucleotides apart from each other, and both start codons contain good Kozak contexts (A-3AAAUGaA+4A-3CAAUGbC+4UA, where underlined AUGa and AUGb are start codons for NB and NA, respectively). Curiously, roughly equal amounts of NA and NB proteins are produced from the mRNA.12 This outcome is readily explained by the RNA-looping hypothesis, but not by the ribosome-scanning hypothesis, which invokes backward excursion of scanning ribosome to explain this seemingly odd phenomenon.11 According to the RNA-looping hypothesis, the probability that a 40S ribosome associated at the 5´ end of this mRNA will interact with the first AUG is almost the same as that for interaction with the second AUG since the AUGs reside very close to each other. Moreover, both AUGs contain favorable contexts10, resulting in little difference in the context effect. Finally, binding of a 40S ribosome to the first or the second AUG is mutually exclusive, since the binding of a 40S ribosome on one AUG blocks the binding of 40S ribosome to the other AUG owing to the large footprint of a ribosome. Therefore, the preferential translation of the first AUG by the clearing effect would not occur in this case, resulting in equal efficiencies of translation from the first and second AUGs. The position effect and clearing effect would become apparent if the 2 AUGs are placed apart from each other, manifesting as preferential translation of the first AUG, as empirically shown.11,12
These three reasons for the preferential usage of the first AUG as a start codon can be applied not only to 5´-cap–dependent translation, but also to translational augmentation by downstream translation-enhancing elements, such as poly(A) tails and CITEs since the effective distance between the first AUG and the 40S ribosome recruitment site in the 3′UTR could be optimal owing to the circularization of mRNA.
The RNA-looping hypothesis des-cribed here could potentially be applied to explain cap-, IRES-, CITE-, and poly(A)-dependent translation. Moreover, the somewhat enigmatic phenomenon of ‘ribosome shunting’ could also be incorporated into this hypothesis. Therefore, all phenomena observed in studies of translation-initiation mechanisms of various eukaryotic mRNAs need to reanalyzed from the perspective of the RNA-looping hypothesis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by a grant of the Bio R&D Program (No. 2012M3A9A9054974) and the BRL (No. 2010-0019706) through the NRF grant funded by the Korea government (MSIP).
References
- 1.Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem Sci 2001; 26:733-40; PMID:11738597; http://dx.doi.org/ 10.1016/S0968-0004(01)01978-8 [DOI] [PubMed] [Google Scholar]
- 2.Paek KY, Hong KY, Ryu I, Park SM, Keum SJ, Kwon OS, Jang SK. Translation initiation mediated by RNA looping. Proc Natl Acad Sci USA 2015; 112:1041-6; PMID:25583496; http://dx.doi.org/ 10.1073/pnas.1416883112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paek KY, Park SM, Hong KY, Jang SK. Cap-dependent translation without base-by-base scanning of an messenger ribonucleic acid. Nucleic Acids Res 2012; 40:7541-51; PMID:22638585; http://dx.doi.org/ 10.1093/nar/gks471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-27; PMID:20094052; http://dx.doi.org/ 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark AT, Robertson ME, Conn GL, Belsham GJ. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J Virol 2003; 77:12441-9; PMID:14610168; http://dx.doi.org/ 10.1128/JVI.77.23.12441-12449.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon AE, Miller WA. 3’ cap-independent translation enhancers of plant viruses. Annu Rev Microbiol 2013; 67:21-42; PMID:23682606; http://dx.doi.org/ 10.1146/annurev-micro-092412-155609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A) tail-promoted translation in yeast. Nature 1998; 392:516-20; PMID:9548259; http://dx.doi.org/ 10.1038/33192 [DOI] [PubMed] [Google Scholar]
- 8.Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5'untranslated mRNAs. Gene 2005; 349:97-105; PMID:15777708; http://dx.doi.org/ 10.1016/j.gene.2004.11.041 [DOI] [PubMed] [Google Scholar]
- 9.Rogozin IB, Kochetov AV, Kondrashov FA, Koonin EV, Milanesi L. Presence of ATG triplets in 5' untranslated regions of eukaryotic cDNAs correlates with a ‘weak’ context of the start codon. Bioinformatics 2001; 17:890-900; PMID:11673233; http://dx.doi.org/ 10.1093/bioinformatics/17.10.890 [DOI] [PubMed] [Google Scholar]
- 10.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986; 44:283-92; PMID:3943125; http://dx.doi.org/ 10.1016/0092-8674(86)90762-2 [DOI] [PubMed] [Google Scholar]
- 11.Matsuda D, Dreher TW. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA 2006; 12:1338-49; PMID:16682564; http://dx.doi.org/ 10.1261/rna.67906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams MA, Lamb RA. Effect of mutations and deletions in a bicistronic mRNA on the synthesis of influenza B virus NB and NA glycoproteins. J Virol. 1989; 63:28-35; PMID:2908923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006; 20:624-36; PMID:16510876; http://dx.doi.org/ 10.1101/gad.1397906 [DOI] [PMC free article] [PubMed] [Google Scholar]


