ABSTRACT
Ribosome profiling (ribo-seq) is a technique that uses high-throughput sequencing to reveal the exact locations and densities of translating ribosomes at the entire transcriptome level. The technique has become very popular since its inception in 2009. Yet experimentalists who generate ribo-seq data often have to rely on bioinformaticians to process and analyze their data. We present RiboGalaxy (http://ribogalaxy.ucc.ie), a freely available Galaxy-based web server for processing and analyzing ribosome profiling data with the visualization functionality provided by GWIPS-viz (http://gwips.ucc.ie). RiboGalaxy offers researchers a suite of tools specifically tailored for processing ribo-seq and corresponding mRNA-seq data. Researchers can take advantage of the published workflows which reduce the multi-step alignment process to a minimum of inputs from the user. Users can then explore their own aligned data as custom tracks in GWIPS-viz and compare their ribosome profiles to existing ribo-seq tracks from published studies. In addition, users can assess the quality of their ribo-seq data, determine the strength of the triplet periodicity signal, generate meta-gene ribosome profiles as well as analyze the relative impact of mRNA sequence features on local read density. RiboGalaxy is accompanied by extensive documentation and tips for helping users. In addition we provide a forum (http://gwips.ucc.ie/Forum) where we encourage users to post their questions and feedback to improve the overall RiboGalaxy service.
KEYWORDS: Galaxy, gene expression, mRNA, protein synthesis, ribosome footprinting, ribosome profiling, ribo-seq, RNA-seq, translation, translatomics
Introduction
Ribosome profiling, also known as ribo-seq, is a high through-put technique where mRNA fragments protected by ribosomes are isolated and sequenced.1 This allows the ribosome density along all mRNA transcripts present in the cell to be measured providing genome-wide information on protein synthesis (GWIPS) in vivo.2 The usages and adaptations of the technique have grown rapidly in recent years (see reviews by 3,4). The pre-processing and alignment of ribo-seq data requires tools which are primarily Linux based and are designed for command-line usage. To visually explore the ribo-seq alignment profiles additional software may be required. If the researcher wishes to compare their ribo-seq alignments to other published ribo-seq data, they would need to download, pre-process and align these data as well. In addition, researchers may wish to carry out analysis that is specific to ribo-seq data, e.g. determine the strength of the triplet periodicity signal and use it for detecting the translated reading frame.5-7 Furthermore, the analysis would likely require access to a robust computational infrastructure with a relatively large storage capacity, facilities that are not always readily available. Even if the relevant resources were at hand, the results by different groups may not be always comparable because of differences in the data processing across studies. To address these challenges, we have developed RiboGalaxy, a Galaxy-based web server8 where researchers can align, analyze and visually explore their ribo-seq data using an internet browser without the need to install numerous software tools and construct analysis pipelines.
Results
Usage of RiboGalaxy is free and does not require user registration. However, to take advantage of certain features such as the published workflows and pages, or upload large datasets using FTP, users will need to create a RiboGalaxy account (a procedure where the user only has to provide a username and password).
Using RiboGalaxy
The typical ribo-seq alignment pipeline requires pre-processing the sequence reads to remove adaptor sequences and filter out any sequence reads that correspond to ribosomal RNA (rRNA). The Pre-processing tool suite on RiboGalaxy hosts Cutadapt9 for adaptor sequence removal and Bowtie10 for rRNA removal. RiboGalaxy currently provides pre-built rRNA indices for 10 model organisms. More rRNA indices will be added over time and users may also build their own indices. As ribosome footprints are derived from mRNA, alignment to a transcriptome is usually appropriate. However, there are cases where alignment to a genome is of interest, for example if a transcriptome is not well characterized. We provide both options on RiboGalaxy. The Align to transcriptome using Bowtie option is available under the Transcriptome Mapping suite. Note that for alignments to a transcriptome, a reference transcriptome FASTA file is required. One option is to obtain a reference FASTA file from the UCSC Main table browser option in Get Data (we explain how to do this on the RiboGalaxy Help page). The Align to genome using Bowtie is available under the GWIPS-viz Mapping suite. Pre-built indices are currently provided for 13 genome assemblies. Genomic ribosome profiles can be created for either RNase I or micrococcal nuclease (MNase) generated data and the profiles for the entire genome can be viewed directly as a custom track in GWIPS-viz.11
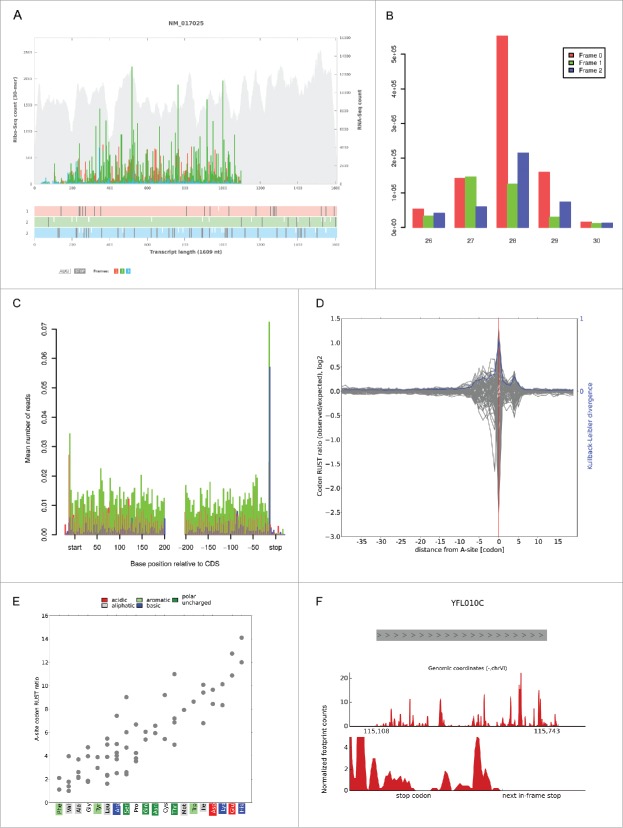
If the researcher wishes to generate mRNA (sub-codon) ribosome profiles, we recommend mapping with Bowtie10 in the Transcriptome Mapping suite followed by riboplot (see Fig. 1A). Running the ribocount tool will provide sub-codon profile counts in spreadsheet format for all transcripts to which ribosome footprints reads were mapped. Triplet periodicity and metagene analysis can also be carried out using the riboSeqR suite of tools12 (see Figs. 1B and 1C for examples). The RUST13 suite of tools allows researchers to check the quality of their ribo-seq data as well as determine the relative impact of mRNA features such as codons, amino acids, dipeptides and tripeptides on the local ribosome profiling read density (see Fig. 1D and 1E). The RiboTools suite14 provides functionality for exploring stop codon readthrough events (Fig. 1F) and translation in alternative reading frames. Differential translation expression analysis can be carried out on ribo-seq and corresponding mRNA-seq data using the baySeq15 tool which is available in the riboSeqR suite on RiboGalaxy.
Figure 1.
(A) A sub-codon ribosome footprint along with the open reading frame (ORF) organization for the rat Ldha gene (NM_017025). The footprint reads are color coded (red, green, blue (see the color version of the figure online)) according to the frame alignment (1, 2, 3). The background gray alignments represent mRNA-seq data for the corresponding transcript. In this profile, the majority of the footprint reads are green indicating that they originate from an ORF in the second reading frame. (B) A triplet periodicity plot generated for protein coding regions of the zebrafish transcriptome showing the frequency of footprint 5′ ends mapping across the 3 frames depending on their nucleotide sequence length. (C) A metagene profile of ribosome density relative to the annotated start and stop codons in the zebrafish transcriptome. (D) A RUST metafootprint profile that reveals the influence of mRNA codons on the read density relative to the decoding center (the A-site codon coordinate is shown as 0). The relative entropy (Kullback-Leibler divergence) across these sites is also provided. (E) The relative enrichment of 61 codons in the A-site assessed as the RUST ratio. Codons are grouped by encoded amino acids that are colored according to their physicochemical properties. (F) A ribo-seq profile for the yeast WWM1 gene (YFL010C). The gray bar shows the location of the gene on chromosome VI. The top red panel shows the footprint alignments across the entire WWM1 gene region while the lower red panel is a zoom into the area around the annotated stop codon showing footprint alignments downstream of the annotated stop codon to the next in-frame stop codon. Panel A was generated using the RiboPlot suite on RiboGalaxy using data from the Andreev DE et al.21 study. Panels B and C were generated using the riboSeqR suite12 on RiboGalaxy using ribo-seq data from the Bazzini AA et al.7 study. Panels C and D were generated using the RUST suite13 on RiboGalaxy using data from the Andreev DE et al.16 study. Panel F was generated using the RiboTools suite14 on RiboGalaxy using ribo-seq data from the Baudin-Baillieu A et al.22 study.
We provide Published pages (available under the Shared Data tab) to illustrate how to use the tools hosted on RiboGalaxy. Data sets from the Andreev et al. study16 were used for this purpose and the raw ribo-seq FASTQ files are available as Data Libraries under the Shared Data tab for testing purposes.
Using the RiboGalaxy published workflows
In RiboGalaxy we provide Published workflows for genome mapping (available under the Shared Data tab). Apart from being time efficient, these published workflows should particularly help researchers who are not familiar with alignment pipelines for ribo-seq data, or who are not familiar with the usage of the individual tools. If the researcher wishes to explore their ribo-seq data as a custom track in GWIPS-viz, with the advantage of comparing their alignment profiles to publicly available ribo-seq and mRNA-seq tracks, then the easiest option is to use the corresponding genome workflow. This will reduce the multi-step pre-processing and alignment process to a minimum of inputs from the user: only the adaptor sequence and the name of the custom track for visualization in GWIPS-viz need to be specified. We currently provide published workflows for the following 13 genome assemblies: Homo sapiens (hg19 and hg38), Mus musculus (mm10), Rattus norvegicus (rn6), Danio rerio (danRer7), Caenorhabditis elegans (ce10), Arabidopsis thaliana (araTha1), Drosophila melanogaster (dm3), Saccharomyces cerevisiae (sacCer3), Escherichia coli (eschcoli_k12), Bacillus subtilis (baciSubt2), Human herpesvirus 5 strain Merlin (HHV5) and Bacteriophage lambda (NC_001416).
On successful completion of an in-built workflow, an email notification is sent to the user. The custom track is visible only to the researcher and not to other public users of GWIPS-viz. All of the alignment results, coverage and profile information can be downloaded from RiboGalaxy while snapshot images of the ribosome profiles can be generated in GWIPS-viz.
We do not provide published workflows for transcriptome alignments. The reason is transcript annotations are updated regularly and it is advisable to obtain the latest reference annotations. We explain on our Help page how to get annotations in FASTA file format using the UCSC Main Table Browser 17 from within RiboGalaxy. The user may also wish to use their own data-specific transcriptome assemblies as reference sequences.
RiboGalalxy help pages and forum
RiboGalaxy is publicly available at http://riboseq.org/ and is accompanied by extensive documentation and tips for helping users. We provide help pages specifically for RiboGalaxy usage under the Help section. For each individual tool, we provide short Tips at the top of each tool page to help the user get started as well as more extensive help support on each tool page. In addition we provide a RiboGalaxy forum (http://gwips.ucc.ie/Forum) where we encourage users to post their questions and feedback to improve the overall RiboGalaxy service.
Discussion
Up to recently, software for the analysis of ribo-seq data were not publicly available. Now, however, several tools have been developed specifically for ribo-seq. We plan to expand the existing repertoire of tools on RiboGalaxy by integrating such tools (e.g., riboDiff18) as well as developing more software for the analysis of ribo-seq data.
Materials and methods
RiboGalaxy uses the Galaxy8 framework for the pre-processing, alignment and analysis pipelines. GWIPS-viz11 is used for the ribo-seq data visualization as the alignment profiles can be explored in conjunction with publicly available ribo-seq and mRNA-seq tracks. Galaxy Trackster19 is also available for data visualization.
RiboGalaxy runs on Ubuntu 14.04.3 LTS, with Apache 2.4.7 and PostgreSQL 9.3.10. The version of each of the tools hosted on RiboGalaxy is provided at the top of the page when you click on the tool.
As RiboGalaxy is a platform dedicated to ribo-seq data analysis, and not a general Galaxy server platform, only tools that are related to ribo-seq data alignment pipelines and analysis are hosted. The pre-processing and alignment software were downloaded from the Galaxy toolshed.20 In house python scripts were used for developing the RiboPlot suite as well as for generating ribosome profiles for visualization in GWIPS-viz while xml wrappers were written for the integration of the riboSeqR12 and the RUST13 suites of tools. The RiboGalaxy implementations of RiboPlot, riboSeqR and RUST are now available in the Galaxy toolshed.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to thank Cormac Fahy for beta testing RiboGalaxy and for all his feedback. We also wish to thank Thomas Hardcastle and Betty Chung for their help in the implementation and testing of riboSeqR in RiboGalaxy and Rachel Legendre and Olivier Namy for their help with the integration of RiboTools.
Funding
This work was supported by Science Foundation Ireland [grant 12/IA/1335 to P.V.B.].
References
- 1.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science (80-) 2009; 324:218-23; PMID:19213877; http://dx.doi.org/23696005 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss RB, Atkins JF. Translation Goes Global. Science (80-) 2011; 334:1509-10; PMID:22174241; http://dx.doi.org/23696005 10.1126/science.1216974 [DOI] [PubMed] [Google Scholar]
- 3.Michel AM, Baranov P V. Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. Wiley Interdiscip Rev RNA 2013; 4:473-90; PMID:23696005; http://dx.doi.org/ 10.1002/wrna.1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet 2014; 15:205-13; PMID:24468696; http://dx.doi.org/ 10.1038/nrg3645 [DOI] [PubMed] [Google Scholar]
- 5.Michel AM, Choudhury KR, Firth AE, Ingolia NT, Atkins JF, Baranov PV. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res 2012; 22(11):2219-29; PMID:22593554; http://dx.doi.org/ 10.1101/gr.133249.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerashchenko M V, Lobanov A V, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci 2012; 109:17394-9; PMID:23045643; http://dx.doi.org/ 10.1073/pnas.1120799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al.. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 2014; 33:981-93; PMID:24705786; http://dx.doi.org/ 10.1002/embj.201488411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blankenberg D, Taylor J, Nekrutenko A. Online resources for genomic analysis using high-throughput sequencing. Cold Spring Harb Protoc 2015; 2015:pdb.top083667; PMID:25655493; http://dx.doi.org/19261174 10.1101/pdb.top083667 [DOI] [PubMed] [Google Scholar]
- 9.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 2011; 17:10; http://dx.doi.org/ 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 10.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25; PMID:19261174; http://dx.doi.org/ 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michel AM, Fox G, M Kiran A, De Bo C, O'Connor PBF, Heaphy SM, Mullan JPA, Donohue CA, Higgins DG, Baranov P V. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res 2014; 42:D859-64; PMID:24185699; http://dx.doi.org/ 10.1093/nar/gkt1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung BY, Hardcastle TJ, Jones JD, Irigoyen N, Firth AE, Baulcombe DC, Brierley IAN. The use of duplex-specific nuclease in ribosome profiling and a user-friendly software package for Ribo-seq data analysis. RNA 2015; 21:1731-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connor PB, Andreev DE, Baranov PV. Surveying the relative impact of mRNA features on local ribosome profiling read density in 28 datasets. bioRxiv doi http://dx.doi.org/ 10.1101/0187622015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legendre R, Baudin-Baillieu A, Hatin I, Namy O. RiboTools: A Galaxy toolbox for qualitative ribosome profiling analysis. Bioinformatics 2015; 31(15):2586-8; PMID:25812744; http://dx.doi.org/20698981 10.1093/bioinformatics/btv174 [DOI] [PubMed] [Google Scholar]
- 15.Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 2010; 11:422; PMID:20698981; http://dx.doi.org/ 10.1186/1471-2105-11-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov P V. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife 2015; 4:1-21; PMID:25621764; http://dx.doi.org/25428374 10.7554/eLife.03971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al.. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 2014; 43:D670-81; PMID:25428374; http://dx.doi.org/ 10.1093/nar/gku1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong Y, Karaletsos T, Drewe P, Sreedharan V, Wendel H, Gunnar R. RiboDiff: Detecting Changes of Translation Efficiency from Ribosome Footprints. bioRxiv doi http://dx.doi.org/101101/017111 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goecks J, Coraor N, Nekrutenko A, Taylor J. NGS analyses by visualization with Trackster. Nat Biotechnol 2012; 30:1036-9; PMID:23138293; http://dx.doi.org/ 10.1038/nbt.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blankenberg D, Von Kuster G, Bouvier E, Baker D, Afgan E, Stoler N, Taylor J, Nekrutenko A. Dissemination of scientific software with Galaxy ToolShed. Genome Biol 2014; 15:403; PMID:25001293; http://dx.doi.org/ 10.1186/gb4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreev DE, O'Connor PB, Zhdanov AV, Dmitriev RI, Shatsky IN, Papkovsky DB, Baranov P V. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol 2015; 16:1-14; PMID:25583448; http://dx.doi.org/ 10.1186/s13059-015-0651-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baudin-Baillieu A, Legendre R, Kuchly C, Hatin I, Demais S, Mestdagh C, Gautheret D, Namy O. Genome-wide Translational Changes Induced by the Prion [PSI+]. Cell Rep 2014; 8:439-48; PMID:25043188; http://dx.doi.org/ 10.1016/j.celrep.2014.06.036 [DOI] [PubMed] [Google Scholar]