Abstract
Vascular endothelial growth factor (VEGF) A is a master regulator of neovascularization and angiogenesis. VEGFA is potently induced by hypoxia and by pathological conditions including diabetic retinopathy and tumorigenesis. Fine-tuning of VEGFA expression by different stimuli is important for maintaining tissue vascularization and organ homeostasis. Here, we tested the effect of the hypoxia mimetic cobalt chloride (CoCl2) on VEGFA expression in human cervical carcinoma HeLa cells. We found that CoCl2 increased the levels of VEGFA mRNA and VEGFA protein without affecting VEGFA mRNA stability. Biotin pulldown analysis to capture the RNA-binding proteins (RBPs) bound to VEGFA mRNA followed by mass spectrometry analysis revealed that the RBP HuR [human antigen R, a member of the embryonic lethal abnormal vision (ELAV) family of proteins], interacts with VEGFA mRNA. VEGFA mRNA-tagging experiments showed that exposure to CoCl2 increases the interaction of HuR with VEGFA mRNA and promoted the colocalization of HuR and the distal part of the VEGFA 3′-untranslated region (UTR) in the cytoplasm. We propose that under hypoxia-like conditions, HuR enhances VEGFA mRNA translation.
Keywords: biotin pulldown, mass spectrometry, post-transcriptional gene regulation, polysome profiling, ribonucleoprotein complex, RNA-binding proteins
Introduction
Vascular endothelial growth factor A (VEGFA), a major endothelial growth factor promoting neovascularization and angiogenesis, plays a role in blood vessel formation during development and helps to maintain vascular tissue homeostasis.1 The expression of VEGFA is subjected to extensive regulation, at both transcriptional and post-transcriptional levels.2 Although the pro-angiogenic isoforms VEGFA121, VEGFA165 and VEGFA189 are highly abundant, an anti-angiogenic isoform called VEGF-Ax was recently described, underscoring the functional complexity of this protein family.3
Hypoxia is one of the most potent triggers of VEGFA expression, acting on VEGFA DNA transcription, VEGFA mRNA stabilization, and VEGFA translation and release.4 Oxygen deficit activates cellular responses that are centrally controlled by the hypoxia inducible factor-1 α (HIF-1α), a transcription factor that regulates hypoxia-inducible genes, including VEGFA, and induces an angiogenic response.5 In particular, VEGFA is also upregulated during early phases of neurodegenerative diseases affecting ocular function such as diabetic retinopathy and age-related macular degeneration, as well as other pathological states such as tumorigenesis.6 The early stages of diabetic retinopathy are characterized by vascular hyperpermeability and by the formation of local microaneurysms. These alterations are followed by microvascular occlusions that lead to progressive retinal ischemia, which in turn causes hypoxia and induces VEGFA synthesis and release.7 Given the pivotal role of VEGF in such pathologies, different approaches have been developed to inhibit VEGF signaling, such as the recombinant protein VEGF-trap,8 and monoclonal antibodies.9
VEGFA expression in pathophysiological states can be induced by post-transcriptional mechanisms directed at the VEGFA mRNA. Changes in transport, stability, and translation rates are robustly regulated by RNA-binding proteins (RBPs).10 Among the vast class of RBPs (which includes many regulatory proteins such as nucleolin, AUF1, TIAR, TIA-1, hnRNP K, and hnRNP L),11 the members of the ELAV/Hu family (HuR/HuA, HuB, HuC, and HuD) are some of the best-characterized RBPs that post-transcriptionally control gene expression in response to specific stimuli. In vertebrates, HuB, HuC and HuD are mainly expressed in neurons, while HuR is ubiquitously expressed.12,13 These proteins preferentially interact with U- and AU-rich sequences present in the 3′-untranslated region (UTR) of mRNA subsets.14 HuR, in particular, can regulate hundreds of target transcripts and is considered to be a key regulator of many cellular functions, including proliferation, cellular stress response, and cell survival.15 Indeed, HuR promotes the expression of general stress-response proteins (HSP70, HO-1, SOD1, p62) and hypoxia-response proteins, including HIF-1α and VEGFA.16-20 Through its influence on subsets of cellular proteins, HuR has been linked to various pathologies, including inflammatory diseases and cancer.12,13 With respect to diabetic retinopathy, the levels of both HuR and VEGFA have been shown to be altered in both in vitro and in vivo models.16,21,22
Given that controlling VEGFA production during hypoxia is important for maintaining tissue function, we set out to examine in detail the effect of a hypoxic mimetic, cobalt chloride (CoCl2), on VEGFA expression. CoCl2 is widely used as a chemical inducer of the hypoxic response,23,24 through the CoCl2-mediated induced expression of the transcription factor HIF-1α,17 which governs the transcriptional induction of many oxygen-dependent genes. In the human cervical carcinoma cell line HeLa, we found that CoCl2 treatment increased VEGFA expression levels but did not affect VEGFA mRNA stability. Mass spectrometry analysis indicated that VEGFA mRNA interacted with HuR, and VEGFA mRNA-tagging experiments revealed that interaction occurred in the cytoplasm. Our data further support a role for HuR in the promotion of VEGFA translation following exposure to CoCl2. We propose that HuR contributes to the regulation of VEGFA translation in hypoxic states, thereby helping to mount an adequate response to low-oxygen challenge.
Materials and Methods
Cell culture, chemicals, transfection and plasmids
HeLa cells were cultured in Dulbecco's modified essential medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum and antibiotics. Control small interfering RNA (Ctrl siRNA or Csi, Qiagen), HuR siRNA (HuRsi, Qiagen), and reporter plasmids were transfected using Lipofectamine 2000 (Invitrogen). Plasmids were transfected at 0.1 μg/ml [pMS2-YFP (expressing a fusion protein bearing MS2-binding protein and yellow fluorescent protein)] or at 0.5 μg/ml [pMS2-RL (Renilla luciferase), pMS2-RL-3′UTR-I, pMS2-RL-3′UTR-II, expressing chimeric RNAs bearing different regions of the VEGFA mRNA and 24 copies of MS2 hairpins]. The RL coding region was included to provide a reporter open reading frame to the MS2 RNA-containing constructs. VEGFA 3′UTR reporter constructs were made by inserting a cDNA corresponding to the proximal (I) or distal (II) part of VEGFA 3′UTR into pMS2. Plasmid pMS2-YFP was described previously.25 For cloning a proximal segment of VEGFA 3′UTR into pMS2-RL (to prepare pMS2-RL-3′UTR-I), primers AAAACTCGAGGGGCAGGAGGAAGGAGCCTCCCTCAGGGTTTCGGG (forward) and AAAACTCGAGTCAGAAGCAGGTGAGAGTAAGCGAAGGCCGCCC (reverse), containing an XhoI site, were used. For cloning the distal part of VEGFA 3′UTR into pMS2-RL (to prepare pMS2-RL-3′UTR-II), the primers used, also containing an XhoI site, were AAAACTCGAGGTTGCCCAGGAGACCACTGGCAGATGTCCCGG (forward) and AAAACTCGAGGAGATCAGAATTAAATTCTTTAATACAAAATG (reverse). HeLa cells were treated with 100 μM or 200 μM CoCl2 for 2 h, 4 h or 8 h. For mRNA stability assays, HeLa cells were treated with actinomycin D (2.5 μg/ml, Sigma Aldrich) to inhibit de novo transcription.
Western blot analysis
Whole-cell lysates were prepared using radioimmunoprecipitation buffer (RIPA), separated by SDS-PAGE (electrophoresis through SDS-containing polyacrylamide gels), and transferred onto nitrocellulose membranes (Invitrogen). Incubations with primary antibodies to detect VEGFA, HuR, α-tubulin, or β-actin (mouse monoclonal, rabbit polyclonal, mouse monoclonal, and mouse monoclonal, respectively all from Santa Cruz Biotechology) were followed by incubations with the appropriate secondary antibodies conjugated with horseradish peroxidase (HRP, GE Healthcare) and by detection using enhanced chemiluminescence (GE Healthcare).
RNA analysis
Total RNA was prepared from whole cells, gradient fractions, or ribonucleoprotein (RNP) immunoprecipitation (IP) samples using TRIzol (Invitrogen). After reverse transcription (RT) of RNA using random hexamers and Maxima reverse transcriptase (ThermoScientific, Fermentas), the abundance of transcripts was assessed by real-time (RT), quantitative Polymerase Chain Reaction (qPCR) analysis using the SYBR green PCR master mix (Kapa Biosystems) and gene-specific primer sets (below). RT-qPCR analysis was performed on Applied Biosystems model 7300 and 7900 instruments. The forward and reverse primers used in this study were, respectively: TATGCGGATCAAACCT-CAC and CTCGGCTTGTCACATTTTTCTTGTCTT for VEGFA, TGCACCACCAACTGCTTAGC and GGCATGGACTGTGGTCATGAG for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), GCGCGAACGACAAGAAAAAGATA and GTGGCAACTGATGAGCAAGC for HIF1A, and CATGTACGTTGCTATCCAGGC and CTCCTTAATGTCACGCACGAT for ACTB (β-actin). For VEGFA pre-mRNA analysis, forward and reverse primers TATGCGGATCAAACCTCAC and ATGCCAAAGGTCACATAGCG were used. RNP IP analysis was performed using as primary antibodies anti-HuR, or control IgG (Santa Cruz Biotech). RNA in the IP samples was extracted using TRIzol and further measured by RT-qPCR analysis using the primers listed above.
Biotin pulldown and mass spectrometry analysis
For biotin pulldown, we used antisense biotinylated oligomers (ASO) CGTCTGACCTGGGGTAGAGA and GCAACGCGAGTCTGTGTTTT, and sense (control) biotinylated oligomers (SO) TGAGGAGTCCAACATCACCA and TCTCTACCCCAGGTCAGACG (IDT Technologies). Whole-cell lysates (1.3 mg per sample) were incubated with 2 μg of biotinylated probes for 1 hour at 4° C, whereupon complexes were isolated with streptavidin-coupled Dynabeads (Invitrogen). The proteins present in the pulldown material were size-separated by SDS-PAGE and stained with Coomassie blue (Invitrogen); gel bands were analyzed by Mass Spectrometry by using high-performance liquid chromatography with tandem mass spectrometric detection (LC/MS/MS, at the Mass Spectrometry and Proteomics Facility, Johns Hopkins University School of Medicine) and the data confirmed by Western blot analysis.
Fractionation of polyribosomes
Polyribosome fractionation assays were carried out as previously explained.26 In short, 48 hours after transfection, cells were incubated with cycloheximide (100 μg/ml for 15 min; Sigma Aldrich); cytoplasmic lysates (900 μl) were fractionated by centrifugation through 10 to 50% linear sucrose gradients and divided into 12 fractions. From each fraction, RNA was extracted using TriPure Isolation Reagent (Roche Applied Science) and used for RT-qPCR analysis to determine the distribution of VEGFA, HIF1A and ACTB mRNAs on the polysome gradients.
Immunocytochemistry
HeLa cells were fixed with 2% formaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 1% bovine serum albumin (BSA). After incubation with a mouse monoclonal primary antibody recognizing HuR (Santa Cruz Biotechnology), an Alexa Fluor 568-conjugated secondary antibody (Invitrogen) was used to detect primary antibody-antigen complexes (red). YFP fluorescence was green and DAPI-stained nuclei were blue (Invitrogen). Images were acquired using Axio Observer microscope (Zeiss) with AxioVision 4.7 Zeiss image processing software or with LSM 510 Meta (Zeiss). Confocal microscopy images were acquired with the Z-sectioning mode with 10-μm spacing and merged using maximum intensity. Brightness and contrast were adjusted using the Best Fit option of the Zen 2012 software (Zeiss).
Data analysis
All statistical analyses were performed by using GraphPad InStat application (GraphPad software, Prism 6, La Jolla, CA, USA). All data were analyzed by using the one-way or 2-way analysis of variance (ANOVA) and, when appropriate, a specific post-hoc test, as indicated in the figure legends. Differences were considered statistically significant when p < 0.05.
Results
Treatment with CoCl2 increases VEGFA mRNA and protein levels but not VEGFA mRNA stability
We tested the effect of treating human cervical carcinoma HeLa cells with the hypoxia mimetic cobalt chloride (CoCl2) on VEGFA expression levels. As shown in Figure 1A, following exposure to 100 μM CoCl2, VEGFA mRNA levels did not change significantly at any of the times investigated, while after 4 h in continuous presence of 200 μM CoCl2 VEGFA mRNA levels increased by fold2-, returning to basal levels by 8 h after exposure. Western blot analysis revealed that intracellular VEGFA abundance increased significantly after 4 h in continuous presence of 200 μM CoCl2 (Fig. 1B); therefore, these treatment conditions were used for all of the subsequent experiments. As VEGFA is a secreted protein, ELISA was employed to measure the concentration of VEGFA released into the medium. A modest but significant increase in VEGFA release was detected by 4 h of treatment (Fig. 1C); higher levels of released VEGFA protein were seen after 8 h of treatment (Fig. S1).
Figure 1.
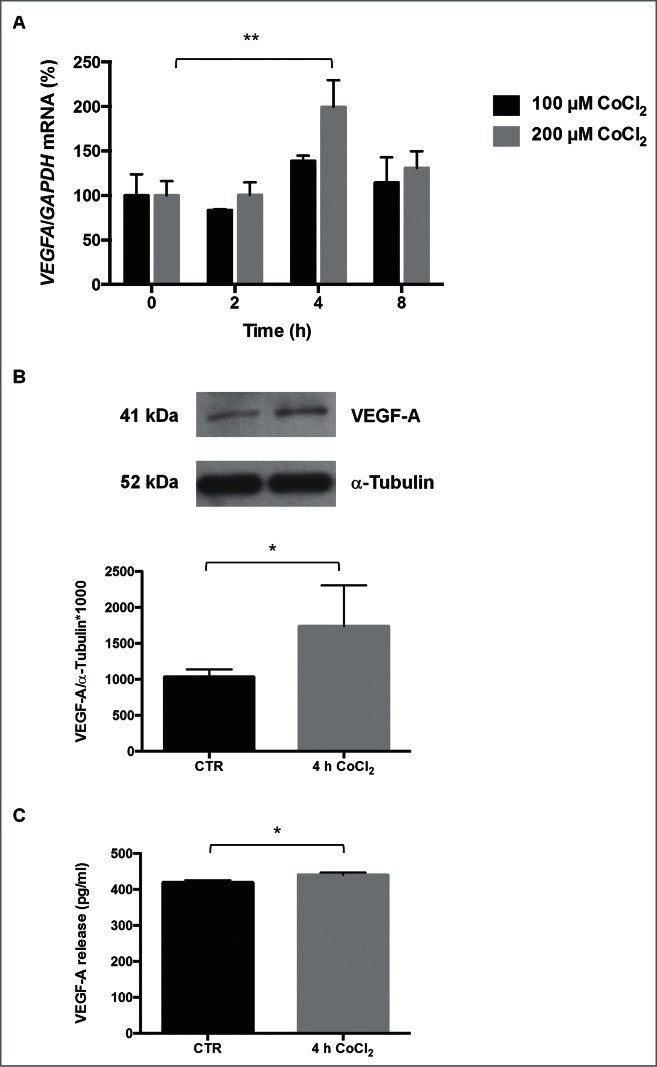
(A) Levels of VEGFA mRNA following exposure to CoCl2. Values are expressed as %. CoCl2 was tested at 100 or 200 μM, for 2, 4 or 8 h. **p < 0.01, Dunnett Multiple Comparisons test, n = 4−5. (B) Levels of intracellular VEGFA protein following treatment with 200 μM CoCl2 for 4 h. Representative Western blot analysis of VEGFA and α-tubulin expression levels; densitometry analysis (means ± SEM) of VEGFA/α-tubulin expressed as arbitrary units. *p < 0.05, Student's t-test, n = 6−8. (C) Levels of released VEGFA protein following treatment with 200 μM CoCl2 for 4 h. The release of VEGFA (pg/ml) was measured by ELISA and expressed as the means ± SEM *p < 0.05, Student's t-test, n = 8.
Since VEGFA mRNA levels can be achieved by transcript stabilization in response to hypoxia,27 we tested if CoCl2 affected the stability of VEGFA mRNA. We measured the half-life of the VEGFA mRNA by quantifying its rate of decay after blocking de novo transcription using actinomycin D. As shown in Figure 2A, treatment with CoCl2 did not significantly change the stability of VEGFA mRNA (˜2.7 h in these cells; Fig. 2A). The levels of a control short-lived mRNA (MYC mRNA, encoding the proto-oncogene MYC) showed no differences in half-life in the presence of CoCl2 (Fig. 2B). The stability of a control stable transcript, GAPDH mRNA, encoding a housekeeping protein, was not influenced by CoCl2 treatment, indicating that not all mRNAs decreased rapidly in the presence of actinomycin D, and only select labile mRNAs displayed reduced stability (Fig. 2C). To test if CoCl2 instead elevated VEGFA transcription, we quantified VEGFA pre-mRNA by using primers spanning intron-exon junctions. As shown in Fig. 2D, VEGFA pre-mRNA levels significantly raised following CoCl2 treatment, supporting the conclusion that CoCl2 increased VEGFA gene transcription but not VEGFA mRNA stability.
Figure 2.
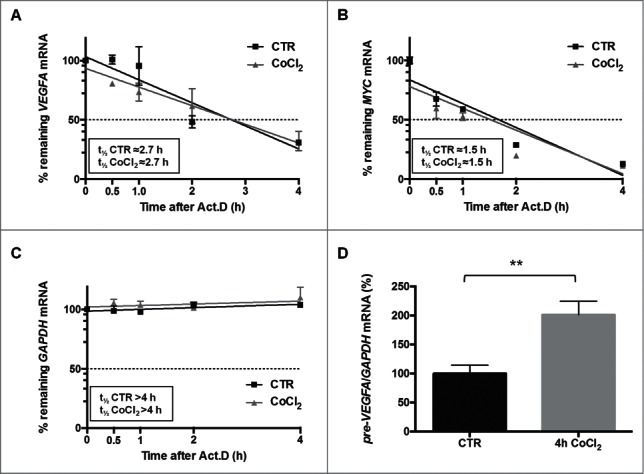
After CoCl2 treatment (200 μM, 4 h), the half-lives of VEGFA mRNA (A), as well as the half-lives of a control labile transcript (MYC mRNA) (B) and a control stable transcript (GAPDH mRNA) (C) were assessed by measuring the time required to achieve a 50% reduction in transcript levels after adding actinomycin D (Act.D). (D) The levels of VEGFA pre-mRNA following CoCl2 treatment were measured using intron-exon-spanning primers. **p < 0.01, Student's t-test, n = 4.
Pulldown of biotinylated VEGFA reveals interacting RNA-binding proteins including HuR
To further study the post-transcriptional regulation of VEGFA expression, we prepared biotinylated DNA oligomers complementary to the VEGFA mRNA for use in pulldown experiments employing streptavidin beads. We then performed mass spectrometry analysis to detect the RNA-binding proteins (RBPs) bound to the endogenous VEGFA mRNA (Fig. 3A). To ensure that the biotin pulldown step was successful, we measured the enrichment in VEGFA mRNA in the pulldown materials; as shown in Figure 3B, far greater levels of VEGFA mRNA were measured in the ASO than in the SO pulldowns (Fig. 3B). Comparison between the 2 SO samples and between the 2 ASO samples did not show any significant differences.
Figure 3.
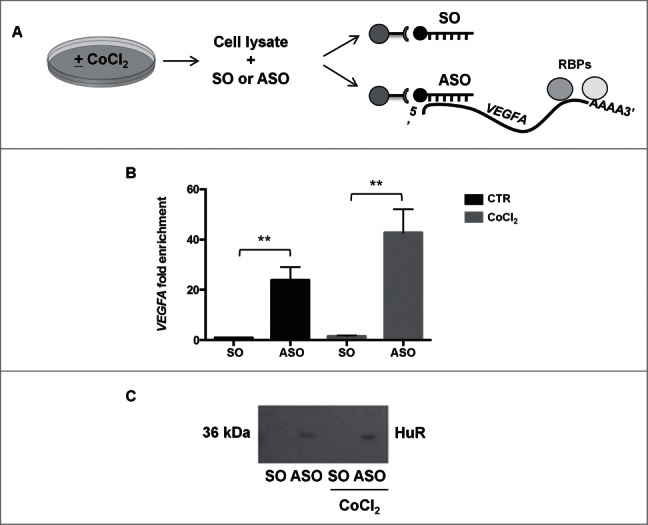
(A) Schematic representation of RNA biotin pulldown using whole-cell lysates that had been prepared from untreated or CoCl2-treated cells. The antisense oligos (ASO) captured VEGFA mRNA along with bound RPBs, while the sense oligos (SO) did not. (B) VEGFA mRNA fold enrichment after biotin pulldown with ASO and SO. **p < 0.01, Student's t-test, n = 10. (C) Representative Western blot analysis of HuR bound to VEGFA mRNA after biotin pulldown.
Mass spectrometry analysis of the bound proteins revealed a number of interacting RBPs (Fig. S2, excel file); among them, we focused our attention on ELAVL1/HuR protein (Fig. S2), an RBP that has been implicated in different forms of post-transcriptional gene regulation. The presence of HuR in the VEGFA mRNA pulldown (ASO) was confirmed by Western blot analysis (Fig. 3C); HuR levels in ASO cultures left untreated was slightly lower (1374 arbitrary units) than in ASO + CoCl2 cultures (1501 arbitrary units). We then performed ribonucleoprotein (RNP) immunoprecipitation (RIP) analysis, using anti-HuR antibody under conditions that preserved RNPs intact, to assess the association of VEGFA mRNA with HuR (Fig. 4A). VEGFA mRNA was enriched more than fold12- in HuR IP samples compared to control IgG IP samples, supporting the existence of [VEGFA mRNA-HuR] complexes (Fig. 4B). After treatment with CoCl2, the levels of [VEGFA mRNA-HuR] complexes increased further (Fig. 4B).
Figure 4.
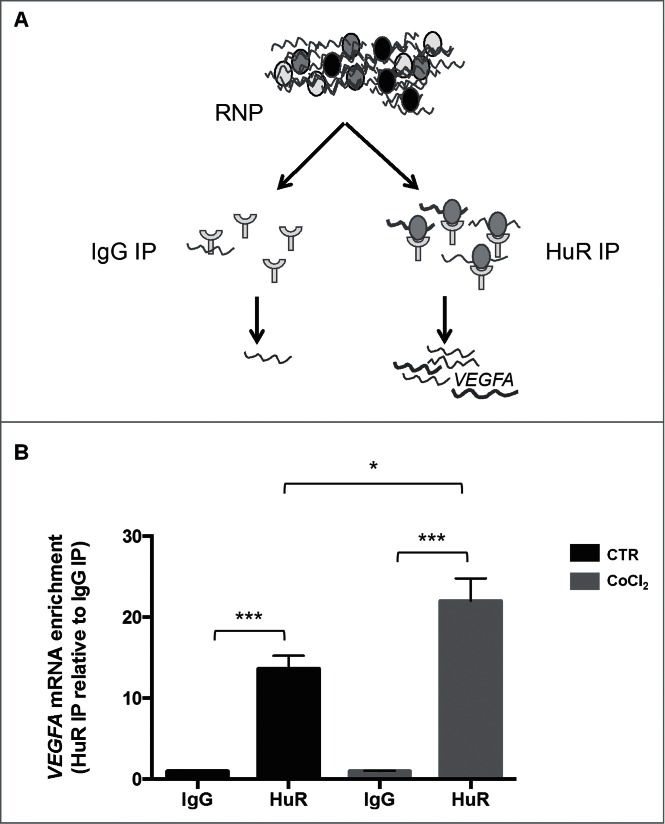
(A) Schematic representation of the RIP assay; RNP, ribonucleoprotein complex. (B) Interaction of VEGFA mRNA with HuR complexes assessed by RIP analysis using anti-HuR antibody followed by RT-qPCR analysis. *p < 0.05, ***p < 0.001, Student's t-test, n = 10.
HuR co-localizes with the distal VEGFA 3′UTR after CoCl2 treatment
Photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) analysis of HuR-bound RNAs revealed that HuR recognizes 4 distinct sites within the VEGFA 3′UTR (Fig. 5A).28 To study this interaction in live cells, the VEGFA mRNA was tagged by adding MS2 hairpins, as described in Srikantan et al.29 We constructed plasmids pMS2-RL-3′UTR-I and pMS2-RL-3′UTR-II, derived from pSL-MS2 (24X), which expressed the Renilla luciferase (RL) coding region fused to the proximal (I) and distal (II) regions of the VEGFA 3′UTR, respectively, along with 24 tandem MS2 RNA hairpins; a control plasmid (pMS2-RL) expressed only the RL coding region fused to MS2 hairpins (Fig. 5B, left panels). HeLa cells were co-transfected with one of these plasmids together with pMS2-YFP, a plasmid that expresses a fusion protein that recognizes MS2 hairpins (named MS2-BP), that is fluorescent (as it contains a YFP domain) and carries a strong nuclear localization signal (NLS) (Fig. 5B, right panels).
Figure 5.
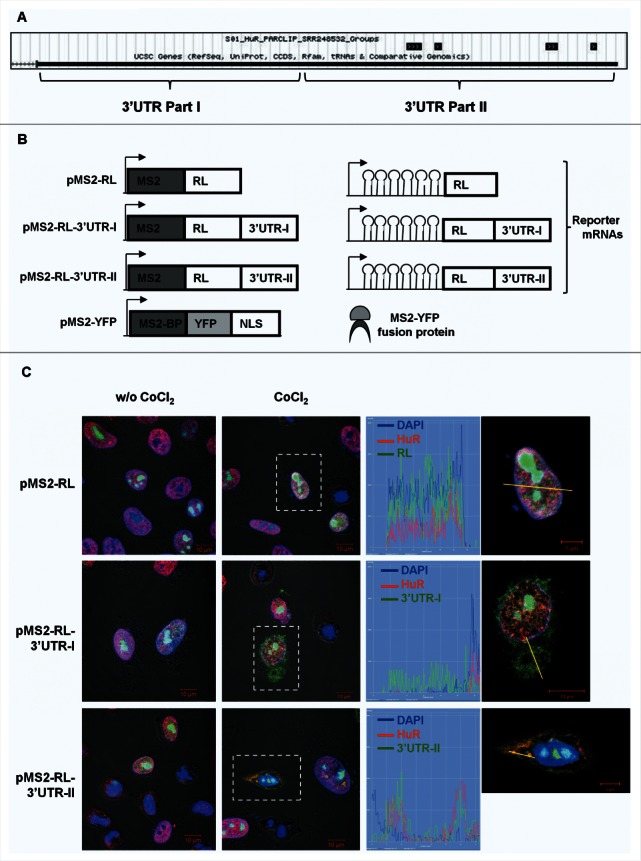
(A) Sites of HuR interaction with VEGFA mRNA as determined by PAR-CLIP analysis (Lebedeva et al., 2011). (B) Schematic representation of the plasmids used for the intracellular tracking of VEGFA mRNA (left panel). pMS2-RL, pMS2-RL-3′UTR-I and pMS2-RL-3′UTR-II were derived from pSL-MS2(24X), and each expressed the Renilla luciferase (RL) coding region and 24 tandem MS2 RNA hairpins; pMS2-RL-3′UTR-I and pMS2-RL-3′UTR-II additionally contained VEGFA 3′UTR. The plasmid pMS2-YFP expressed a fusion fluorescent protein (MS2-YFP) capable of binding MS2-containing RNA (right panel). NLS, nuclear localization signal. (C) Representative confocal fluorescence micrographs of HeLa cells to visualize HuR (using anti-HuR antibody, red) and nuclei (using DAPI, blue). YFP fluorescence is green. Merged panels show co-localization of MS2-tagged RNAs and HuR, or MS2-tagged RNAs in the cytoplasm. Magnification, 40×. Untreated (w/o CoCl2) and CoCl2-treated cells are shown. Graphs depict the quantification of single-cell signal intensities in the segment indicated (yellow lines).
Merged confocal fluorescence microscopy showed that YFP fluorescence (green) was almost exclusively nuclear in all of the cells, due to the presence of the NLS in the fusion protein MS2-YFP protein (Fig. 5C, left panels). However, when CoCl2 was added, a substantial amount of cytoplasmic fluorescence was detected in cells transfected with pMS2-RL-3′UTR-I and pMS2-RL-3′UTR-II, but not in those transfected with pMS2-RL (middle and right panels). The fluorescent signal coming from the distal VEGFA 3′UTR (pMS2-RL-3′UTR-II transfection group) overlapped with the signals of cytoplasmic HuR; the overlaps are displayed in orange color in the merged images and in the graph (right panels). These findings indicate that HuR co-localizes with the distal part of VEGFA 3′UTR following exposure to CoCl2. Single-channel images are displayed in the Supplemental Figure 3.
HuR affects VEGFA translation
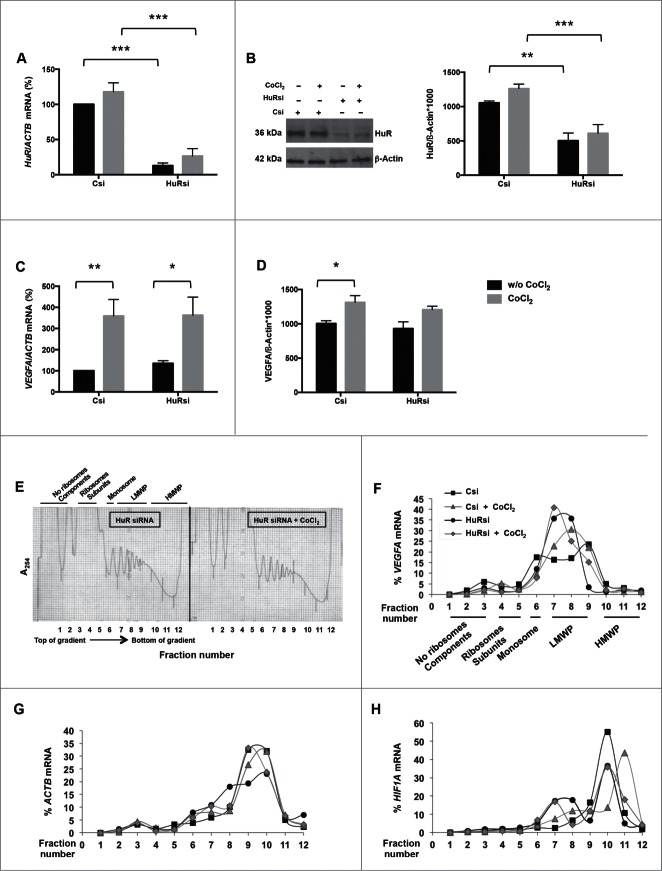
Since the stability of VEGFA mRNA was not affected by CoCl2 treatment, we sought to study if HuR influenced the translation of VEGFA mRNA. To test this possibility, we silenced HuR by using specific HuR-directed small interfering RNA (HuRsi) and scrambled small interfering RNA (Csi) in control transfection groups. As expected, 48 h later HuR silencing strongly decreased HuR mRNA (Fig. 6A) and protein (Fig. 6B) both in the presence and absence of CoCl2 (w/o CoCl2). HuR silencing did not alter basal or CoCl2-upregulated VEGFA mRNA levels (Fig. 6C). HuR silencing also failed to decrease VEGFA protein levels in basal conditions (Fig. 6D). However, CoCl2 treatment significantly increased VEGFA protein levels in control cells but not in HuR-silenced cells (Fig. 6D), suggesting that HuR influenced the translation of VEGFA mRNA only under hypoxic conditions.
Figure 6.
For figure legend, see page 8.
To test this possibility directly, we monitored the distribution of VEGFA mRNA on polysome gradients in Csi and HuRsi cells. No major differences in global polysome profiles were seen among the 4 groups [Csi relative to Csi+CoCl2 (Fig. S4); HuRsi relative to HuRsi+CoCl2 (Fig. 6E)] 48 h after transfection. In these gradients, fractions 1–4 comprised mRNAs not associated with components of the translation machinery and hence not translated, fractions 5–7 included mRNAs bound to single ribosomes (monosomes) or forming polysomes of low molecular weight (translated at low-to-moderate levels), and fractions 8–10 included mRNAs that were associated with polysomes of high molecular weight, and thus considered to be actively translated. After isolating RNA from each gradient fraction, the levels of VEGFA mRNA were measured to assess the size of polysomes present under each condition. This analysis revealed that the relative size of VEGFA mRNA polysomes increased slightly after CoCl2 treatment in Csi cells, with an increased percentage of the messenger in the actively translating fractions of polysomes (Fig. 6F). Importantly, however, HuR silencing provoked a leftward shift of the curve, consistent with the appearance of smaller polysomes and hence predicting weaker translation; this shift was only partially restored by addition of CoCl2 (Fig. 6F). As a positive control, we analyzed HIF1A mRNA, which is also a target of HuR,17 and encodes the transcription factor HIF-1α, the oxygen-regulated subunit of HIF-1, that controls transcription of hypoxia-inducible genes and is induced by CoCl2.24 HIF1A mRNA showed more extensive association with the actively translating polysome fraction in cells treated with CoCl2 (Fig. 6H); but in HuRsi cells, HIF1A mRNA was found in smaller polysomes regardless of the presence or absence of CoCl2 (Fig. 6H). As expected, the distribution of the mRNA encoding the housekeeping protein β-Actin (ACTB mRNA) was comparable among all of the groups (Fig. 6G). Taken together, these results support the view that HuR promotes the translation of VEGFA mRNA in response to CoCl2 treatment.
Discussion
VEGFA is involved in several functions, such as angiogenesis, vasculogenesis, and endothelial cell growth. VEGFA expression is tightly regulated under different physiologic and pathologic conditions.30 In the present study, we employed CoCl2, a widely used chemical compound that mimics hypoxia in mouse models and in cultured cells,31,32 to study the regulation of VEGFA expression in HeLa cells. Analysis of the dose and time dependence of this treatment revealed that 4 hours of exposure to 200 μM CoCl2 increased the levels of VEGFA mRNA as well as the levels of intracellular VEGFA protein. After 4 h of CoCl2 treatment, the extracellular release of VEGFA was modest (Fig. 1), but it increased at later time points. After 8 h of continuous exposure to CoCl2 the amount of released VEGFA increased, supporting the notion that the intracellular VEGFA pool decreased and favored an increase of the extracellular VEGFA pool (Fig. S1). Given that small increases in VEGFA protein abundance can trigger robust cellular responses in tissues in vivo,33,34 these modest elevations in VEGF production and secretion were deemed to be physiologically important. Moreover, the levels of circulating VEGFA can also vary moderately with disease states such as neuropathy or diabetic retinopathy.35,36 In general, hormones, interleukins and chemokines may trigger a response even if their circulating amount is low.37,38 Hypoxia induced by treatment with 1% O2 has been shown to elevate VEGFA mRNA stability;27 however, in our current model, the half-life of the VEGFA mRNA was not affected by the chemical inducer CoCl2, while transcription was moderately increased (Fig. 2), perhaps through a CoCl2-elicited increase in HIF-1α expression levels.17 The differences between our results and the results obtained by Zhou and colleagues27 likely stem from differences in triggering hypoxic stress using 1% O2 instead of CoCl2.
VEGF expression levels can be influenced post-transcriptionally by numerous RNA-binding proteins (e.g., nucleolin, AUF1, TIAR, TIA-1, HuD, HuC, and hnRNP K), which bind to VEGF mRNA and form ribonucleoprotein complexes that regulate all steps of VEGF production, from VEGF pre-mRNA splicing to translation.11 In addition, VEGFA 3′UTR may also undergo a conformational change mediated by the RBP hnRNP L and the GAIT complex, the latter involved in the silencing of inflammatory genes in response to environmental signals. During hypoxia, this conformational switch in the VEGFA 3′UTR overrides the repressive effect of the GAIT complex allowing high levels of VEGFA translation.39
To gain further insight into the post-transcriptional regulation of VEGFA mRNA in our system, biotinylated DNA oligomers complementary to the VEGFA mRNA were used to pull down bound RBPs (Fig. 3). Mass spectrometry analysis revealed numerous RBPs that associated with the endogenous VEGFA mRNA (Fig. S2, excel file), HuR among them.40,41 In the cell model studied here, VEGFA mRNA was highly enriched in HuR ribonucleoprotein complexes, and this interaction increased further after CoCl2 treatment (Fig. 4). Among the other RPBs that were found to bind VEGFA mRNA (Fig. S2), some of them have already been shown to regulate VEGFA mRNA fate, including polypyrimidine tract-binding protein 1 isoform b (PTB) and TIA-1-related protein (TIAR)],42 and others have been also tested under hypoxia conditions [e.g., heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), hnRNP A2/B1, and hnRNP L].43-45 To our knowledge, the impact on VEGFA regulation by some of the RBPs identified in our screen [for example, GTP-binding nuclear protein Ran (Fig. S2)] has never been studied. Their function will be analyzed as our studies progress.
Exposure to the hypoxia mimetic CoCl2 did not elevate significantly HuR levels in the cell (Fig. 6), in line with previous reports using the same cell line.17 It is likely that HuR undergoes post-translational modifications, including phosphorylation, by different CoCl2-regulated kinases which can modulate HuR binding to the mRNA.46 In this regard, a recent study described an in vitro model of blood retinal barrier constituted by pericytes and endothelial cells in coculture, wherein phosphorylation by protein kinase C modulated HuR function and consequently VEGFA protein expression.22 The same cascade was investigated in the retina of diabetic rats.21 However, CoCl2 did not trigger detectable changes in HuR phosphorylation in our system (not shown).
Further insight into HuR regulation of VEGFA expression in response to CoCl2 treatment was gained from HuR silencing, an intervention that did not affect the magnitude of VEGFA mRNA upregulation following CoCl2 treatment (Fig. 6). Similarly, HuR silencing did not decrease VEGFA protein levels in basal conditions. However, we observed an important difference in VEGFA expression following CoCl2 treatment, as HuR was essential for triggering a significant increase in VEGFA levels following CoCl2, supporting the view that HuR is needed for inducing VEGFA protein levels after a hypoxic stimulus. In line with these findings, polysome analysis revealed that VEGFA mRNA was associated with larger polysomes (actively translating polysomes) after CoCl2 treatment in control cells (Fig. 6), but this increase in polysome size was reduced after silencing HuR. In other words, the weaker translation of VEGFA mRNA in HuR-silenced cells was only partially restored by the addition of CoCl2, further supporting the notion that HuR promotes VEGFA mRNA translation under hypoxia-like conditions.
The function of HuR is strongly regulated by its shuttling between the nucleus and the cytoplasm.47,48 Interestingly, we found that the predominantly nuclear HuR co-localizes in the cytoplasm with VEGFA mRNA after treatment with CoCl2, suggesting that hypoxia, after promoting the export of HuR protein in the cytoplasm, enhances its interaction with VEGFA mRNA in this cellular compartment (Fig. 5). In this regard, the cytoplasmic translocation of HuR has been reported in response to other stress conditions, including DNA damage and glucose deprivation.49,50 Given our findings, modulating HuR levels or activity may constitute a rational pharmacological approach to manipulate VEGFA in disorders where it is desirable to inhibit neovascularization, such as diabetic retinopathy and cancer.
Funding
JLM, JK, XY, CAM, FEI, KA, and MG were supported by the NIA-IRP, NIH. CO was supported by an EMBO short-term fellowship and by the Fondazione Banca del Monte di Lombardia fellowship.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Prof. Paul Fox for the VEGFA 3′UTR plasmid constructs. The authors thank prof. Mariaclara Cuccia and Dr. Chiara Boiocchi for the technical support.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Tammela T, Enholm B, Alitalo K, Paavonen K. The biology of vascular endothelial growth factors. Cardiovasc Res 2005; 65:550-63; PMID:15664381; http://dx.doi.org/ 10.1016/j.cardiores.2004.12.002 [DOI] [PubMed] [Google Scholar]
- 2.Arcondéguy T, Lacazette E, Millevoi S, Prats H, Touriol C. VEGF-A mRNAprocessing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res 2013; 41:7997-8010; http://dx.doi.org/ 10.1093/nar/gkt539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eswarappa SM, Potdar AA, Koch WJ, Fan Y, Vasu K, Lindner D, Willard B, Graham LM, DiCorleto PE, Fox PL. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell 2014; 157:1605-18; PMID:24949972; http://dx.doi.org/ 10.1016/j.cell.2014.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pages G, Pouyssegur J. Transcriptional regulation of the Vascular Endothelial Growth Factor gene-a concert of activating factors. Cardiovasc Res 2005; 65:564-73; PMID:15664382; http://dx.doi.org/ 10.1016/j.cardiores.2004.09.032 [DOI] [PubMed] [Google Scholar]
- 5.Kurihara T, Westenskow PD, Friedlander M. Hypoxia-inducible factor (HIF)/vascular endothelial growth factor (VEGF) signaling in the retina. Exp Med Biol 2014; 801:275-81; http://dx.doi.org/ 10.1007/978-1-4614-3209-8_35 [DOI] [PubMed] [Google Scholar]
- 6.Carmeliet P. Angiogenesis in health and disease. Nat Med 2003; 9:633-60; PMID:12778148; http://dx.doi.org/ 10.1038/nm0603-653 [DOI] [PubMed] [Google Scholar]
- 7.Bhisitkul RB. Vascular endothelial growth factor biology: clinical implications for ocular treatments. Br J Ophthalmol 2006; 90:1542-7; PMID:17114590; http://dx.doi.org/ 10.1136/bjo.2006.098426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michael IP, Westenskow PD, Hacibekiroglu S, Greenwald AC, Ballios BG, Kurihara T, Li Z, Warren CM, Zhang P, Aguilar E, et al.. Local acting Sticky-trap inhibits vascular endothelial growth factor dependent pathological angiogenesis in the eye. EMBO Mol Med 2014; 6:604-23; PMID:24705878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis LM. Bevacizumab. Nat Rev Drug Discov 2005; S8-S9; PMID:15962523; http://dx.doi.org/ 10.1038/nrd1727 [DOI] [PubMed] [Google Scholar]
- 10.Yao P, Potdar AA, Ray PS, Eswarappa SM, Flagg AC, Willard B, Fox PL. The HILDA complex coordinates a conditional switch in the 3′-untranslated region of the VEGFA mRNA. PLoS Biol 2013; 11:e1001635; PMID:23976881; http://dx.doi.org/ 10.1371/journal.pbio.1001635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv Exp Med Biol 2014; 825:1-55; PMID:25201102; http://dx.doi.org/ 10.1007/978-1-4939-1221-6_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srikantan S, Gorospe M. HuR function in disease. Front Biosci 2012; 17:189-205; http://dx.doi.org/ 10.2741/3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pascale A, Govoni S. The complex world of post-transcriptional mechanisms: is their deregulation a common link for diseases? Focus on ELAV-like RNA-binding proteins. Cell Mol Life Sci 2012; 69:501-17; PMID:21909784; http://dx.doi.org/ 10.1007/s00018-011-0810-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolognani F, Perrone-Bizzozero NI. RNA–protein interactions and control of mRNA stability in neurons. J Neurosci 2008; 86:481-9 [DOI] [PubMed] [Google Scholar]
- 15.Govindaraju S, Lee BS. Adaptive and maladaptive expression of the mRNA regulatory protein HuR. World J Biol Chem 2013; 4:111-8; PMID:24340134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amadio M, Scapagnini G, Lupo G, Drago F, Govoni S, Pascale A. PKCbetaII/HuR/VEGF: A new molecular cascade in retinal pericytes for the regulation of VEGF gene expression. Pharmacol Res 2008; 57:60-6; PMID:18206386; http://dx.doi.org/ 10.1016/j.phrs.2007.11.006 [DOI] [PubMed] [Google Scholar]
- 17.Galbán S, Kuwano Y, Pullmann R Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, et al.. RNA-binding proteinsHuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol 2008; 28:93-107; http://dx.doi.org/ 10.1128/MCB.00973-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milani P, Amadio M, Laforenza U, Dell'Orco M, Diamanti L, Sardone V, Gagliardi S, Govoni S, Ceroni M, Pascale A, et al.. Posttranscriptional regulation of SOD1 gene expression under oxidative stress: Potential role of ELAV proteins in sporadic ALS. Neurobiol Dis 2013; 60:51-60; PMID:23969235; http://dx.doi.org/ 10.1016/j.nbd.2013.08.005 [DOI] [PubMed] [Google Scholar]
- 19.Amadio M, Scapagnini G, Davinelli S, Calabrese V, Govoni S, Pascale A. Involvement of ELAV RNA-binding proteins in the post-transcriptional regulation of HO-1. Front Cell Neurosci 2015; 8:459; PMID:25642166; http://dx.doi.org/ 10.3389/fncel.2014.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R, Rilla K, Akhtar S, Provenzani A, D'Agostino VG, et al.. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One 2013; 8:e69563; PMID:23922739; http://dx.doi.org/ 10.1371/journal.pone.0069563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amadio M, Bucolo C, Leggio GM, Drago F, Govoni S, Pascale A. The PKCbeta/HuR/VEGF pathway in diabetic retinopathy. Biochem Pharmacol 2010; 80:1230-7; PMID:20599775; http://dx.doi.org/ 10.1016/j.bcp.2010.06.033 [DOI] [PubMed] [Google Scholar]
- 22.Amadio M, Osera C, Lupo G, Motta C, Drago F, Govoni S, Pascale A. Protein kinase C activation affects, via the mRNA-binding Hu-antigen R/ELAV protein, vascular endothelial growth factor expression in a pericytic/endothelial coculture model. Mol Vis 2012; 18:2153-64; PMID:22879735 [PMC free article] [PubMed] [Google Scholar]
- 23.Park H, Lee DS, Yim MJ, Choi YH, Park S, Seo SK, Choi JS, Jang WH, Yea SS, Park WS, et al.. Three,3′-Diindolylmethane inhibits VEGF expression through the HIF-1α and NF-κB pathways in human retinal pigment epithelial cells under chemical hypoxic conditions. Int J Mol Med 2015; [Epub ahead of print]; 36(1):301-8 [DOI] [PubMed] [Google Scholar]
- 24.Piret JP, Mottet D, Raes M, Michiels C. CoCl2, a chemical inducer of hypoxia-inducible factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell line HepG2. Ann N Y Acad Sci 2002; 973:443-7; PMID:12485908; http://dx.doi.org/ 10.1111/j.1749-6632.2002.tb04680.x [DOI] [PubMed] [Google Scholar]
- 25.Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol 2011; 31:4219-31; PMID:21859890; http://dx.doi.org/ 10.1128/MCB.05955-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci U S A 2008; 105:20297-302; PMID:19088191; http://dx.doi.org/ 10.1073/pnas.0809376106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou S, Gu L, He J, Zhang H, Zhou M. MDM2 regulates vascular endothelial growth factor mRNA stabilization in hypoxia. Mol Cell Biol 2011; 31:4928-37; PMID:21986500; http://dx.doi.org/ 10.1128/MCB.06085-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 2011; 43:340-52; PMID:21723171; http://dx.doi.org/ 10.1016/j.molcel.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 29.Srikantan S, Abdelmohsen K, Lee EK, Tominaga K, Subaran SS, Kuwano Y, Kulshrestha R, Panchakshari R, Kim HH, Yang X, et al.. Translational control of TOP2A influences doxorubicin efficacy. Mol Cell Biol 2011; 31:3790-801; PMID:21768308; http://dx.doi.org/ 10.1128/MCB.05639-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular-specific growth factors and blood vessel formation. Nature 2000; 407:242-8; PMID:11001067; http://dx.doi.org/ 10.1038/35025215 [DOI] [PubMed] [Google Scholar]
- 31.Nagineni CN, Raju R, Nagineni KK, Kommineni VK, Cherukuri A, Kutty RK, Hooks JJ, Detrick B. Resveratrol Suppresses Expression of VEGF by Human Retinal Pigment Epithelial Cells: Potential Nutraceutical for Age-related Macular Degeneration. Aging Dis 2014; 5:88-100; PMID:24729934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou H, Chen X, Zhang WM, Zhu LP, Cheng L. HIF-1α inhibition reduces nasal inflammation in a murine allergic rhinitis model. PLoS One 2012; 7:e48618; PMID:23133644; http://dx.doi.org/ 10.1371/journal.pone.0048618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinoda K, Ishida S, Kawashima S, Wakabayashi T, Matsuzaki T, Takayama M, Shinmura K, Yamada M. Comparison of the levels of hepatocyte growth factor and vascular endothelial growth factor in aqueous fluid and serum with grades of retinopathy in patients with diabetes mellitus. Br J Ophthalmol 1999; 83:834-7; PMID:10381671; http://dx.doi.org/ 10.1136/bjo.83.7.834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penel N, Ray-Coquard I, Bal-Mahieu C, Chevreau C, Le Cesne A, Italiano A, Bompas E, Clisant S, Baldeyrou B, Lansiaux A, et al.. Low level of baseline circulating VEGF-A is associated with better outcome in patients with vascular sarcomas receiving sorafenib: an ancillary study from a phase II trial. Target Oncol 2014; 9:273-7; PMID:24218035; http://dx.doi.org/ 10.1007/s11523-013-0299-0 [DOI] [PubMed] [Google Scholar]
- 35.Briani C, Dalla Torre C, Lessi F, Cavallaro T, Scarlato M, Ferrari S, Campagnolo M, Lucchetta M, Cabrini I, Morbin M, et al.. Pentraxin-3 and VEGF in POEMS syndrome: a 2-year longitudinal study. J Neuroimmunol 2014; 277:189-92; PMID:25447599; http://dx.doi.org/ 10.1016/j.jneuroim.2014.09.021 [DOI] [PubMed] [Google Scholar]
- 36.Mahdy RA, Nada WM, Hadhoud KM, El-Tarhony SA. The role of vascular endothelial growth factor in the progression of diabetic vascular complications. Eye 2010; 24:1576-84; PMID:20508651; http://dx.doi.org/ 10.1038/eye.2010.86 [DOI] [PubMed] [Google Scholar]
- 37.Molica S, Vitelli G, Levato D, Levato L, Dattilo A, Gandolfo GM. Clinico-biological implications of increased serum levels of interleukin-8 in B-cell chronic lymphocytic leukemia. Haematologica 1999; 84:208-11; PMID:10189383 [PubMed] [Google Scholar]
- 38.Ruige JB, Bekaert M, Lapauw B, Fiers T, Lehr S, Hartwig S, Herzfeld de Wiza D, Schiller M, Passlack W, Van Nieuwenhove Y, et al.. Sex steroid-induced changes in circulating monocyte chemoattractant protein-1 levels may contribute to metabolic dysfunction in obese men. J Clin Endocrinol Metab 2012; 97:E1187-91; PMID:22523336; http://dx.doi.org/ 10.1210/jc.2011-3069 [DOI] [PubMed] [Google Scholar]
- 39.Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature 2009; 457:915-9; PMID:19098893; http://dx.doi.org/ 10.1038/nature07598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011; 43:327-39; PMID:21723170; http://dx.doi.org/ 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy AP, Levy NS, Wegner S, Goldberg MA. Transcriptional regulation of the rat vascular endothelial growth factor gene by hypoxia. J Biol Chem 1995; 270:13333-40; PMID:7768934; http://dx.doi.org/ 10.1074/jbc.270.22.13333 [DOI] [PubMed] [Google Scholar]
- 42.Lu L, Wang S, Zheng L, Li X, Suswam EA, Zhang X, Wheeler CG, Nabors LB, Filippova N, King PH. Amyotrophic lateral sclerosis-linked mutant SOD1 sequesters Hu antigen R (HuR) and TIA-1-related protein (TIAR): implications for impaired post-transcriptional regulation of vascular endothelial growth factor. J Biol Chem 2009; 284:33989-98; PMID:19805546; http://dx.doi.org/ 10.1074/jbc.M109.067918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vumbaca F, Phoenix KN, Rodriguez-Pinto D, Han DK, Claffey KP. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol Cell Biol 2008; 28:772-83; PMID:18039850; http://dx.doi.org/ 10.1128/MCB.02078-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamilton BJ, Nichols RC, Tsukamoto H, Boado RJ, Pardridge WM, Rigby WF. hnRNP A2 and hnRNP L bind the 3′UTR of glucose transporter 1 mRNA and exist as a complex in vivo. Biochem Biophys Res Commun 1999; 261:646-51; PMID:10441480; http://dx.doi.org/ 10.1006/bbrc.1999.1040 [DOI] [PubMed] [Google Scholar]
- 45.Jafarifar F, Yao P, Eswarappa SM, Fox PL. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J 2011; 30:1324-34; PMID:21343907; http://dx.doi.org/ 10.1038/emboj.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim HH, Abdelmohsen K, Gorospe M. Regulation of HuR by DNA Damage Response Kinases. J Nucleic Acids 2010; 8 p. 981487; PMID:20798862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu C, Xin W, Zhen J, Liu Y, Javed A, Wang R, Wan Q. Human antigen R mediated post-transcriptional regulation of epithelial-mesenchymal transition related genes in diabetic nephropathy. J Diabetes 2014; 7:562-72; doi: 10.1111/1753-0407.12220. [DOI] [PubMed] [Google Scholar]
- 48.Scheiba RM, de Opakua AI, Díaz-Quintana A, Cruz-Gallardo I, Martínez-Cruz LA, Martínez-Chantar ML, Blanco FJ, Díaz-Moreno I. The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets. RNA Biol 2014; 11:1250-61; PMID:25584704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 2000; 20:760-9; PMID:10629032; http://dx.doi.org/ 10.1128/MCB.20.3.760-769.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burkhart RA, Pineda DM, Chand SN, Romeo C, Londin ER, Karoly ED, Cozzitorto JA, Rigoutsos I, Yeo CJ, Brody JR, et al.. HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer. RNA Biol 2013; 10:1312-23; PMID:23807417; http://dx.doi.org/ 10.4161/rna.25274 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.