Abstract
Recent evidence has shown that the ribosome itself can play a highly regulatory role in the specialized translation of specific subpools of mRNAs, in particular at the level of ribosomal proteins (RP). However, the mechanism(s) by which this selection takes place has remained poorly understood. In our recent study, we discovered a combination of unique RNA elements in the 5′UTRs of mRNAs that allows for such control by the ribosome. These mRNAs contain a Translation Inhibitory Element (TIE) that inhibits general cap-dependent translation, and an Internal Ribosome Entry Site (IRES) that relies on a specific RP for activation. The unique combination of an inhibitor of general translation and an activator of specialized translation is key to ribosome-mediated control of gene expression. Here we discuss how these RNA regulatory elements provide a new level of control to protein expression and their implications for gene expression, organismal development and evolution.
Keywords: specialized ribosome, 5′UTR, RNA element, translation, IRES, translational inhibitory element
Introduction
The regulatory logic and circuitry for how the genetic code is translated to give rise to the remarkable diversity of cell types poses one of the greatest challenges in gene regulation. Emerging evidence suggests that in additional to transcription, an important additional layer of gene expression control may be conferred through a more regulatory function of the ribosome itself.1-5 The eukaryotic ribosome is composed of 80 ribosomal proteins (RP) and 4 rRNAs. RP expression has been observed to vary between different tissues, suggesting that there is a dynamic regulation of ribosome activity.2,6 Thereby, “specialized ribosomes,” harboring a unique composition of RPs, can be fine-tuned to translate specific subpools of mRNAs. While it has been speculated that there may be cis-regulatory elements on mRNAs that are recognized by specialized ribosomes, the nature of these elements and the ribosome recruitment mechanism have remained largely a mystery.
The primary purpose of an mRNA is to code for a protein. However, much information is also encoded in the untranslated regions (UTR), including sequences and structures that affect the translation, localization and stability of the mRNA.7 We recently uncovered a mechanism used by a group of mRNAs encoding homeobox (Hox) genes to recruit specialized ribosomes containing RPL38.8 Hox genes are crucial genes in patterning the developing embryo and their expression needs to be tightly regulated in both time and space.9 We found that many Hox mRNAs contain cis-regulatory elements in the 5′UTRs, which serve as a link between the mRNA and ribosome specificity. In particular, these mRNAs contain a new 5′UTR RNA regulatory element we have termed the Translation Inhibitory Element (TIE), which potently inhibits general cap-dependent translation. The TIE allows these Hox mRNAs to be regulated independently from other mRNAs in the cell. In addition, these Hox mRNAs also contain an Internal Ribosome Entry Site (IRES) that recruits the ribosome through a cap-independent mechanism (Fig. 1). Due to the presence of the TIE, these mRNAs can only be translated through the IRES and require more specialized regulation by the ribosome itself, as revealed by the unique requirement for a single RP, RPL38, to promote Hox IRES activity. Here we discuss the implications for newly discovered 5′UTR cis-acting RNA regulatory elements, which similar to transcriptional enhancers and silencer, act in concert toward the translation of a single transcript in time and space.
Figure 1.
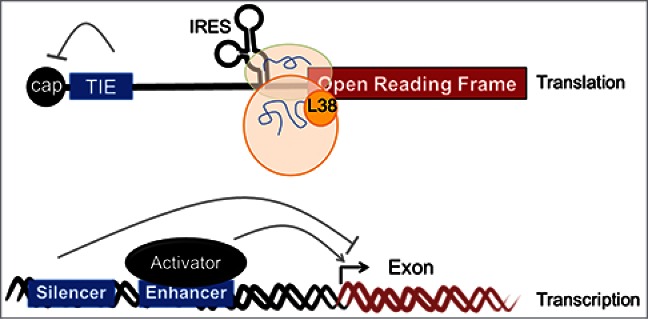
A Translation Inhibitory Element (TIE) in the 5′UTRs of certain mRNAs inhibits cap-dependent translation and frees the mRNA from cap-dependent translational regulation. An additional cis-regulatory element, the Internal Ribosome Entry Site (IRES), can recruit the ribosome through a cap-independent mechanism. Translation from the IRES enables specialized regulation by the ribosome itself. For example, RPL38 is required for the translation of several TIE and IRES-containing Hox transcripts. These RNA elements are analogous to transcriptional enhancers and silencers (bottom), which act together to regulate gene expression.
Elements That Inhibit General Cap-Dependent Translation
The predominant mode of translation initiation in the eukaryotic cell is cap-dependent. Cap-dependent translation relies on the eIF4F complex, which is recruited to the mRNA cap and in turn facilitates the recruitment of the preinitiation complex or 43S, a complex that includes the small subunit of the ribosome.10 We have found that many Hox mRNAs contain a TIE located near the 5′ cap.8 In a chimeric reporter construct where the TIE is placed upstream of the β-globin 5′UTR, the TIE is able to reduce translation of the reporter by more than 50 fold. By inhibiting cap-dependent translation, these mRNAs can be differentially regulated, and become exquisitely sensitized to translational regulation by the ribosome. In the several Hox mRNAs that contain TIE elements, there is no obvious sequence similarity among these TIE elements and it is unknown how they mechanistically function to achieve such a pronounced effect on inhibition of cap-dependent translation. It is also unclear if TIE activity can be regulated, for example to turn cap-dependent translation of specific mRNAs on or off in different tissues. Here we discuss some of the mechanisms that the TIE may employ to inhibit cap-dependent translation.
Previous studies have found elements in 5′UTRs that inhibit translation by blocking 43S scanning. These include upstream open reading frames (uORFs)11 and hairpins with high thermodynamic stability. In the case of uORFs, ribosome dissociation after translation of uORFs makes continued scanning to the main ORF difficult. Some of these elements are highly regulated under select conditions. For example, GCN4 is a transcriptional activator of amino acid biosynthesis in yeast and contains 4 uORFs in its 5′UTR. These uORFs act as amino acid sensors and their translation affects whether GCN4 is translated.12 Global translation of uORFs has also been observed to decrease upon differentiation of mouse embryonic stem cells,13 suggesting that uORF-mediated translation is regulated and can play an instructive role in the control of gene expression. Strong hairpins can also inhibit translation by physically blocking passage of the scanning 43S. The 5′UTRs of ODC 14 and TGFB1 15 for example, have high GC contents and form stable hairpins that inhibit cap-dependent translation. Curiously, the TIEs in Hox mRNAs do not have high GC contents and their inhibitory activity is not due to the presence of uORFs.8 However, one possible mechanism may be that the TIE could fold into a highly specific RNA structure that blocks 43S scanning. Therefore, future studies will be required to determine on a comparative basis whether specific structural features may be shared between the TIEs first identified within subsets of Hox 5′UTRs.
Another mechanism to inhibit cap-dependent translation is blocking 43S recruitment. The histone H4 mRNA, for instance, contains a cap-binding pocket formed by the mRNA that sequesters the cap from interacting with eIF4E, the major cap-binding protein and initiation factor.16 Without alternative mechanisms to recruit initiation factors and the ribosome, sequestering the cap can be a potent mode of inhibiting cap-dependent translation. Recent studies have also shown that miRNAs can inhibit translation by dissociating eIF4A from the eIF4F complex, which in turn blocks recruitment of the 43S.17,18 The TIE may use similar mechanisms to prevent the formation or promote the dissociation of the eIF4F complex.
A final possible mechanism for inhibiting an mRNA's translation is by changing the localization of the mRNA. mRNA export from the nucleus is highly regulated and involves a myriad of proteins. Certain subsets of mRNAs require specific export adaptor proteins to target them for mRNA export receptors.19 For example, some mRNAs are transported through the CRM1 nuclear pore receptor. CRM1 does not bind RNA and requires different RNA binding proteins to target specific mRNAs.20 An RNA element that masks an adaptor protein binding site may prevent a transcript from ever reaching the cytoplasm to be translated. Alternatively, an RNA regulon such as the TIE may be responsible for targeting the mRNA into an RNA granule in the cytoplasm. It is known that stress granules and P bodies are formed under certain types of stress.21 RNAs found in such granules are translationally inactive. These granules are thought to temporarily store mRNAs that the cell is not equipped to translate. More physiologically, RNA granules are found in neurons as transport vehicles for mRNAs from the cell body to the dendrites.22 These mRNAs are kept in a translationally repressed state by the accompanying proteins until they reach their destinations. Therefore, the TIE element may preferentially target mRNAs into cytoplasmic granules as a mechanism for translational inhibition.
Future studies will reveal more mechanistic details of TIE-mediated translational inhibition. Furthermore, identification of TIE-containing mRNAs genome-wide holds promise for delineating mRNAs that rely on direct interactions with the ribosome to achieve dynamic control of translation in a regulated manner in time and space.
Elements That Recruit Specific Ribosomes
Blocking general cap-dependent translation of a transcript offers an mRNA the possibility to be selectively translated by specialized translation machinery. Since cap-dependent translation has been inhibited, an alternative, cap-independent mode of recruiting the ribosome is required to promote translation initiation of such transcripts. Many viruses, especially ones that shut off the host's general translational machinery, have evolved IRES elements that can initiate cap-independent translation. IRES elements are RNA elements within the 5′UTR that recruit translation initiation factors or the ribosome itself directly to the mRNA, without the need for the mRNA cap.10 A number of cellular mRNAs also contain IRES elements and can initiate translation using a reduced set of initiation factors. The majority of known IRES-containing cellular mRNAs belong to stress-response genes, where the IRES element is thought to maintain their translation under stress conditions, when global cap-dependent translation is suppressed.23-25 However, the role of IRES elements in the context of normal development, cell physiology and gene regulation has remained poorly understood.
Our findings have revealed that an IRES element can be a regulatory platform or “landing pad” to recruit specialized ribosomes. We found that a subset of Hox mRNAs, in addition to possessing TIEs within their 5′UTRs that block cap-dependent translation, also contains IRES elements, which require RPL38 for their activation.2,8 Although many cellular IRESs only function in conditions when cap-dependent translation is down-regulated such as in mitosis26,27 or under stress,24,25 IRES-dependent translation is the predominant mode of translation initiation for these genes under normal physiological conditions. By creating the first targeted knockout of an IRES, we found that the Hoxa9 IRES is necessary for normal protein production in development. HOXA9 is translationally regulated by RPL38 and is usually expressed in the neural tube and somites, where RPL38 is also highly expressed.2 In Hoxa9 IRES knockout mice, production of the HOXA9 protein is blocked within the neural tube and somites, and the resulting mice display a characteristic Hox loss of function phenotype- a homeotic transformation. IRES activity is dependent on RPL38 as knocking down RPL38 decreases IRES activity.8 Hence, translation of these Hox mRNAs can only occur when the IRES element is active and RPL38 is present on ribosomes. In addition, RPL38 is not required for translation of viral IRESs such as the Hepatitis C Virus (HCV) IRES. Together, this shows that RPL38 is responsible for translating specific IRES-containing mRNAs. RPL38 is not alone in being involved in IRES-dependent translation. Previous studies have found that many viral and cellular IRESs depend on RPS25 for translational activation.28,29 Knocking down RPS25 causes a reduction in the IRES activity of these mRNAs, and ribosomes lacking RPS25 are unable to bind to the IRES.28,30 This suggests that different types of specialized ribosomes and, in particular, the individual activities of specific RPs, may promote IRES-mediated translational activation of subpools of mRNAs.
To date, IRES elements are identified through experimentation, mostly employing bicistronic reporter assays, where the first reporter reports for cap-dependent translation and the second is translated only if the preceding RNA sequence can recruit ribosomes in a cap-independent manner. However, care must be taken to ensure that the construct does not contain a cryptic splice site or promoter, which will produce a false positive result. Some control experiments we have performed include confirming that both reporters are found in a similar ratio at the RNA level between all our constructs. We also used an shRNA against one reporter and observed a knockdown of both reporters.8 These assays strongly suggest that both reporters are found on the same mRNA. While some groups advocate for the use of an m7G cap versus an A-capped monocistronic constructs to determine the relative contributions of cap vs. IRES-dependent translation in cellular IRESs, we have taken a less artificial approach. First we deleted the IRES in a capped monocistronic reporter and showed that it is required for translation. Then we deleted the IRES in the endogenous locus in vivo and observe a similarly important effect of the IRES in protein production in a developing mouse embryo. Notably, the majority of the Hoxa9 transcript in Hoxa9 IRES knock-out embryos accumulates in pre-polysomal fractions, reflecting an accumulation of mRNA not bound by translationally active ribosomal subunits. Together these experiments show that Hox IRES elements have a major contribution in the translation of these mRNAs.
Known cellular IRES elements to date do not have a consensus sequence or structure and hence it has been challenging to predict for and identify cellular IRES elements. We have functionally identified a number of IRES elements in Hox genes. With these newfound IRESs, one can attempt to extract common features, such as their RNA structures or RNA binding proteins. We have shown that Hoxa9 requires its RNA structure for IRES function. In particular, Hoxa9 contains an asymmetric bulge that introduces a ∼90° bend to the RNA structure. A similar asymmetric bulge is present in Hoxa5 8 and in domain II of the HCV IRES.31 Structural studies have shown that the bulge in HCV allows the IRES to bend and adopt an L-shape conformation in solution and also when bound to the ribosome. 31,32 The bend in HCV is crucial for 80S assembly 33 and it will be interesting to determine if the Hox IRES elements, which also recruit both the 40S and 60S, do so employing a similar structural element. With the rise in methods for genome-wide RNA structure determination,34 it will be informative to have a global perspective on structural features of mammalian 5′UTRs and identify common structural features, such as those shared between Hox IRES elements and other 5′ UTRs to pinpoint structural requirements for IRES elements. This can shed light on how cellular IRES elements interact with the ribosome.
Our unexpected finding of crucial IRES elements in Hox mRNAs raises many interesting questions. For example, how is the IRES regulated? Does its activity change in space and time during embryonic development? Most cellular IRESs require RNA binding proteins known as IRES Trans-Acting Factors (ITAFs) for ribosome recruitment and function.35,36 What are the RNA binding protein and ITAF requirements for Hox IRES elements? More genome-wide, how many other IRES elements are there? Future studies will be able to address these and other exciting questions.
Implications for a Combination of TIE and IRES Elements
We have described a powerful way of achieving ribosome-mediated translational specificity. By using a TIE to inhibit general cap-dependent translation of a specific mRNA, specialized ribosomes can exert gene-specific regulatory control of translation through the IRES. Translation of these mRNAs requires both the TIE and IRES to be present in cis and specialized ribosome activity in trans, providing additional controls over gene expression. As RP expression changes between tissues,2 we believe that the IRES activity and translation efficiency will change as well. This can provide much greater variation for how a transcript can be read out into a functional protein for multicellular organismal development.
A fascinating question is how and when such a regulatory mechanism for gene regulation evolved. The cluster of mRNAs that we focused on, the Hox genes, is an ancient group of genes found in all bilateria.37 In Drosophila melanogaster, Hox genes such as Ultrabithrox and Antennapedia appear to contain IRES elements. The 5′UTRs of these mRNAs also contain multiple uORFs which may function to inhibit cap-dependent translation.38,39 It will be interesting to determine if RPL38 also regulates the translation of Drosophilia Hox genes. While the Hox IRESs are highly conserved in vertebrates, the TIEs are only found in mammals. One speculation is that the IRESs arose earlier as a fail-safe mechanism to ensure the expression of key developmental regulators. As embryogenesis takes place in a more controlled environment in mammals, mammals can evolve additional RNA elements such as the TIE to increase regulation.
It is also interesting to think about the co-evolution of 5′UTR RNA regulons and the ribosome. RPL38 and RPS25 are both eukaryotic inventions, as are Hox genes. Knockouts of either of these RPs are viable in Saccharomyces cerevisiae with mild phenotypes,40 while a knockout of RPL38 in the mouse is embryonic lethal.41 This suggests that these RPs may have evolved additional specificity functions in multicellular organisms. It remains to be determined how many other RPs may confer such specificity on translational control.
The Hox genes have been a hotbed for discovery of regulatory mechanisms. Regulatory elements including chromatin modifiers,42,43 micro-RNAs,44 and long non-coding RNAs 45 were found and studied in Hox loci. We have uncovered a unique set of mRNA elements that allows for ribosome-mediated specificity. Using a combination of RNA elements to regulate translation is likely to be a common mechanism in the genome. Analogous to transcriptional enhancers and silencers, these elements can add a brand new layer of post-transcriptional regulation to gene expression (Fig. 1).
Funding
This work was supported by the Agency of Science, Technology and Research of Singapore (S.X.), NIH Director's New Innovator Award 7DP2OD00850902 (M.B.), Alfred P. Sloan Research Fellowship (M.B.), Mallinckrodt Foundation Award (M.B.) and Pew Scholars Award (M.B.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 2012; 13:355-69; PMID:22617470; http://dx.doi.org/ 10.1038/nrm3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 2011; 145:383-97; PMID:21529712; http://dx.doi.org/ 10.1016/j.cell.2011.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee AS, Burdeinick-Kerr R, Whelan SPJ. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci U S A 2013; 110:324-9; PMID:23169626; http://dx.doi.org/ 10.1073/pnas.1216454109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina ACC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 2011; 147:147-57; PMID:21944167; http://dx.doi.org/ 10.1016/j.cell.2011.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell 2007; 131:557-71; PMID:17981122; http://dx.doi.org/ 10.1016/j.cell.2007.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bortoluzzi S, D'Alessi F, Romualdi C, Danieli GA. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics 2001; 17:1152-7; PMID:11751223; http://dx.doi.org/ 10.1093/bioinformatics/17.12.1152 [DOI] [PubMed] [Google Scholar]
- 7.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol 2002; 3:REVIEWS0004; PMID:11897027; http://dx.doi.org/ 10.1186/gb-2002-3-3-reviews0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 2015; 517:33-8; PMID:25409156; http://dx.doi.org/ 10.1038/nature14010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol 2009; 25:431-56; PMID:19575673; http://dx.doi.org/ 10.1146/annurev.cellbio.042308.113423 [DOI] [PubMed] [Google Scholar]
- 10.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/ 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A 2009; 106:7507-12; PMID:19372376; http://dx.doi.org/ 10.1073/pnas.0810916106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller PF, Hinnebusch AG. cis-Acting sequences involved in the translational control of GCN4 expression. Biochim Biophys Acta 1990; 1050:151-4; PMID:2207139; http://dx.doi.org/ 10.1016/0167-4781(90)90157-W [DOI] [PubMed] [Google Scholar]
- 13.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011; 147:789-802; PMID:22056041; http://dx.doi.org/ 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzella JM, Blackshear PJ. Regulation of rat ornithine decarboxylase mRNA translation by its 5′-untranslated region. J Biol Chem 1990; 265:11817-22; PMID:2365701 [PubMed] [Google Scholar]
- 15.Jenkins RH, Bennagi R, Martin J, Phillips AO, Redman JE, Fraser DJ. A conserved stem loop motif in the 5′untranslated region regulates transforming growth factor-β(1) translation. PLoS One 2010; 5:e12283; PMID:20865036; http://dx.doi.org/ 10.1371/journal.pone.0012283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin F, Barends S, Jaeger S, Schaeffer L, Prongidi-Fix L, Eriani G. Cap-assisted internal initiation of translation of histone h4. Mol Cell 2011; 41:197-209; PMID:21255730; http://dx.doi.org/ 10.1016/j.molcel.2010.12.019 [DOI] [PubMed] [Google Scholar]
- 17.Fukao A, Mishima Y, Takizawa N, Oka S, Imataka H, Pelletier J, Sonenberg N, Thoma C, Fujiwara T. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol Cell 2014; 56:79-89; PMID:25280105; http://dx.doi.org/ 10.1016/j.molcel.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 18.Fukaya T, Iwakawa H, Tomari Y. MicroRNAs block assembly of eIF4F translation initiation complex in Drosophila. Mol Cell 2014; 56:67-78; PMID:25280104; http://dx.doi.org/ 10.1016/j.molcel.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 19.Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 2007; 8:761-73; PMID:17786152; http://dx.doi.org/ 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- 20.Siddiqui N, Borden KLB. mRNA export and cancer. Wiley Interdiscip Rev RNA 2012; 3:13-25; PMID:21796793; http://dx.doi.org/ 10.1002/wrna.101 [DOI] [PubMed] [Google Scholar]
- 21.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 2009; 10:430-6; PMID:19461665; http://dx.doi.org/ 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- 22.Holt CEE, Schuman EMM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 2013; 80:648-57; PMID:24183017; http://dx.doi.org/ 10.1016/j.neuron.2013.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holcik M, Sonenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet 2000; 16:469-73; PMID:11050335; http://dx.doi.org/ 10.1016/S0168-9525(00)02106-5 [DOI] [PubMed] [Google Scholar]
- 24.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell 2008; 100:27-38; PMID:18072942; http://dx.doi.org/ 10.1042/BC20070098 [DOI] [PubMed] [Google Scholar]
- 25.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J 2010; 29:1865-76; PMID:20453831; http://dx.doi.org/ 10.1038/emboj.2010.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev 2001; 15:2083-93; PMID:11511540; http://dx.doi.org/ 10.1101/gad.889201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyronnet S, Pradayrol L, Sonenberg N. Alternative splicing facilitates internal ribosome entry on the ornithine decarboxylase mRNA. Cell Mol life Sci 2005; 62:1267-74; PMID:15905964; http://dx.doi.org/ 10.1007/s00018-005-5020-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 2009; 23:2753-64; PMID:19952110; http://dx.doi.org/ 10.1101/gad.1832209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hertz MI, Landry DM, Willis AE, Luo G, Thompson SR. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol Cell Biol 2013; 33:1016-26; PMID:23275440; http://dx.doi.org/ 10.1128/MCB.00879-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, Spahn CMT. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res 2011; 39:5264-75; PMID:21378123; http://dx.doi.org/ 10.1093/nar/gkr114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol 2003; 10:1033-8; PMID:14578934; http://dx.doi.org/ 10.1038/nsb1004 [DOI] [PubMed] [Google Scholar]
- 32.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 2001; 291:1959-62; PMID:11239155; http://dx.doi.org/ 10.1126/science.1058409 [DOI] [PubMed] [Google Scholar]
- 33.Paulsen RB, Seth PP, Swayze EE, Griffey RH, Skalicky JJ, Cheatham TE, Davis DR. Inhibitor-induced structural change in the HCV IRES domain IIa RNA. Proc Natl Acad Sci U S A 2010; 107:7263-8; PMID:20360559; http://dx.doi.org/ 10.1073/pnas.0911896107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat Rev Genet 2014; 15:469-79; PMID:24821474; http://dx.doi.org/ 10.1038/nrg3681 [DOI] [PubMed] [Google Scholar]
- 35.Faye MD, Holcik M. The role of IRES trans-acting factors in carcinogenesis. Biochim Biophys Acta – Gene Regul Mech 2015; 1849:887-97; PMID:25257759; http://dx.doi.org/ 10.1016/j.bbagrm.2014.09.012 [DOI] [PubMed] [Google Scholar]
- 36.Komar AA, Hatzoglou M. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 2011; 10:229-40; PMID:21220943; http://dx.doi.org/ 10.4161/cc.10.2.14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrier DEK, Holland PWH. Ancient origin of the Hox gene cluster. Nat Rev Genet 2001; 2:33-8; PMID:11253066; http://dx.doi.org/ 10.1038/35047605 [DOI] [PubMed] [Google Scholar]
- 38.Ye X, Fong P, Iizuka N, Choate D, Cavener DR. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol Cell Biol 1997; 17:1714-21; PMID:9032298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh S, Scott MP, Sarnow P. Homeotic gene Antennapedia mRNA confer translational initiation by internal ribosome binding. Genes Dev 1992; 6:1643-53; PMID:1355457; http://dx.doi.org/ 10.1101/gad.6.9.1643 [DOI] [PubMed] [Google Scholar]
- 40.Steffen KK, McCormick MA, Pham KM, MacKay VL, Delaney JR, Murakami CJ, Kaeberlein M, Kennedy BK. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 2012; 191:107-18; PMID:22377630; http://dx.doi.org/ 10.1534/genetics.111.136549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paterson HF. In vivo and in vitro studies on the early embryonic lethal tail-short (Ts) in the mouse. J Exp Zool 1980; 211:247-56; PMID:7373273; http://dx.doi.org/ 10.1002/jez.1402110214 [DOI] [PubMed] [Google Scholar]
- 42.Duncan IM. Polycomblike: a gene that appears to be required for the normal expression of the bithorax and antennapedia gene complexes of Drosophila melanogaster. Genetics 1982; 102:49-70; PMID:6813190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature 1978; 276:565-70; PMID:103000; http://dx.doi.org/ 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- 44.Yekta S, Shih I-H, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science 2004; 304:594-6; PMID:15105502; http://dx.doi.org/ 10.1126/science.1097434 [DOI] [PubMed] [Google Scholar]
- 45.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al.. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129:1311-23; PMID:17604720; http://dx.doi.org/ 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]


