Abstract
The Spot 42 RNA is a 109 nucleotide long (in Escherichia coli) noncoding small regulatory RNA (sRNA) encoded by the spf (spot fourty-two) gene. spf is found in gamma-proteobacteria and the majority of experimental work on Spot 42 RNA has been performed using E. coli, and recently Aliivibrio salmonicida. In the cell Spot 42 RNA plays essential roles as a regulator in carbohydrate metabolism and uptake, and its expression is activated by glucose, and inhibited by the cAMP-CRP complex. Here we summarize the current knowledge on Spot 42, and present the natural distribution of spf, show family-specific secondary structural features of Spot 42, and link highly conserved structural regions to mRNA target binding.
Keywords: gamma proteobacteria, non-coding RNA, pirin, sRNA, small RNA, Spot 42, spf
Introduction
The spf gene is highly conserved in Escherichia, Shigella, Klebsiella, Salmonella and Yersinia (genera) within the Enterobacteriacea family.1 In E. coli the spf gene is flanked by polA (upstream) and yihA (downstream),2,3 and a CRP binding sequence and -10 and -35 promoter sequences are found upstream of spf. spf is also highly conserved within the Vibrionaceae family, and was recently identified in 76 Vibrionaceae genomes that were available at that time (e.g., Vibrio, Aliivibrio, Photobacterium and Grimontia genera).4 In e.g., Vibrio cholerae, Vibrio vulnificus, Aliivibrio fischeri and A. salmonicida the spf gene is flanked by polA (upstream) and a sRNA gene encoding the novel VSsrna24 RNA (downstream).
Spot 42 was first described in 1973 as an unstable RNA species of 109 nucleotides in E. coli.5,6 It was discovered by polyacrylamide gel electrophoresis and 2-D fingerprinting in an attempt to study the accumulation of small RNAs in E. coli during amino acid starvation. In these experiments the electrophoretic mobility of Spot 42 was similar to that of 5S rRNA. In 1979, Spot 42 was reported to accumulate under growth in the presence of glucose (i.e., when adenosine 3′,5′-cyclic monophosphate (cAMP) is low).7,8 During growth with a non-glucose carbon source (i.e., when cAMP concentrations are high) Spot 42 concentrations were significantly lower. Later experiments showed that overexpression of Spot 42 (tenfold increase) resulted in impaired growth and lowered ability to adapt to shifts to richer media.9 Further, shift from glucose to succinate as the carbon source resulted in a long lag period and slow growth rate, the reason for the abnormal responses was caused by an elevated number of excessive Spot 42 RNA gene products rather than excess of the gene itself. A deletion study of spf in E. coli cells resulted in viable spf null mutants, which indicated that Spot 42 was non-essential, at least under controlled lab conditions.10
It was for some years unclear if the function of Spot 42 was mediated through the 109 nucleotide RNA itself or if the function was mediated through the 14 amino acids long peptide which is hypothetically encoded from within the sRNA sequence. This confusion was based on the observation that Spot 42 contains structural features similar to other non-coding RNAs found in E. coli (such as 6S RNA and lambda bacteriophage), as well as features that are typically found in mRNAs (i.e., polypurine sequence followed by AUG, 14 amino acids codons and an UGA termination codon).7 Using a filter binding assay and other methods Rice et al. showed that Spot 42 is not an mRNA.11 In this approach the affinity between Spot 42 and the 70S ribosome was tested. Here, Spot 42 showed very inefficient binding to purified 70S ribosomes, which lead to the conclusion that the function of Spot 42 is mediated by the RNA itself.
The direct responsiveness of Spot 42 levels to glucose and cAMP is due to repression of spf expression by a cAMP-CRP (cAMP-receptor protein) complex.2 The reduction of Spot 42 in cells grown in secondary carbon sources is a result of binding of the cAMP-CRP complex to the spf promoter, which negatively regulates transcription of Spot 42. Later, the proximity of spf to polA (gene encoding DNA polymerase I) led Dahlberg and co-workers to test whether the products of these genes could influence each other.12 They found that by reducing levels of Spot 42, either by deletion of spf or by manipulating the growth conditions, the DNA pol A activity was reduced. The underlying mechanism for this observation remains however unknown.
Spot 42 can interact directly with mRNA targets through base pairing. The first Spot 42 target was discovered by Møller et al., who showed that Spot 42 specifically binds to a short complementary region at the translation initiation region of galK (encodes a galactokinase) mediated through binding of the posttranscriptional regulator Hfq.1 galK is the third gene in the galactose operon, which contains 4 genes (galETKM) and produces a polycistronic mRNA. Spot 42 mediates discoordinate expression of the gal operon (i.e., the individual genes in the operon are not similarly expressed) by binding to the galK Shine-Dalgarno region, thereby blocking ribosome binding and translation of the galK gene. The physiological significance of the discoordinate expression is unclear, but suggests that Spot 42 plays a role in fine-tuning gene expression to optimize the utilization of carbon sources. Recently, Wang et al. showed that Spot 42 represses expression of galK through direct binding to the 5´ end of the galK mRNA , and also mediates transcription termination of galT in the galT-galK junction.13
Beisel and Storz demonstrated with microarray analysis and reporter fusions that Spot 42 plays a broader role in metabolism by regulating at least 14 operons.14 These operons contain a number of genes involved in uptake and catabolism of non-favored carbon sources. During overexpression of Spot 42 16 different genes showed consistently twofold reduced or elevated levels of mRNA. The identified reduced genes are mostly involved in central and secondary metabolism, as well as uptake and catabolism of non-preferred carbon sources and oxidation of NADH. In 2012 Beisel et al. performed computational target analysis using the 3 conserved regions of Spot 42 as input. Compared to when using full-length Spot 42 sequence as input the target identification was improved and additional targets were revealed.15 The target analysis combined with assays of reporter fusions identified 7 novel Spot 42 mRNA targets, all involved in catabolite repression. Mutational analysis showed that the interactions of the 3 conserved regions of Spot 42 are critical in target regulation and that regulation through multiple conserved regions of Spot 42 as well as increased base-pairing in these regions strengthen the target regulation.
The evolution of sRNAs in E. coli and their regulatory interactions with mRNAs was recently studied using computational methods.16 Compared to cis-acting sRNA and other non-coding RNA (housekeeping RNA), trans-acting sRNA was the latest to appear in evolution. Furthermore, after Enterobacteriales diverged into a separate lineage within gamma-proteobacteria, the trans-acting sRNAs likely appeared in relatively high numbers compared to the cis-acting sRNAs that evolved more evenly among all orders within gamma-proteobacteria. The evolutionary age of 15 sRNAs and 49 corresponding sRNA-mRNA interactions were examined. Here, Spot 42 was found to be the most ancient sRNA. Of the 6 Spot 42 mRNA targets considered, only 2 (xylF and galK) evolved before Spot 42, albeit all the complementary mRNA binding sites appeared after Spot 42.
The observation that A. salmonicida contains the spf gene (which encodes the Spot 42 RNA), but lacks the galK operon (the natural Spot 42 target in E. coli), have inspired scientists to study the role of Spot 42 in this fish pathogen.4 A. salmonicida is unable to utilize galactose (lacks gal operon) in minimal medium and addition of galactose has little effect on the growth rate. When cells are grown in glucose the level of Spot 42 is increased 16–40 fold, but is in contrast decreased threefold when cAMP is added, indicating that Spot 42 have similar roles as in E. coli, i.e., in carbohydrate metabolism. It has been hypothesized that Spot 42 works in concert with a novel sRNA gene, called VSsrna24, located 262 nt downstream of spf. The VSsrna42 RNA is approximately 60 nt in length and has an expression pattern opposite to that of Spot 42. Furthermore, in a spf deletion mutant a gene encoding a pirin-like protein was upregulated 16 fold. Pirin has key roles in the central metabolism by regulating the activity of pyruvate dehydrogenase E1 and therefore select whether pyruvate will be fermented, or subjected to respiration through the TCA cycle and electron transport.
Although the Spot 42 RNA was discovered more than 40 y ago there are still a number of unanswered questions related to this highly interesting RNA, e.g.: What is the natural distribution of the Spot 42 gene (spf) in Bacteria? What is the complete set of biological roles of Spot 42, and does Spot 42 play the proposed key role in the central metabolism? How does Spot 42 interact with its apparently many mRNA targets? In this work we have summarized the current literature on Spot 42, and extended this knowledge by surveying the known natural distribution of spf, we have identified family-specific structural features of Spot 42, and evaluated if highly conserved structural regions can be linked to mRNA binding.
Results
spf is restricted to 5 orders of gamma-proteobacteria
The distribution of spf in nature is shown in Figure 1. The basis for the figure was available nucleotide sequences of spf included in the Rfam database (677 sequences), and spf sequences identified in this study by using the Blastn server and spf sequences from selected taxa as queries. All previously known cases of spf originate from gamma-proteobacteria, and after fruitless searches in all other domains of Bacteria we therefore concentrated our efforts on specific searches within gamma-proteobacteria, both by using spf sequences from the closest neighbors, and by manual inspection of the known genic location of spf, i.e., in the intergenic region between polA and engB. The result of our search was finally mapped onto a phylogenetic tree generated using the iTOL web service.
Figure 1.
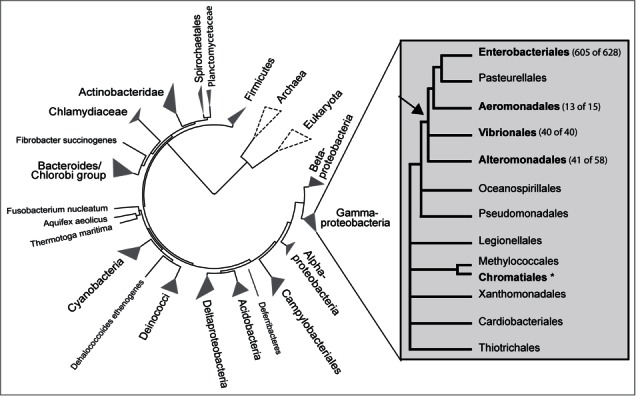
The natural distribution of spf. spf is restricted to 5 orders of gamma-proteobacteria (shown in bold letters), 4 of which share the same closest common ancestor (indicated by an arrow). The circular phylogenetic tree (made using the iTol web tool) shows all major branches of Bacteria. The gamma-proteobacteria phylogeny in the right panel is based on Gao et al.25 Here, numbers in parentheses indicate the number of complete genomes that contain spf (first number) and the total number of available complete genomes (second number) in each order. In addition, spf is found in 8 Chromatiales draft genomes (asterisk).
The result show that spf is exclusively found in 5 orders of gamma-bacteria, i.e., in Enterobacteriales, Aeromonadales, Alteromonadales, Vibrionales and Chromatiales. These orders, except Chromatiales, share the same closest common ancestor (arrow in Fig. 1), and constitutes a clade. spf has still not been found in Pasteurellales, which is likely due to that Pasteurellales genomes are underrepresented in the European Nucleotide Archive (ENA) compared to e.g., the sister Enterobacteriales. We suspect that spf will be discovered in Pasteurellales as more genomes are being sequenced. In addition to known cases of spf our Blastn search revealed previously unreported cases within genera of Enterobacteriales and Alteromonadales. In Enterobacteriales spf was identified in the genera Morganella and Raoultella, as well as in draft genomes of Budvicia, Cedecea, Hafnia, Leminorella, Plesimonas and Yokenella. And, in genera where spf was already known to occur, spf was in this work identified in Enterobacter radicincitans and Escherichia blattae. Similarly, in Alteromonadales spf is found in the 5 families Ferrimonadaceae, Shewanellaceae, Moritellaceae, Pseudoalteromonadaceae and Alteromonadaceae, and spf was in this study identified in the 3 genera Glaciecola, Alteromonas and Pseudoalteromonas by our blast searches, whereas spf was found in Moritella viscosa by manual inspection of the intergenic region polA/engB. Interestingly, in Chromatiales, spf is exclusively found in the genera Rheinheimera and Arsukibacterium, which is represented in ENA by 6 and 2 available draft genomes, all containing spf. Given that the phylogeny as shown in Figure 1 is correct then it is tempting to speculate that spf was acquired by lateral transfer, perhaps from a donor within the clade marked by an arrow in Figure 1.
We also wanted to answer the following question: Is spf optional or ubiquitous within the individual orders and families? Spot 42 appears to play central roles in the carbohydrate metabolism, and we therefore hypothesized that it might be present in all representatives of the same order, family or genus once it has been identified in one genome. To answer this question we used the list of complete bacterial genomes found at the NCBI Genomes resource (http://www.ncbi.nlm.nih.gov/genome/), and searched for presence of spf in all representatives of the current orders, families and genera. Our result show that spf is found in 699 of 741 complete genomes distributed among 34 genera (a detailed list is provided in Table S1). spf is missing in representatives of the 2 genera Glaciecola and Pseudoalteromonas of Alteromonadales. In both of these genera spf is found in one of 3 complete genomes. All three genomes of Glaciecola have the same genic organization with polA and engB as neighbors (spf is usually located between these 2 genes). In Pseudoalteromonas, spf is only found in one genome, i.e., in Pseudoalteromonas atlantica, where polA and engB are located next to each other. The two other genomes with no spf have a different genic organization (synteny) at this region. Finally, spf has not been found in any of the complete genomes within the following genera: Buchnera, Candidatus Moranella, Candidatus Riesia and Wigglesworthia (from Enterobacteriales), Oceanimonas and Tolumonas (from Aeromonadales), Marinobacter, Sacchrophagus, Colwellia, Idiomarina and Psychromonas (from Alteromonadales), and all genera of Chromatiales (i.e., spf found in 6 draft genomes of the genus Rheinheimera and 2 draft genomes of Arsukibacterium). In summary, of a total of 741 genomes from the 5 orders Enterobacteriales, Aeromonadales, Alteromonadales, Chromatiales and Vibrionales, 699 complete genomes contain spf, whereas 42 lack spf. The result is in agreement with conserved, but not necessarily indispensable roles of spf.
The Spot 42 RNA consensus secondary structure
We next mapped the level of identity among all known spf sequences (120 in total when redundant sequences have been removed) onto a consensus secondary structure model of Spot 42 (based on structure probing by Møller et al.1) to find clues to possible structural regions that might be important for target identification and interaction, in general (Fig. 2). The Spot 42 RNA consists of one long hairpin structure located at the 5` end (from now on referred to as the 5′ hairpin; 45−59 nt in length), and a second smaller hairpin separated from the 5′ hairpin by a 9 - 20 nt long single-stranded region. In addition, a rho-independent terminator is located immediately downstream of the second hairpin. Structural regions of Spot 42 from the families Vibrionaceae, Aeromonadaceae and Shewanellaceae differ from the general "consensus" and are shown in separate boxes in Figure 2. The sRNA gene is, in general, highly conserved with 76 of 108 positions (when using the "consensus" sequence as the reference) being 80−100% identical across all orders (shown as uppercase bold letters in Fig. 2). Notably, the 5` hairpin is highly conserved, i.e., 80−100% identity from positions 1−41, which indicate that these positions are interesting candidates for having general roles in target binding, perhaps with the terminal loop functioning as the seed sequence. The single-stranded region separating the 5` hairpin and the second hairpin is less conserved, with 80−100% identity in 3 positions and 60−79% identity in 6 positions, and is therefore perhaps less likely to have general roles in target recognition. spf is as expected most conserved within families. The Shewanellaceae spf differs most from the "consensus." Here, the 5` hairpin contains 2 bulges with 8 additional nt (inserted between pos. 39 and pos. 47). The Vibrionaceae and Aeromonadaceae sequences also differ to some extent from the "consensus." In summary, Spot 42 is a highly conserved sRNA across 5 orders. The 5´ hairpin represents the most conserved region and is therefore expected to have general roles in target recognition and interaction.
Figure 2.
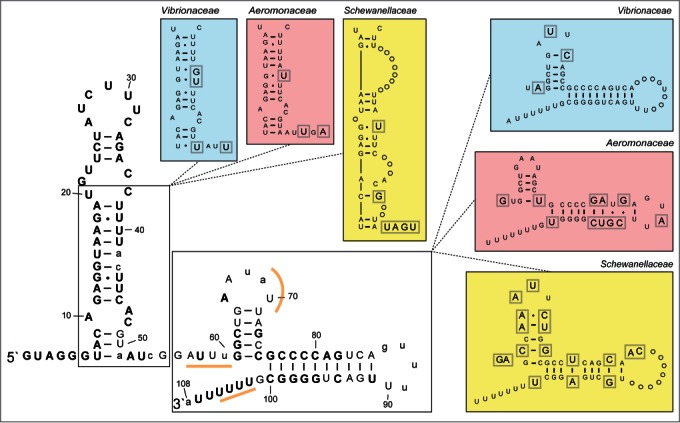
Secondary structure consensus model of the Spot 42 RNA. The structure model was made by aligning all known spf sequences, and by mapping the consensus sequence onto a secondary structure model of the E. coli Spot 42 (based on Møller et al.1). The structure consists of a relatively long 5` hairpin, a 9 nt long single-stranded region followed by a second hairpin and a rho-independent terminator. Level of identity is shown using different type of letters in the structure. Uppercase bold letters indicate 80–100 % identity, uppercase regular letters indicate 60–79% identity, and lowercase letters indicate <60% identity. Structural segments with family-specific (i.e., Vibrionaceae, Aeromonadaceae and Shewanellaceae) variations are shown in separate colored boxes. Here, circles indicate U or A insertions (compared to the "consensus"). Gray square around a letter symbolizes aberration from the consensus structure.
Spot 42 structure conservation and potential base pairing with targets
We next wanted to investigate if the highly conserved nucleotide positions of Spot 42 (as described above) are implicated in target binding (i.e., base-pairing between Spot 42 and mRNA target). Interactions between Spot 42 and galK mRNA has been determined using structure probing,1 whereas potential base-pairing to other targets is based on bioinformatics predictions followed by experimental work.4,14,15
Figure 3 shows schematically potential base-pairing between Spot 42 and experimentally verified mRNA targets for the following genes: galK, pirin, fucI, xylF, sthA, gltA, srlA, nanC, paaK, ascF, caiA, fucP, atoD, puuE and nanT. Interestingly, for all except 2 genes (i.e., sthA and fucP) the most conserved region of the 5` hairpin (i.e., pos. 1–41) can potentially participate in extensive base-pairing with the corresponding mRNAs. This suggests that the 5` hairpin, is essential for target recognition and binding. Moreover, the first 6 positions of Spot 42 (5' single stranded region) can potentially base-pair with 10 of 15 targets (galK, pirin, fucI, xylF, gltA, nanC, paaK, ascF, atoD and nanT), and the terminal loop of the 5` hairpin can base-pair with 8 of 15 targets (galK, pirin, fucI, xylF, srlA, caiA, puuE and nanT). The second hairpin is only partly conserved. In agreement with this observation base-pairing with targets are rarer and only observed for 2 targets (galK and pirin). This is in agreement with results from Beisel et al.15 Using 3 unstructured regions (the 5' single stranded region, the 5` hairpin and the single-stranded region separating the hairpins) as input during computational target identification, they improved identification of direct targets, compared to when using the full-length sequence of Spot 42. In summary, highly conserved nucleotide positions of Spot 42 have the potential to participate in extensive base-pairing with known mRNA targets.
Figure 3.
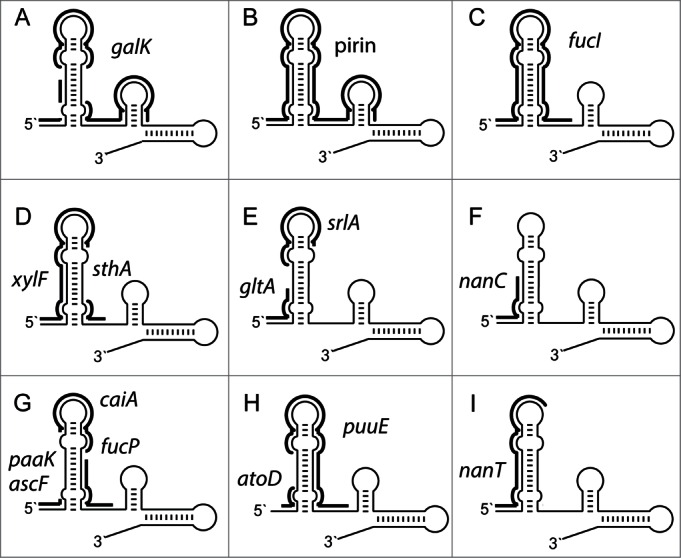
Potential base-pairing between the Spot 42 RNA and experimentally verified mRNA targets from the following genes: (A) galK, (B) pirin, (C) fucI, (D) xylF and sthA, (E) gltA and srlA and (F) nanC, (G) paaK, ascF, caiA and fucP, (H) atoD and puuE and (I) nanT. Fig. 3 is based on data from Møller et al.,1 Hansen et al.,4 Beisel and Storz,14 and Beisel et al.15
sRNA genes in the intergenic region downstream of polA
Interestingly, spf is not the only sRNA gene located in the intergenic region downstream of polA (see Fig. 4). In Vibrionaceae a gene encoding the sRNA VSsrna24 is located approximately 600 nt downstream of spf. Expression of VSsrna24 is repressed by glucose, and is hypothesized to have roles in the central carbohydrate metabolism.4 The sRNAs sX13,17 ErsA18 and Smr7C,19,20 are found in Xanthomonadaceae, Pseudomonas and Rhizobialez, respectively, but neither has the same function or structure as Spot 42. sX13 and Smr7C share secondary structure features comprising 3 stem-loops with C-rich motifs and are Hfq-independent.17,21 ErsA is Hfq-mediated and regulated by sigma factor 22, in contrast to Spot 42 that is dependent on sigma factor 70. If any of these 4 sRNA genes originates from a common ancestral gene or not is currently unknown.
Figure 4.
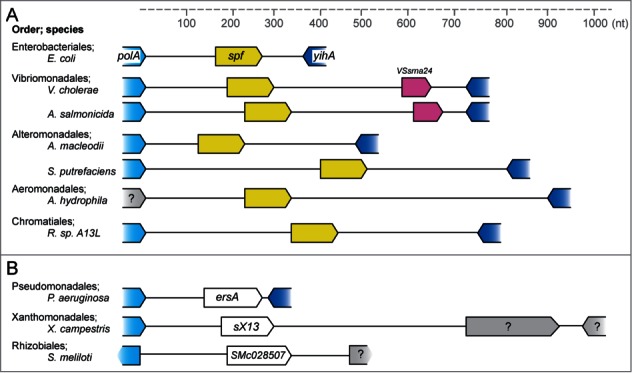
sRNA genes in the intergenic region downstream of polA. The figure shows currently known sRNA genes which have been found in the same intergenic region as spf. The scale bar shows distance in nucleotides. (A) Representative species containing spf are shown. The VSsrna24 sRNA gene is located downstream of spf in V. cholerae and A. salmonicida. Question mark denotes hypothetical protein. (B) Genomic location of the sRNA genes ersA in Pseudomonas aeruginosa, sX13 in Xanthomonas campestris and SMc02857 in Sinorhizobium meliloti.
Concluding Remarks
We have conducted a survey on Spot 42 RNA in order to learn about its natural distribution, conservation patterns, and mRNA target recognition. We demonstrated that Spot 42, which was first identified in E. coli (Enterobacteriales), is also common in 4 other orders, i.e., Aeromonadales, Alteromonadales, Chromatiales and Vibrionales. Using blastn analysis we discovered novel spf sequences. Of a total of 741 complete genomes from the 5 orders Enterobacteriales, Aeromonadales, Alteromonadales, Chromatiales and Vibrionales, 699 genomes contain spf. Furthermore, a total of 30 draft genomes distributed among 11 genera (from all orders except Aeromonadales) contain spf. As shown in Figure 1, within gamma-proteobacteria, Aeromonadales, Alteromonadales, Enterobacteriales and Vibrionales share the same last common ancestor, whereas Chromatiales does not, which suggest that spf was introduced into Chromatiales by lateral transfer by a donor from the clade marked by an arrow. We made a consensus secondary structure model of Spot 42 based on all known spf sequences and compared this to a schematically figure showing potential base-pairing between Spot 42 and known mRNA targets. Our results show that highly conserved nucleotide positions, in general, have potential to participate in extensive base-pairing with target mRNAs. This is in agreement with an earlier study by Beisel et al. which suggested that the strength of Spot 42 regulation is directly dependent on the number of nucleotides and the number of highly conserved structural regions which are involved in base-pairing between Spot 42 and its target.15
It is intriguing to us that although Spot 42 was discovered more than 40 years ago, there are still many unanswered questions. As more sequence data are being produced from high-throughput sequencing techniques and better tools and search algorithms are being developed, the known natural distribution of spf will certainly expand to new orders, families and genera (and perhaps phyla). And detailed knowledge on target recognition (other than galK) and roles in cellular processes will come from functional and bioinformatics studies. One particularly interesting aspect of Spot 42 is its apparent central role (via pirin) in the central metabolism by directing pyruvate toward fermentation or respiration through the tricarboxylic acid (TCA) cycle and electron transport.
Materials and Methods
Homology search
All previously known spf sequences were retrieved from Rfam (http://rfam.sanger.ac.uk/family/RF00021).22 Blastn searches in all domains of Bacteria were performed using spf sequences from 43 selected taxa as query sequences. All complete bacterial genomes found at the NCBI Genomes resource (http://www.ncbi.nlm.nih.gov/genome/) were checked for the presence of spf. More thorough blastn searches were performed in gamma-proteobacteria, as spf were exclusively found in this bacterial class. This was done as follows: Representative spf sequences from all spf-containing genera were used as queries in blast searches. All blast "hits" had a low E-value (i.e., high statistical support; typically below 1e-11). In other words, spf was identified with a high degree of confidence, or, spf was not found. In one case a hit with a poor E-value was found (0.65). Here, we did a manual inspection to decide the presence/absence of spf. First, the NCBI Sequence Viewer (http://www.ncbi.nlm.nih.gov/projects/sviewer/) was used to locate the intergenic region between polA and engB (genes that are known to flank spf). Next, a manual text search revealed the presence of highly conserved 5` hairpin, and thereafter the entire spf. The presence/absence of spf in all complete genomes from gamma-proteobacteria is provided in Table S1. The presence of spf was next mapped on the tree of life, which was produced using the iTol web tool.23
Alignments and nucleotide diversity
The sequences from the Rfam list and the newly discovered sequences of spf were automatically aligned and manually examined using Jalview.24 An alignment containing only one version of each nucleotide variation of spf (no redundant spf sequences) was used to examine the variations on nucleotide level between families, genera and species. A consensus spf sequence was made based on the alignment and was mapped onto an E. coli secondary structure (Fig. 2).1 The spf alignment in Rfam includes the first 10 nucleotide upstream of the 5' end of spf. However, the promoter region of spf was not considered in this work, and was not included in the alignment. Existing literature on experimentally verified mRNA targets of Spot 42 were used to map mRNA targets onto the secondary structure of Spot 42 (Fig. 3).4,14, 15
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by internal grants from UiT- The Arctic University of Norway.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev 2002; 16:1696-706; PMID:12101127; http://dx.doi.org/ 10.1101/gad.231702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polayes DA, Rice PW, Garner MM, Dahlberg JE. Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J Bacteriol 1988; 170:3110-4; PMID:2454912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joyce CM, Grindley ND. Identification of two genes immediately downstream from the polA gene of Escherichia coli. J Bacteriol 1982; 152:1211-9; PMID:6183253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen GA, Ahmad R, Hjerde E, Fenton CG, Willassen NP, Haugen P. Expression profiling reveals Spot 42 small RNA as a key regulator in the central metabolism of Aliivibrio salmonicida. BMC Genomics 2012; 13:37; PMID:22272603; http://dx.doi.org/ 10.1186/1471-2164-13-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikemura T, Dahlberg JE. Small ribonucleic acids of Escherichia coli. I. Characterization by polyacrylamide gel electrophoresis and fingerprint analysis. J Biol Chem 1973; 248:5024-32; PMID:4577761 [PubMed] [Google Scholar]
- 6.Ikemura T, Dahlberg JE. Small ribonucleic acids of Escherichia coli. II. Noncoordinate accumulation during stringent control. J Biol Chem 1973; 248:5033-41; PMID:4577762 [PubMed] [Google Scholar]
- 7.Sahagan BG, Dahlberg JE. A small, unstable RNA molecule of Escherichia coli: spot 42 RNA. I. Nucleotide sequence analysis. J Mol Biol 1979; 131:573-92; PMID:390161; http://dx.doi.org/ 10.1016/0022-2836(79)90008-1 [DOI] [PubMed] [Google Scholar]
- 8.Sahagan BG, Dahlberg JE. A small, unstable RNA molecule of Escherichia coli: spot 42 RNA. II. Accumulation and distribution. J Mol Biol 1979; 131:593-605; PMID:229230; http://dx.doi.org/ 10.1016/0022-2836(79)90009-3 [DOI] [PubMed] [Google Scholar]
- 9.Rice PW, Dahlberg JE. A gene between polA and glnA retards growth of Escherichia coli when present in multiple copies: physiological effects of the gene for spot 42 RNA. J Bacteriol 1982; 152:1196-210; PMID:6183252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatfull GF, Joyce CM. Deletion of the spf (spot 42 RNA) gene of Escherichia coli. J Bacteriol 1986; 166:746-50; PMID:2940230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice PW, Polayes DA, Dahlberg JE. Spot 42 RNA of Escherichia coli is not an mRNA. J Bacteriol 1987; 169:3850-2; PMID:2440852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polayes DA, Rice PW, Dahlberg JE. DNA polymerase I activity in Escherichia coli is influenced by spot 42 RNA. J Bacteriol 1988; 170:2083-8; PMID:2452153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Ji SC, Jeon HJ, Lee Y, Lim HM. Two-level inhibition of galK expression by Spot 42: Degradation of mRNA mK2 and enhanced transcription termination before the galK gene. Proc Natl Acad Sci U S A 2015; 112(24):7581-6; PMID:26045496; http://dx.doi.org/ 10.1073/pnas.1424683112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell 2011; 41:286-97; PMID:21292161; http://dx.doi.org/ 10.1016/j.molcel.2010.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beisel CL, Updegrove TB, Janson BJ, Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J 2012; 31:1961-74; PMID:22388518; http://dx.doi.org/ 10.1038/emboj.2012.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peer A, Margalit H. Evolutionary patterns of Escherichia coli small RNAs and their regulatory interactions. RNA 2014; 20:994-1003; PMID:24865611; http://dx.doi.org/ 10.1261/rna.043133.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidtke C, Abendroth U, Brock J, Serrania J, Becker A, Bonas U. Small RNA sX13: a multifaceted regulator of virulence in the plant pathogen Xanthomonas. PLoS Pathog 2013; 9(9):e1003626; PMID:24068933; http://dx.doi.org/ 10.1371/journal.ppat.1003626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrara S1, Carloni S, Fulco R, Falcone M, Macchi R, Bertoni G. Post-transcriptional regulation of the virulence-associated enzyme AlgC by the σ(22) -dependent small RNA ErsA of Pseudomonas aeruginosa. Environ Microbiol 2015; 17(1):199-214; PMID:25186153; http://dx.doi.org/ 10.1111/1462-2920.12590 [DOI] [PubMed] [Google Scholar]
- 19.del Val C, Rivas E, Torres-Quesada O, Toro N, Jiménez-Zurdo JI. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol Microbiol 2007; 66(5):1080-91; PMID:17971083; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05978.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valverde C, Livny J, Schlüter JP, Reinkensmeier J, Becker A, Parisi G. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics 2008; 9:416; PMID:18793445; http://dx.doi.org/ 10.1186/1471-2164-9-416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres-Quesada O1, Oruezabal RI, Peregrina A, Jofré E, Lloret J, Rivilla R, Toro N, Jiménez-Zurdo JI. The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa. BMC Microbiol 2010; 10:71; PMID:20205931; http://dx.doi.org/ 10.1186/1471-2180-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 2013; 41:D226-232; PMID:23125362; http://dx.doi.org/ 10.1093/nar/gks1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 2011; 39:W475-478; PMID:21470960; http://dx.doi.org/ 10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009; 25:1189-91; PMID:19151095; http://dx.doi.org/ 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao B, Mohan R, Gupta RS. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int J Syst Evol Microbiol 2009; 59:234-47; PMID:19196760; http://dx.doi.org/ 10.1099/ijs.0.002741-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


