SUMMARY
Autism spectrum disorder is a heritable, common neurodevelopmental disorder with diverse genetic causes. Several studies have implicated protein synthesis as one among several of its potential convergent mechanisms. We originally identified janus kinase and microtubule-interacting protein 1 (JAKMIP1) as differentially expressed in patients with distinct syndromic forms of ASD, Fragile X Syndrome and 15q duplication syndrome. Here, we provide multiple lines of evidence that JAKMIP1 is a component of polyribosomes and an RNP translational regulatory complex that includes fragile X mental retardation protein, DEAD box helicase 5, and poly(A) binding protein, cytoplasmic 1. JAKMIP1 loss dysregulates neuronal translation during synaptic development, affecting glutamatergic NMDAR signaling, and results in social deficits, stereotyped activity, abnormal postnatal vocalizations, and other autistic-like behaviors in the mouse. These findings define an important and novel role for JAKMIP1 in neural development and further highlight pathways regulating mRNA translation during synaptogenesis in the genesis of neurodevelopmental disorders.
INTRODUCTION
Autism spectrum disorder (ASD) is a pervasive, heritable neurodevelopmental disorder (Abrahams and Geschwind, 2008; Berg and Geschwind, 2012) that manifests during infancy or early childhood by impaired social communication and restrictive and repetitive behaviors (DSM-V, 2013). A growing number of risk genes have been identified, and similar to other complex genetic conditions, it is estimated that hundreds of genes may contribute to ASD risk (Iossifov et al., 2014). This complexity has spawned focus on identifying pathways where multiple ASD risk genes may converge, including the RNA-binding proteins FMRP and RBFOX1 (De Rubeis et al., 2014; Fogel et al., 2012; Steinberg and Webber, 2013). Other convergent pathways related to ASD include mTOR, which itself is not a ASD risk gene, but is recognized as regulating a key pathway impacted by ASD risk genes, such as PTEN and TSC1/2 (Sawicka and Zukin, 2012).
JAKMIP1 is an RNA binding protein that is conserved on the vertebrate lineage (Couve et al., 2004; Steindler et al., 2004) and expressed highly in glutamatergic neurons during brain development (genepaint.org) (Cahoy et al., 2008; Vidal et al., 2009). We found that JAKMIP1 was differentially expressed in patients with two syndromic forms of ASD, Fragile X and (dup)15q11–13 syndrome, and upon RBFOX1 knockdown (Fogel et al., 2012; Nishimura et al., 2007). To date, eleven ASD subjects have been identified with copy number variations that contain JAKMIP1(Autism Genome Project et al., 2007; Kaminsky et al., 2011; Poultney et al., 2013; Tzetis et al., 2012). But, its interactions or function in the CNS, especially during brain development, are largely unknown.
Here, we use an unbiased proteomic approach to identify JAKMIP1’s protein interactome during its peak expression in vivo and observe that JAKMIP1’s binding partners are remarkably enriched for proteins involved in translation. JAKMIP1 binds a complex including FMRP, multiple proteins known to interact with FMRP (Kanai et al., 2004; Villace et al., 2004), and its translational targets (Fernandez et al., 2013; Santoro et al., 2012). We show that JAKMIP1 is expressed in fractions containing mRNPs, monosomes, and polyribosomes and demonstrate a functional role for JAKMIP1 in translation by showing that Jakmip1 knockout reduces new translation in neurons. Jakmip1 knockout in mouse leads to ASD related behaviors including motor and neurological stereotypies, social abnormalities, abnormal ultrasonic vocalizations, reduced anxiety/increased impulsivity, and motor impairments as well as glutamatergic NMDAR signaling deficits that are predicted by some of its targets. These data demonstrate an important role for JAKMIP1 in translational regulation during development, coinciding with the peak period of cortical synaptogenesis. These observations also strengthen the link between neuronal translation and behavior, an emerging theme in the pathophysiology of ASD (Gkogkas et al., 2013; Kelleher and Bear, 2008; Santini et al., 2013).
RESULTS
Defining JAKMIP1’s protein interactome during neocortex development in vivo
To determine JAKMIP1 function in the CNS in an unbiased manner, we employed Multidimensional Protein Identification Technology (MudPIT) (Wohlschlegel, 2009) to investigate JAKMIP1’s proteomic interactome during its peak expression in mouse neocortical development (p8-p14; Figures 1A and 1B). Using conservative criteria, we identified a core set of 33 JAKMIP1 interactors (Table 1), including eleven genes involved in translation and YWHAG, a previously identified interactor, which serves as a positive control (Jin et al., 2004). Pathway analysis of JAKMIP1’s top binding partners revealed that they coalesce into two primary networks that share ‘protein synthesis’ as the common denominator and the most significant molecular and cellular function (Figure 1C).
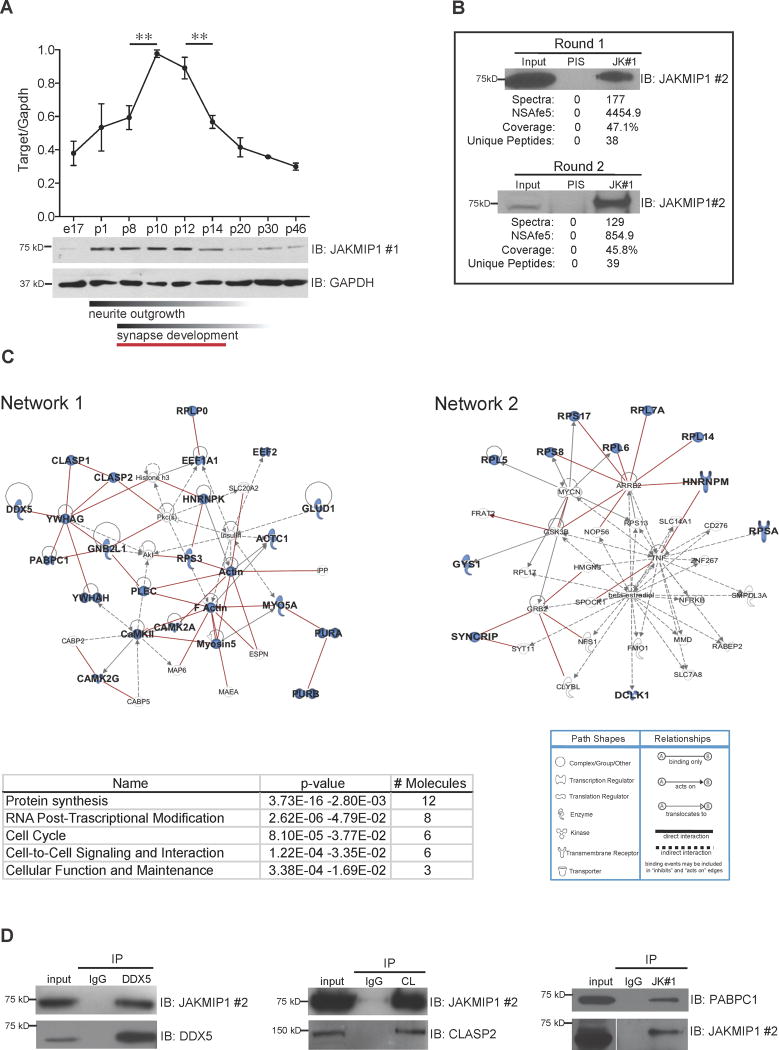
Figure 1. JAKMIP1 Associates with Proteins Involved in Translation.
(A) JAKMIP1 increases in expression during the second postnatal week in mouse neocortex, peaking from P8 to P14. Upper graph displays relative JAKMIP1 protein levels between P1 and P46. JAKMIP1 expression in embryonic day 17 (E17) whole brain (far left) is also included. The y axis represents within-blot GAPDH normalized values from Image J densitometry analysis of JAKMIP1. A representative western blot (IB) is shown below (65% increase from P8 to P10, p = 0.007, two-tailed unpaired t test; 36% decrease from P12 to P14, p = 0.01, two-tailed unpaired t test; n = 3 for each time point). Red bar below denotes period of JAKMIP1 expression change. Data are shown as mean ± SEM. (B) JAKMIP1 was immunoprecipitated from two pools of postnatal WT mouse neocortex (round 1 and round 2). PIS is rabbit preimmune serum (negative control). JAKMIP1 pull-down was confirmed by immunoblotting (IB). MudPIT readouts are listed below. (C) Upper left: Ingenuity network 1, ‘‘cell to cell signaling and interaction, nervous system development and function, protein synthesis.’’ Lower left: a list of the most significant molecular and cellular functions of JAKMIP1 interactors. Upper right: Ingenuity network 2, ‘‘protein synthesis, RNA post-transcriptional modification, cell death.’’ Lower right: Ingenuity pathway analysis network legend. Proteins highlighted in blue are JAKMIP1 protein interactors. Red lines indicate known PPIs. (D) JAKMIP1 or target protein was immunoprecipitated from mouse postnatal neocortex or differentiated mouse neural progenitor cells (PABPC1) followed by immunoblotting with target protein or JAKMIP1. JK #1 (JAKMIP1 #1) is J269–286 (Steindler et al., 2004), JAKMIP1 #2 is rabbit anti JAKMIP1 (Proteintech Group), and CL is rabbit anti CLASP2. See also Figure S1.
Table 1. JAKMIP1’s in vivo developmental proteomic interactome.
| MudPIT run #1 | MudPIT run #2 | MudPIT run #1 | MudPIT run #2 | |||||
|---|---|---|---|---|---|---|---|---|
| Entrez Gene Name | Symbol | NSAFe5 | NSAFe5 | Spectral counts | Spectral counts | PatternLab ACFold (p-value) | Networks | Location |
| janus kinase and microtubule interacting protein 1 | JAKMIP1 | 4455/0 | 855/0 | 177/0 | 129/0 | 4.42 E-26 | NA | Cytoplasm |
| actin, alpha, cardiac muscle 1 | ACTC1 | 585/0 | 1255/0 | 14/0 | 114/0 | NS | 1 | Cytoplasm |
| calcium/calmodulin-dependent protein kinase II alpha | CAMK2A | 264/0 | 260/0 | 8/0 | 30/0 | NS | 1 | Cytoplasm |
| calcium/calmodulin-dependent protein kinase II gamma | CAMK2G | 208/0 | 118/0 | 7/0 | 15/0 | 0.028 | 1 | Cytoplasm |
| cytoplasmic linker associated protein 1 | CLASP1 | 554/0 | 149/0 | 54/0 | 55/0 | 2.65 E-09 | 1 | Cytoplasm |
| cytoplasmic linker associated protein 2 | CLASP2 | 881/127 | 132/0 | 72/6 | 41/0 | 2.028 E-06 | 1 | Cytoplasm |
| CAP-GLY domain containing linker protein family, member 4 | CLIP4 | 895/0 | 194/0 | 40/0 | 33/0 | 2.56 E-06 | NA | unknown |
| doublecortin-like kinase 1 | DCLK1 | 229/0 | 181/0 | 11/0 | 33/0 | NS | 2 | Cytoplasm |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | DDX5 # | 436/0 | 41/0 | 17/0 | 6/0 | 0.023 | 1 | Nucleus |
| eukaryotic translation elongation factor 1 alpha 1 | EEF1A1 # | 102/0 | 871/0 | 3/0 | 97/0 | NS | 1 | Cytoplasm |
| eukaryotic translation elongation factor 2 | EEF2 | 202/0 | 174/0 | 11/0 | 36/0 | NS | 1 | Cytoplasm |
| glutamate dehydrogenase 1 | GLUD1 | 819/195 | 37/0 | 29/4 | 5/0 | NS | 1 | Cytoplasm |
| guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | GNB2L1 | 696/257 | 275/0 | 14/3 | 21/0 | NS | 1 | Cytoplasm |
| glycogen synthase 1 (muscle) | GYS1 | 534/0 | 11/0 | 25/0 | 2/0 | 0.012 | 2 | Cytoplasm |
| heterogeneous nuclear ribonucleoprotein K | HNRNPK # | 306/235 | 116/0 | 9/4 | 13/0 | NS | 1 | Nucleus |
| heterogeneous nuclear ribonucleoprotein M | HNRNPM | 108/0 | 61/0 | 5/0 | 10/0 | NS | 2 | Plasma Membrane |
| myosin VA (heavy chain 12, myoxin) | MYO5A*# | 51/0 | 16/0 | 6/0 | 7/0 | NS | 1 | Cytoplasm |
| poly(A) binding protein, cytoplasmic 1 | PABPC1*# | 124/0 | 46/0 | 5/0 | 7/0 | NS | 1 | Cytoplasm |
| plectin | PLEC | 118/0 | 10/0 | 35/0 | 11/0 | 3.99 E-04 | 1 | Cytoplasm |
| purine-rich element binding protein A | PURA*# | 147/0 | 65/0 | 3/0 | 5/0 | NS | 1 | Nucleus |
| purine-rich element binding protein B | PURB # | 584/0 | 51/0 | 12/0 | 4/0 | NS | 1 | Nucleus |
| ribosomal protein L14 | RPL14 | 871/0 | 76/0 | 12/0 | 4/0 | NS | 2 | Cytoplasm |
| ribosomal protein L5 | RPL5 | 955/0 | 70/0 | 18/0 | 5/0 | 0.023 | 2 | Cytoplasm |
| ribosomal protein L6 | RPL6 # | 586/0 | 70/0 | 11/0 | 5/0 | NS | 2 | Cytoplasm |
| ribosomal protein L7a | RPL7A | 829/0 | 31/0 | 14/0 | 2/0 | NS | 2 | Cytoplasm |
| ribosomal protein, large, P0 | RPLP0 # | 845/0 | 92/0 | 17/0 | 7/0 | 0.020 | 1 | Cytoplasm |
| ribosomal protein S17 | RPS17 | 1634/0 | 154/0 | 14/0 | 5/0 | 0.046 | 2 | Cytoplasm |
| ribosomal protein S3 | RPS3 | 1167/0 | 102/0 | 18/0 | 6/0 | NS | 1 | Cytoplasm |
| ribosomal protein S8 | RPS8 | 1212/522 | 100/0 | 16/4 | 5/0 | NS | 2 | Cytoplasm |
| ribosomal protein SA | RPSA | 694/0 | 56/0 | 13/0 | 4/0 | NS | 2 | Plasma Membrane |
| RUN and FYVE domain containing 3 | RUFY3 | 235/174 | 88/0 | 7/3 | 10/0 | NS | NA | unknown |
| synaptotagmin binding, cytoplasmic RNA interacting protein | SYNCRIP # | 253/0 | 27/0 | 10/0 | 4/0 | NS | 2 | Nucleus |
| tubulin, alpha 4a | TUBA4A | 1512/0 | 2695/0 | 43/0 | 291/0 | NS | NA | Cytoplasm |
| tyrosine 3-monooxygenase/tryptophan 5- monooxygenase activation protein, gamma polypeptide | YWHAG | 191/0 | 739/0 | 3/0 | 44/0 | NS | 1 | Cytoplasm |
| tyrosine 3- monooxygenase/tryptophan 5- | YWHAH | 128/0 | 641/0 | 2/0 | 38/0 | NS | 1 | Cytoplasm |
| monooxygenase activation protein, eta polypeptide | ||||||||
| 1810049H1 9Rik | 1112/0 | 895/0 | 18/0 | 55/0 | 2.56 E-06 | NA | NA | |
| Igkv1-117 | 1787/0 | 122/0 | 27/0 | 7/0 | NS | NA | NA |
Columns 3–6 from left are JAKMIP1 IP values/Control IP values;
Did not meet spectral criteria, but did meet 2 of 3 screening criteria;
FMRP interactor; NA: Not Assigned, NS: Not significant.
We next cross-referenced the JAKMIP1 interactors in network 1 with the Human Protein Reference Database (HPRD) to externally validate these networks and to assess their conservation between mouse and human (Prasad et al., 2009). We identified 17 protein interactions that were observed in the MudPIT from mouse tissue within the HPRD protein interaction list, demonstrating a statistically significant enrichment of known protein-protein interactions (PPIs) (Permutation test; p-value =3.4 e–12). To experimentally validate the identified protein associations, we performed co-immunoprecipitation (Co-IP), confirming DDX5, CLASP2, PABPC1 and CAMK2G using either JAKMIP1 or target protein pull-down (Figure 1D; Figure S1A–D). These results indicate that JAKMIP1 binds an evolutionarily conserved protein interactome related to neural translation during postnatal cortical development.
JAKMIP1 and its protein interactome are strongly associated with the cellular translational machinery
That the majority of JAKMIP1 protein interactors are involved in translation or associated with ribosomal function suggested that JAKMIP1 itself might also be ribosome associated. To test this, we determined if JAKMIP1 binds to polyribosomes by Co-IP using a BacTRAP N2A cell line in which RPL10A, an integral part of the 60S ribosome subunit, is fused to an enhanced green fluorescent protein tag (Heiman et al., 2008). We found that JAKMIP1, DEAD box helicase 5 (DDX5), and poly(A) binding protein, cytoplasmic 1 (PABPC1) all Co-IP with RPL10A, which was not observed in the controls (Figure S2A), confirming JAKMIP1 interactions with ribosomal components.
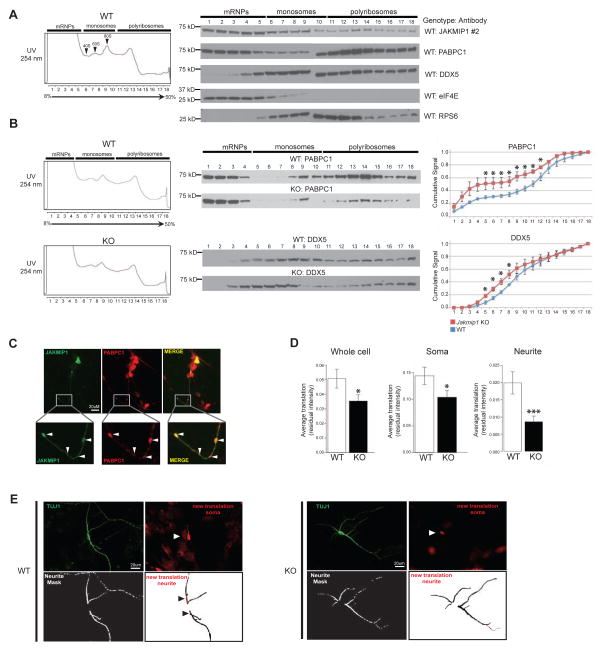
To further explore JAKMIP1’s involvement with translation in vivo, we tested its co-expression with messenger ribonucleoprotein particles (mRNPs), monosomes and polyribosomes. Polyribosome profiling from mouse brain revealed that JAKMIP1 was expressed alongside its confirmed MudPIT interactors DDX5 and PABPC1, a polyribosome-enriched control (Gebauer and Hentze, 2004), in mRNP, monosome and polyribosome fractions (Figure 2A). Fractionation controls, RPS6, eIF4E, and PABPC1, all showed the expected protein expression distribution. To further test whether JAKMIP1 is important for the integrity of the translational machinery, we repeated these experiments in Jakmip1 −/− (KO) mice (Figure S2B–F). We observed that in the absence of JAKMIP1, both PABPC1 and DDX5 proteins showed a statistically significant shift from polyribosome fractions to monosome and mRNP fractions (Figure 2B). This in vivo data indicates that JAKMIP1 is expressed alongside mRNPs, monosomes, and polyribosomes and that JAKMIP1 contributes to the association of PABPC1 and DDX5 with polyribosomes.
Figure 2. JAKMIP1 is an integral component of translation-associated RNP granules and regulates neuronal translation.
(A) Left: A representative neocortex polyribosome fractionation profile from WT postnatal mice. Right: Representative western blots demonstrating JAKMIP1, PABPC1, DDX5, eukaryotic translation initiation factor 4E (eIF4E), and ribosomal protein S6 (RPS6) expression in each corresponding fraction (1–18). (B) Left: Representative polyribosome fractionation profiles of postnatal WT or Jakmip1 KO mouse neocortex. Middle: Representative western blots demonstrating PABPC1 and DDX5 protein expression in corresponding fractions (1–18). Right: Quantification of polyribosome protein shifts. Graphs are the cumulative ratio of protein signal (Y-axis) per fraction (X-axis) calculated within sample. Red represents Jakmip1 KO signal, while blue represents WT signal [PABPC1 (WT: N=3, KO: N=3), DDX5 (WT: N=4, KO: N=4)]. Significance was calculated using a two sample, one-tailed t-test. Data are shown as mean +/− SEM. For (A) and (B), fractions 1–9 and 10–18 span two blots and were run simultaneously. (C) JAKMIP1 colocalizes with PABPC1-positive granules in vitro (arrows). Human JAKMIP1 N-ter protein (left) and human PABPC1-dsRED fusion protein (middle) are coexpressed in granules (right) of differentiated neurons from Jakmip1 KO mice. To match signal intensity across channels, brightness and contrast was adjusted for each channel independently. (D), Nascent protein synthesis, defined as the mean signal intensity (total pixel intensity/area measured) is decreased in whole cell and soma (WT, N =199; KO, N= 224: p=0.049 and 0.045, respectively) and neurites (WT, N=227; KO, N=264 : p=0.00125, N refers to number of images) of neurons lacking Jakmip1. Significance across three trials was estimated using mixed effects regression with a fixed effect covariate for imaging date and a random intercept for litter. Residual intensity after correction for date and littermate set is displayed. Data are shown as mean +/− SEM. (E) Representative images used for FUNCAT analysis, where greater pixel intensity (red, Azide 555) demonstrates increased translation. To facilitate translation visualization, brightness of the red image was increased equally for both genotypes. Brightness was adjusted slightly for TUJ1 images. Panel descriptions: Upper left, TUJ1; Upper right, Azide 555 (new translation, cell soma); Lower left, neurite mask (space analyzed); Lower right (nascent translation from cell neurite analysis space shown. Neurite mask is inverted to black show translation space). White arrows indicate cell soma translation, while black arrows indicate neurite translation. See also Figure S2 and Figure S4.
JAKMIP1 associates with an FMRP-containing RNA transport granule
Transport RNA-protein granules involved in translational control have been identified that contain FMRP, kinesin and several JAKMIP1 binding partners identified here by MudPIT, including PABPC1 (Villace et al., 2004) and DDX5 (Kanai et al., 2004). To further explore JAKMIP1’s relationship with RNA granules, we tested for overlap of JAKMIP1 interactors identified here with known FMRP-containing RNA-transport granules and found significant overlap with JAKMIP1 interactors (Kanai et al., 2004) (Hypergeometric probability; p-value =9.7 e–08; Table 2). A similar overlap was observed between JAKMIP1’s protein interactome and an FMRP RNA-transport granule identified by hStaufen pull-down (Villace et al., 2004) (Hypergeometric probability; p-value =1.3 e–11; Table 2).
Table 2. MudPIT-identified JAKMIP1 protein interactors show overlap with previously identified FMRP-containing RNA-transporting granules involved in translation.
JAKMIP1’s protein interactome overlaps with an FMRP/kinesin RNA transport granule (A, Kanai et al. 2004) and with an FMRP/Staufen RNA transport granule (B, Villace et al. 2004).
| A. | ||
|---|---|---|
| Protein name | JAKMIP1 binder | MudPIT |
| FMR1 | Co-IP evidence | |
| FXR1 | X | 1 |
| FXR2 | - | |
| PURA* | X | 2 |
| PURB* | X | 2 |
| Staufen | - | |
| EF-1a | X | 2 |
| eIF2a | - | |
| eIF2b | - | |
| eIF2y | - | |
| Hsp70 | - | |
| RPL3 | X | 1 |
| DDX1* | X | 1 |
| DDX3* | X | 1 |
| DDX5 | X | 2 |
| hnRNPA/B | - | |
| hnRNPA0 | - | |
| hnRNPA1 | - | |
| hnRNPD | X | 1 |
| hnRNPU* | X | 2 |
| ARF-GEP100/BRAG2 | - | |
| ALY* | - | |
| CIRBP | - | |
| EWS | - | |
| NONO* | X | 1 |
| Nucleolin | X | 1 |
| PSPC1 | - | |
| PSF* | - | |
| RTCD1 | - | |
| RNA binding motif | - | |
| protein 3 | ||
| SYNCRIP* | X | 2 |
| TLS* | - | |
| Ser/Thr kinase receptor associated protein | - | |
| TRIM2 | - | |
| TRIM3 | X | 1 |
| B. | ||
|---|---|---|
| Protein name | JAKMIP1 binder | MudPIT |
| β-5 Tubulin | - | |
| α-Tubulin | X | 2 |
| Tau | - | |
| hStaufen isoform 2 | - | |
| ACTB | - | |
| Myosin heavy chain | X | 2 |
| RNA-dependent RNA | - | |
| helicase A | ||
| Nucleolin | X | 1 |
| hnRNPU | X | 2 |
| PABP1 | X | 2 |
| α-Internexin | X | 1 |
| Dynein intermediate chain | X | 1 |
| Kinesin | X | 1 |
| p-Associated protein kinase II | - | |
| Ras GAP | - | |
| Rac1 | - | |
| Cdc42 | - | |
| IQGAP1 | - | |
| FMRP | Co-IP evidence | |
| RPLP0 | X | 2 |
| RPS4 | X | 2 |
| RPS6 | X | 1 |
| RPL6 | X | 2 |
| RPL28 | - | |
Most conservative granule members; Proteins listed under ‘Protein name’ are part of the RNP complex; X under JAKMIP1 binder denotes a MudPIT-identified JAKMIP1 interactor; Numbers listed under MudPIT are the number of MudPIT runs that identified the protein.
DDX5 is a member of the FMRP-kinesin transport RNP granule and we confirmed its JAKMIP1 binding (Figure 1D and Figure S1A). To further explore this interaction, we performed Co-IP experiments and found that DDX5 co-immunoprecipitates with JAKMIP1, PABPC1, and FMRP in vitro and in vivo (Figure S3A–C). We observed that binding of DDX5 to JAKMIP1 and FMRP depends on single, but not double stranded, RNA (Figure S3C), consistent with its participation in a RNA-protein complex. Based on these data, we hypothesized that JAKMIP1 is a likely member of PABCP1-containing RNP granules. To test this directly, we overexpressed human JAKMIP1 and PABPC1 in differentiated mouse neural progenitor cells lacking Jakmip1 and assessed localization by immunocytochemistry. We observed that JAKMIP1 co-localizes with PABPC1-positive RNP granules, providing evidence for its membership in these complexes (Figure 2C).
JAKMIP1 binds to FMRP mRNA targets and modulates neuronal translation
These MudPIT, Co-IP, in vitro and in vivo data provide multiple lines of evidence for JAKMIP1’s interaction with mRNPs, monosomes, and polyribosomes, its binding to protein-RNA granules containing FMRP, and JAKMIP1’s role in the composition of polyribosomes. Given these data, we hypothesized that JAKMIP1 plays a role in neuronal translation. To test this, we measured translation in neurons from Jakmip1 KO versus WT mice using Fluorescent Non-Canonical Amino Acid Tagging (FUNCAT) (Dieterich et al., 2010), where newly synthesized proteins are measured by incorporation of a fluorescently labeled methionine analog (Figure S4). We observed a significant reduction in nascent whole cell, soma and neurite translation in Jakmip1 KO mouse neurons compared to matched WT neurons (Figure 2D and 2E). These data demonstrate that JAKMIP1 is not only present in RNP granules and polyribosomes, but that it regulates neuronal translation.
We next considered whether JAKMIP1 might interact with FMRP by performing Co-IP of either JAKMIP1 or FMRP. We observed that JAKMIP1 interacts with FMRP in postnatal neocortex from mouse (Figure 3A, top panels). Furthermore, the maintenance of this association in the presence of RNAse demonstrates that the JAKMIP1-FMRP interaction is independent of double or single stranded RNA (Figure 3A, middle panels).
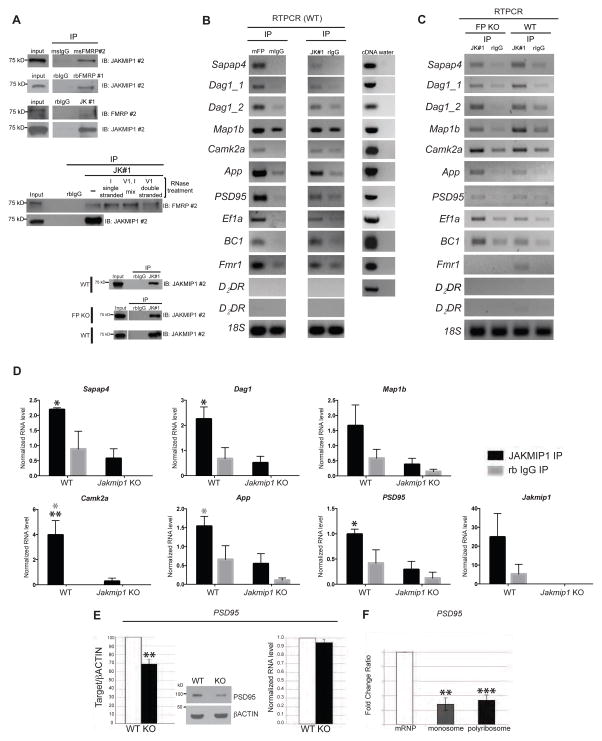
Figure 3. JAKMIP1 immunoprecipitates with FMRP and associated mRNA and regulates PSD95 expression.
(A) Top panels: JAKMIP1 and FMRP Co-IP in mouse postnatal neocortex. FMRP #1 is rabbit anti FMRP (Santa Cruz Biotechnology, Dallas, TX), and FMRP #2 is mouse anti FMRP (Millipore, Billerica, MA). Middle panels: JAKMIP1’s association with FMRP does not depend on RNA. JAKMIP1 IP reactions were conducted using mouse postnatal brain in the presence of RNases and immunoblotted for FMRP (top panel) and JAKMIP1 (bottom panel). Lanes labelled ‘single stranded,’ ‘mix,’ ‘double stranded’, and ‘(−)’ are IPs conducted in the presence of RNase I, RNase I and V1, RNaseV1, or without the RNases, respectively. Bottom panels: confirmation of JAKMIP1 IP for IPs conducted in (B, top) and (C, bottom). (B) JAKMIP1 immunoprecipitates with FMRP mRNA targets in mouse postnatal neocortex. Left column is RNA immunoprecipitating with FMRP (mFP, FMRP #2, positive control) or mouse IgG (mIgG, negative control). Middle column is RNA immunoprecipitating with JAKMIP1 or rabbit IgG (rIgG, negative control). The right column demonstrates that all mRNA targets are present in postnatal cortex. D2DR, not regulated by FMRP, serves as a negative control. 18S serves as a loading control. (C) JAKMIP1 immunoprecipitates with FMRP mRNA targets in the absence of FMRP in mouse postnatal neocortex. From left to right: RNA immunoprecipitating with JAKMIP1 (lane 1) or rabbit IgG (lane 2) in Fmr1 KO (FP KO) mouse neocortex, and JAKMIP1 (lane 3) or rabbit IgG (lane 4) in WT mouse neocortex. RT-PCR targets are analyzed at 30 or 35 cycles. D2DR signal is shown at both cycles in (B) and (C). (D) qRT-PCR of FMRP mRNA targets extracted from postnatal brain protein (WT, n = 3: Jakmip1 KO, n = 3) by JAKMIP1 (JAKMIP1 #1) or rabbit IgG immunoprecipitation. D2DR RNA was not present in immunoprecipitation reactions (not shown). Statistical significance was determined using 2way repeated measures ANOVA and Sidak multiple comparisons test. *p <= 0.05, (across genotype), ** p <= 0.01 (across genotype), *p <= 0.05 (within genotype, gray). (E) Reduction of PSD95 protein (left), but not RNA (right) in the identical cortex of postnatal Jakmip1 KO mice shown as a percentage from WT littermate (protein) or fold change (RNA). WT, N=3; KO, N=4, as one litter contains two KOs. Statistical significance is determined using a two-tailed, one sample t-test (p=0.012). A representative western blot is shown in the middle panel. (F) PSD95 mRNA is reduced in the monosome and polyribosome fractions of Jakmip1 KO mice (N= 4; Monosomes, p=0.002; Polyribosome, p=0.001; one-tailed unpaired t-tests. For D-F, data are shown as mean +/− SEM. See also Figure S5 and Figure S6.
To determine whether JAKMIP1 also binds mRNA, we conducted RNA immunoprecipitation (RIP) experiments by immunoprecipitating JAKMIP1 from WT postnatal neocortex followed by RTPCR (Figure 3B and C) and from WT and Jakmip1 KO postnatal brain (negative control) followed by quantitative RTPCR (Figure 3D, Figure S5A) for FMRP targets (De Rubeis and Bagni, 2011; Dictenberg et al., 2008; Santoro et al., 2012). FMRP immunoprecipitation was run alongside as a positive control (Figure S5B). We observed that JAKMIP1 bound Sapap4, Dag1, Camk2a, App, and PSD95 mRNAs (Figure 3B–D). Since JAKMIP1 and FMRP interact, we next tested whether their RNA binding was dependent on one another by performing JAKMIP1 RIP in Fmr1 KO mouse neocortex and FMRP RIP from Jakmip1 KO mouse neocortex. We observed no change in RNA binding in either condition (Figure 3C; Figure S5C and D), showing that JAKMIP1 and FMRP binding of mouse synaptic protein mRNAs is independent of each other.
Given our finding that JAKMIP1 binds to multiple FMRP mRNA targets and regulates protein translation, we next asked if JAKMIP1 modulates the translation of these specific targets in vivo (Figure 3E and Figure S6A–C). We observed that PSD95, a major component of the post synaptic density, showed a statistically significant decrease in protein, but not RNA expression, with Jakmip1 loss (Figure 3E), consistent with a role for JAKMIP1 in regulating PSD95 translation. Protein or mRNA for APP, CAMK2A, SAPAP4 and DAG1 did not change (Figure S6A–C). Comparison of the distribution of PSD95 mRNA in the Jakmip1 KO versus WT mice revealed a reduction of PSD95 mRNA in the monosome and polyribosome fractions from Jakmip1 KO mice (Figure 3F). These data demonstrate that JAKMIP1 regulates the translation of PSD95, a major postsynaptic density (PSD) component (Kim and Sheng, 2004).
JAKMIP1 KO mice display ASD associated behaviors
Recent work, including knockout of the eukaryotic translation initiation factor 4E-binding protein 2 (4E-BP2) (Gkogkas et al., 2013) and overexpression of the eukaryotic translation initiation factor 4E (eIF4E) (Santini et al., 2013), suggests that disrupting mRNA translation can lead to ASD-like behaviors in the mouse. We developed a line of knockout mice (Figure S2) and characterized multiple ASD-related behavioral domains including repetitive behavior, social abnormalities, and altered vocal communication, as well as other associated phenotypes including increased impulsivity and motor abnormalities in these Jakmip1 KO mice.
Jakmip1 KO mice showed striking motor stereotypies during home cage behavior including significantly increased grooming and repetitive jumping behavior with over 90% penetrance (Movie S1; Figure 4A and 4B). Jakmip1 KO mice also show increased perseveration in the T maze, displaying significantly fewer alterations than WT mice (Figure 4C). These behaviors are considered mouse analogues of restrictive and repetitive behaviors observed in ASD (Silverman et al., 2010). Probing social behavior with the three chamber task also revealed social dysfunction in Jakmip1 KO mice (Figure 4D and 4E). This social deficit was not due to disrupted olfaction, as Jakmip1 KO mice showed normal olfaction in the buried food test (Figure S7A).
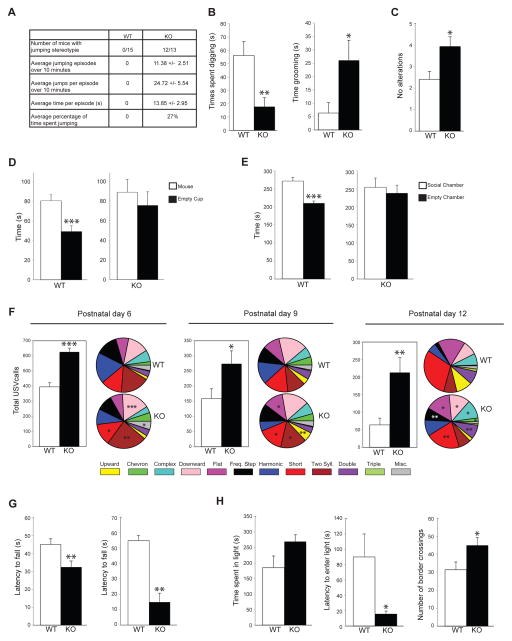
Figure 4. Jakmip1 loss leads to ASD associated behaviors.
(A and B) Jakmip1 KO mice show repetitive and perseverative behavior in the Home Cage Behavior Test. (A) Characteristics of repetitive jumping stereotypy in Jakmip1 KO mice. Rightmost column displays mean +/− SEM (B) Time spent digging (left) or grooming (right) within a 10 minute period. P values were calculated using a two-tailed unpaired t-test (Time spent digging, p=0.0063, Time spent grooming, p=0.025, WT, N=15; KO, N=13). (C) T maze spontaneous alternation test. Number of alterations are shown. P value was calculated using a two-tailed unpaired t-test (WT, N=15; KO, N=13, p=0.015). (D and E) Jakmip1 KO mice show impaired social behavior in the three chamber social test. (D) Time spent sniffing a sex-matched novel mouse or an empty cup over a 10 minute period. P value calculated using a two-tailed paired t-test (WT, N=15; KO, N=13: WT, p=0.00051; KO, p=0.51). (E) Time spent in the social chamber containing a novel mouse or in the chamber containing an empty cup. P value calculated using a two-tailed paired t-test (WT, N=15; KO, N=13: WT, p=0.00064; KO, p=0.73). (F) Number of USV calls emitted from postnatal day 6, 9, and 12 pups after being separated from their mother (5 minute period). Pie charts to the right of graphs show call type distribution. Statistical significance is denoted on the KO pie charts with all significant call types increased in KO mice [(P6, WT, N=15; KO, N=15: p=1.2 E-6), (P9, WT, N=17; KO, N=17: p=0.038), (P12, WT, N=15; KO, N=15: p=0.0034)]. P values were calculated using a two-tailed unpaired t-test. (G) Jakmip1 KO mice show impaired motor coordination and decreased strength. Left: Accelerating rotarod. Y axis is latency to fall from the rotarod. Maximum time of trial is 180 seconds. P value calculated using a two-tailed unpaired t-test (WT, N=15; KO, N=13: p=0.011). Right: Wire Hang Test. Y axis is latency to fall from an inverted wire cage lid. Maximum time of trial is 60 seconds. P value calculated using the non-parametric Mann-Whitney test (WT, N=9; KO, N=7: p=0.002). (H) Jakmip1 KO mice show increased impulsivity/reduced anxiety in the light-dark box test. P values were calculated using a two-tailed unpaired t-test. Left: Time spent in the bright compartment over a 10 minute period (WT, N=15; KO, N=13: p=0.075). Middle: Time before the mouse first enters the bright compartment (WT, N=15; KO, N=13: p=0.029). Right: Number of times the mouse crosses compartments over a 10 minute period (WT, N=15; KO, N=13: p=0.034). Data are shown as mean +/− SEM. See also Figure S7.
To determine if Jakmip1 KO mice display disruptions in vocal communication, we tested ultrasonic vocalizations in postnatal WT and KO mice upon separation from their mother. These distress calls are thought to model early vocalization abnormalities in ASD infants and are observed in several mouse models of ASD, including Fmr1 KO mice (Lai et al., 2014; Nakatani et al., 2009; Penagarikano et al., 2011; Scattoni et al., 2011). We found that Jakmip1 KO mice showed a significant increase in vocalizations and various call types across all time points (Figure 4F), not related to weight (Figure S7B). Jakmip1 KO mice also displayed significant deficits in motor function and impulsivity, which, although not core diagnostic features of ASD, are frequently observed in patients, including those with Fragile X syndrome (De Rubeis and Bagni, 2011; Moon et al., 2006).
Jakmip1 KO mice performed poorly on both the accelerating rotarod and wire hang test, indicating abnormal motor function (Figure 4G). In the light/dark box test for anxiety, Jakmip1 KO mice showed a trend toward spending more time in the light compartment compared to the dark compartment, a significant reduction of latency to enter the light compartment, as well as increased border crossings (Figure 4H), suggestive of either reduced anxiety or increased impulsivity (Zaichenko et al., 2011).
To test for learning impairments, we conducted auditory fear conditioning, which measures hippocampal and amygdala dependent learning (Kim and Fanselow, 1992; Phillips and LeDoux, 1992). Jakmip1 KO mice were able to learn to freeze to a tone after tone-shock pairings during the acquisition phase of fear conditioning, but showed significant decreases in freezing during the second and third intertone intervals as compared to WT mice (Figure S7C, top left) not due to impaired sensitivity to shock stimuli (Figure S7C, top right). Jakmip1 KO mice showed normal context dependent fear conditioning, indicative of preserved hippocampal dependent learning (Figure S7C, bottom left, context A). They also performed normally in a test of generalized fear assessment when placed in a new context (Figure S7C, bottom left, context B). However, Jakmip1 KO mice displayed decreased noise-cued fear response (Figure S7C, bottom right), suggesting disrupted amygdala/auditory pathway function (Phillips and LeDoux, 1992).
Marble burying is a naturalistic repetitive behavior that is often reduced in WT C57BL/6 mice treated with anxiolytics or selective serotonin reuptake inhibitors (Kedia and Chattarji, 2014; Njung'e and Handley, 1991). Jakmip1 KO mice buried fewer marbles than WT mice (Figure S7D), consistent with the increased impulsivity/decreased anxiety suggested by their performance in the light dark box. They also showed decreased digging, another form of naturalistic repetitive behavior (Figure 4B). Jakmip1 KO mice performed indistinguishably from WT mice on tests of sensory acuity (Figure S7E), nesting (Figure S7F), and showed normal open field activity (Figure S7G).
JAKMIP1 regulates NMDAR associated glutamatergic signaling in the postnatal striatum
One of the most salient behaviors displayed by the Jakmip1 KO mouse is restrictive and repetitive jumping. Previous work indicates that this behavior is mediated by the striatal glutamatergic system (Presti et al., 2004). Additionally, structural imaging of ASD mouse models implicates basal ganglia and cortex circuits in repetitive behavior (Ellegood et al., 2015). To first assess gross morphology in implicated brain structures, we conducted live scan MRI imaging of 2 and 10 month old Jakmip1 KO and WT male mice. We found a significant reduction in caudate and putamenal volume, along with reductions in cortex and cerebellum volume. Absence of a strain by age effect in any of the structures analyzed indicates that this is most likely developmental microcephaly rather than neurodegeneration (Table 3; Table 3 Supplemental Images).
Table 3. Total Volume, Cortex, Caudate Putamen, and Cerebellar volume are decreased in Jakmip1 KO mice.
| A. TotalVol, total volume; CaudoPut, caudate putamen; HippF, hippocampal formation; Dien, diencephalon; Vent, ventricles; Cereb, cerebellum; OlfBulb,olfactory bulb.
| ||||||
|---|---|---|---|---|---|---|
| Trait | mean | age effect | strain effect | strain x age | Rsq | Fstat |
| TotalVol | −0.22 | −0.03 (0.49) | 1.6 (0.0012) | −0.097 (0.14) | 0.553 | 10.08 (0.00034) |
| Cortex | −0.3 | −0.018 (0.72) | 1.7 (0.0029) | −0.12 (0.13) | 0.462 | 7.29 (0.0019) |
| CaudoPut | 0.048 | −0.11 (0.0031) | 1.3 (0.00037) −0.0089 (0.85) | 0.785 | 27.84 (3.7e–07) | |
| HippF | 0.24 | −0.046 (0.43) | 0.26 (0.65) | 0.037 (0.66) | −0.001 | 0.99 (0.42) |
| Dien | −0.17 | −0.016 (0.8) | 0.98 (0.13) | −0.046 (0.62) | 0.097 | 1.79 (0.18) |
| Vent | 0.14 | −0.029 (0.69) | 0.27 (0.71) | −0.024 (0.83) | −0.11 | 0.27 (0.85) |
| Cereb | −0.15 | −0.047 (0.28) | 1.5 (0.0025) | −0.054 (0.4) | 0.566 | 10.57 (0.00026) |
| OlfBulb | −0.38 | 0.1 (0.17) | 0.24 (0.75) | −0.12 (0.27) | 0.015 | 1.11 (0.37) |
| B. TotalVol, total volume; CaudoPut, caudate putamen; HippF, hippocampal formation; Dien, diencephalon; Vent, ventricles; Cereb, cerebellum; OlfBulb, olfactory bulb.
| ||||||
|---|---|---|---|---|---|---|
| Trait | WT_mu | KO_mu | effect size | conf int | t | p |
| TotalVol | 1.12 | −0.28 | −1.4 | −2.17 to −0.62 | −4.04 | 2.5e-03 |
| Cortex | 1.11 | −0.34 | −1.44 | −2.36 to −0.52 | −3.58 | 6.6e-03 |
| CaudoPut | 1.16 | −0.17 | −1.32 | −1.82 to −0.82 | −5.97 | 1.8e-04 |
| HippF | 0.48 | 0.15 | −0.33 | −1.31 to 0.64 | −0.75 | 4.7e-01 |
| Dien | 0.68 | −0.2 | −0.88 | −2.06 to 0.29 | −1.68 | 1.2e-01 |
| Vent | 0.31 | 0.08 | −0.22 | −1.22 to 0.78 | −0.49 | 6.4e-01 |
| Cereb | 1.1 | −0.24 | −1.34 | −2.09 to −0.59 | −4.16 | 3.4e-03 |
| OlfBulb | −0.17 | −0.17 | 0.01 | −0.73 to 0.74 | 0.02 | 9.8e-01 |
Summary table of ANCOVA results. Estimates from the linear regression are shown in each row/column, and the p-values for each estimate are shown in parentheses.
Summary table of t-test results for 2 month-olds. The mean for each trait and strain is shown in the second and third column. Trait values have been standardized to mean=0, SD=1 prior to testing. The column labeled conf int shows the 95 percent confidence interval for the difference in means.
These findings, coupled with Jakmip1’s high levels of expression in the striatum both during brain development (Figure S2F) and adulthood (Allen Mouse Brain Atlas and the Allen Human Brain Atlas, http://mouse.brain-map.org and http://brainspan.org), led us to hypothesize that JAKMIP1’s loss would disrupt striatal function via reduction in target PSD protein mRNAs. We first tested JAKMIP1’s ability to bind to high confidence FMRP RNA targets known to be involved in glutamatergic signaling in the striatum. We conducted RNA immunoprecipitation (RIP) experiments by immunoprecipitating JAKMIP1 from postnatal WT and Jakmip1 KO striatum (negative control) and observed that JAKMIP1 bound Grin2a, Grin2b, and Shank2, known FMRP RNA targets (Figure 5A). Taf13 mRNA (negative control), a gene expressed in postnatal neurons and not identified as an FMRP target (Darnell et al., 2011) was not present in immunoprecipitation reactions (not shown).
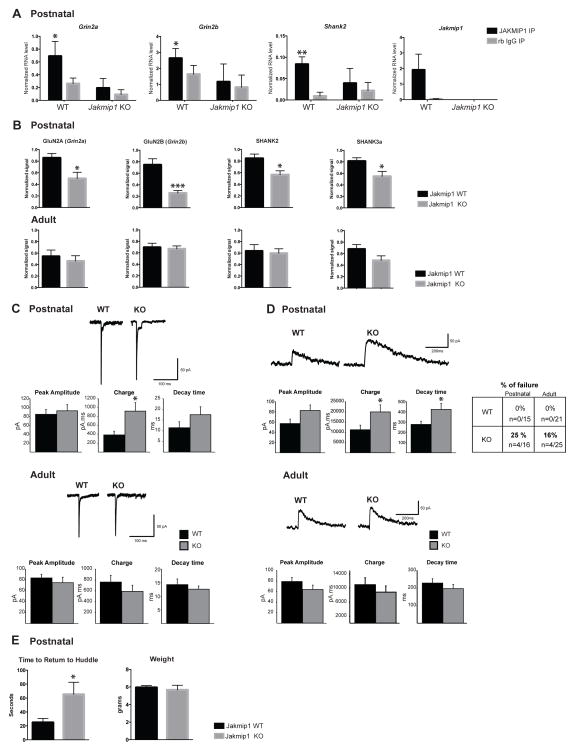
Figure 5. JAKMIP1 Regulates Glutamatergic Signaling in the Postnatal Striatum through an NMDA Receptor Complex.
(A) JAKMIP1 immunoprecipitates with FMRP mRNA targets involved in glutamatergic signaling and ASD in mouse postnatal striatum. qRT-PCR of FMRP mRNA targets extracted from postnatal striatum (WT, N=4: Jakmip1 KO, N=4: all male) by JAKMIP1 (JAKMIP1 #1) or rabbit IgG immunoprecipitation. Taf13 RNA (negative control) was not present in immunoprecipitation reactions (not shown). Statistical significance was determined using 2way repeated measures ANOVA and Sidak multiple comparisons test. *p <= 0.05, *p <= 0.001 (within genotype). (B) Top panels: GluN2A (Grin2a, P=0.019), GluN2B (Grin2b, P=0.0012), SHANK2 (P=0.018), and SHANK3a (P=0.029), are reduced in postnatal striatal synapses of Jakmip1 KO mice (N=7) compared to WT control (N=7). Bottom panels: GluN2A (Grin2a), GluN2B (Grin2b), SHANK2, and SHANK3a are not changed in adult striatal synapses of Jakmip1 KO mice (N=7) compared to WT control (N=7). Statistical significance was determined using two tailed unpaired t-tests. Signal was normalized to corresponding βactin loading control and then to 1 within blot. (C) AMPAR-mediated synaptic responses. Representative traces showing evoked inward currents in MSNs from postnatal and adult Jakmip1 KO and WT mice (Postnatal, WT N=15, KO N=16; adult, WT N=21, KO N=25). Graphs show mean +/− SEM of peak amplitude, charge and decay time. Charge in MSNs from postnatal animals is increased compared to WT (t-test, p<0.05). (D) NMDAR- mediated synaptic responses. Left: Representative traces showing outward evoked currents in cells from Jakmip1 KO and WT mice in postnatal (WT N=15, KO N=12) and adult MSNs (WT N=21, KO N=21) with graphs showing mean ± SEM of peak amplitude, charge and decay time. The charge and decay time are significantly increased compared to WT responses (t-test, p<0.05) in postnatal Jakmip1 KO MSNs. In the adult, no significant changes were observed (WT N=21, KO N=21). Table to the right displays the percentage of cells that showed an AMPAR- but not NMDAR-mediated response. (E) Left: Jakmip1 KO mice take a significantly longer time to return to social huddle compared to WT controls after separation from the huddle litter (WT, N=16; KO, N=12, p=0.02). Right: Weights of mice used in the return to huddle test (WT, N=15; KO, N=12). P value calculated using a two-tailed unpaired t-test. All data are shown as mean ± SEM.
We next asked if the corresponding proteins were decreased in striatal synaptosomal membranes of Jakmip1 KO mice. We found that GluN2A (Grin2a), GluN2B (Grin2b), SHANK2, and SHANK3a, all members of an NMDAR scaffolding complex, were reduced in striatal synapses of Jakmip1 KO mice compared to controls during postnatal development, but not adulthood (Figure 5B). We found no change in AMPA receptor subunits throughout development (data not shown). Of note, GluN2A, SHANK2, and SHANK3a, but not GluN2B, were also reduced in synaptosomal membranes from postnatal neocortex of Jakmip1 KO mice compared to controls, consistent with regional differences (Figure S6D).
We hypothesized that decreased expression of the NMDAR complex in Jakmip1 KO mice would result in abnormal NMDAR- but not AMPAR-mediated signaling. To test this, we conducted electrophysiological recordings of early postnatal and adult dorsolateral medium-sized spiny neurons (MSNs) from Jakmip1 KO and WT mice. At a holding potential of −70 mV, electrical stimulation evoked an AMPAR-mediated inward current with a fast-rising component followed by a slowly-desensitizing component, which is characteristic of juvenile AMPAR currents in the striatum. The amplitude (WT 84.8 ± 10.8 pA, KO 92.7 ± 13.2 pA) and the decay time (WT 11.4 ± 2.8 ms, KO 17.8 ± 3.7 ms) of the AMPAR-mediated responses were similar between groups. However, the charge was significantly increased in Jakmip1 KO MSNs compared to WT MSNs (WT 385.5 ± 83 pA.ms, KO 902.2 ± 191.6 pA.ms, t-test p<0.05) due to an increased duration of the slowly-desensitizing component. In adults, no differences in amplitude, charge and decay time were observed between WT and Jakmip1 KO AMPAR-mediated responses (Figure 5C).
A proportion of Jakmip1 KO MSNs (postnatal 25%, adult 16%) showing an AMPAR-mediated response failed to display an NMDAR-mediated response, whereas all WT MSNs responded (Figure 5D, right upper panel). The population of responding postnatal Jakmip1 KO MSNs showed a greater NMDAR-mediated response than WT MSNs. The charge (WT 11263.5 ± 2213.9 pA.ms, KO 19945.6 ± 3466.9 pA.ms, t-test p<0.05) and the decay time (WT 277.1 ± 30 ms, KO 423.6 ± 58 ms, t-test p<0.05) were significantly increased. In adults, no changes in amplitude, charge and decay time were observed between WT and KO NMDAR-mediated responses in MSN consistent with no change in NMDAR subunit protein expression in adults (Figure 5D, left lower panels).
Finally, we asked if Jakmip1 KO mice display an increased time to return to huddle, a postnatal social behavior observed in mice with a GluN2B to 2A substitution (Wang et al., 2011), which would be consistent with the physiological findings related to GluN2B dysfunction. We found that Jakmip1 KO mice take longer to return to huddle during the postnatal period than WT controls (Figure 5E). Taken together, these results show that loss of JAKMIP1 disrupts striatal NMDAR signaling, contributing to the disruption of their behavior.
DISCUSSION
A critical goal of research in etiologically complex neurodevelopmental disorders such as ASD is to identify potentially convergent molecular pathways (Parikshak et al., 2013; Voineagu et al., 2011). A salient example of such convergence is at the protein mTOR, which although not known to be mutated in ASD, functions as a key modulator of multiple ASD risk genes that regulate mRNA translation pathways (Sawicka and Zukin, 2012). Based on our original findings that JAKMIP1 expression is regulated downstream of three ASD risk loci, FMRP, (dup)15q11-13 and RBFOX1(Fogel et al., 2012; Nishimura et al., 2007), we hypothesized that JAKMIP1, a highly conserved neuronal mRNA binding protein, would have a role in regulating mRNAs important for synaptic function and ASD pathophysiology. We show for the first time that JAKMIP1 regulates neuronal translation and binds FMRP protein, directly or indirectly, as well as many known FMRP protein interactors and its mRNA binding targets. JAKMIP1’s polyribosome profile suggests that it is expressed in polyribosomes, but also in mRNPs, not unlike FMRP (Zalfa et al., 2003). Loss of JAKMIP1 in vivo disrupts the polyribosome expression profile of PABPC1 and DDX5, the former of which is required for translation initiation, suggesting that JAKMIP1 may be important in stabilizing FMRP-containing polyribosomes. The recognition here for the first time that JAKMIP1 binds to mRNAs that are important for synaptic function and members of an FMRP-related protein complex known to regulate translation during the peak period of cortical synaptogenesis provide a strong motivation for future detailed investigation of its biochemical mechanisms.
JAKMIP1 also regulates the expression of a subset of FMRP mRNA targets that are implicated in ASD (PSD95, Grin2a, Grin2b, Shank2, and Shank3) at postsynaptic membranes (Fernández et al., 2013; Santoro et al., 2012). Mutations in both SHANKs and GRIN2B have been shown to contribute to ASD (De Rubeis et al., 2014; Iossifov et al., 2014; Jiang and Ehlers, 2013). Interestingly, our data suggests that FMRP and JAKMIP1 can bind their mRNA targets independently of one another, implying the two proteins might work in parallel. Remarkably, mouse models in which either SHANK2 or SHANK3 expression is reduced show behavioral or physiologic abnormalities parallel to those observed in the Jakmip1 KO mouse including repetitive behaviors, decreased social interaction and similar changes in glutamatergic signaling (Peca et al., 2011; Won et al., 2012).
JAKMIP1’s regulation of the NMDAR receptor subunits GluN2A and GluN2B is another area of potential convergence. Recent exome-sequencing studies identify Grin2b as among two dozen genes with the most compelling genetic evidence for involvement in ASD (De Rubeis et al., 2014; Iossifov et al., 2014; O'Roak et al., 2012). We found that JAKMIP1 binds both Grin2a and Grin2b mRNA in striatum and that loss of JAKMIP1 decreases the levels of their protein products at striatal synapses. A percentage of MSNs from the postnatal and adult striatum of Jakmip1 KO mice showed no NMDAR evoked response, but had preserved AMPAR evoked response, corroborating the biochemical findings. The greater NMDA receptor current shown by the responding Jakmip1 KO MSNs may be due to compensatory mechanisms, including receptor subunit changes, transporter modification, and/or glutamate spill over. Consistent with lowered GluN2B levels, we observed an increase in return to huddle time of Jakmip1 KO mice during postnatal development, a social deficit also observed in mice with a GluN2B to 2A substitution (Wang et al., 2011). That JAKMIP1 itself has limited genetic evidence for contribution to ASD genetic risk, but some of its mRNA targets have significant genetic support for their role in ASD biology is reminiscent of mTOR, which regulates multiple cellular functions including protein synthesis, and whose pathways family members, but not mTOR itself, have strong genetic evidence for contributing to ASD risk (Lipton and Sahin, 2014; Sawicka and Zukin, 2012).
In this regard, our observations connecting disruption of neuronal protein translation by JAKMIP1 knockout with ASD-like behaviors is worth highlighting (Kelleher and Bear, 2008). Jakmip1 KO mice show repetitive jumping, increasing grooming and reduced alternation in the T-maze, as well as social deficits, the two core features in ASD. Other allied, non-core ASD deficits, such as motor dysfunction and changes in USV were also observed, but hyperactivity was not. Consistent with this last observation, nest building, which is related to dopaminergic function and can be associated with changes in motor activity in mice (Deacon, 2006a; Szczypka et al., 2001) was also normal. Testing of hippocampal memory by contextual fear conditioning revealed no gross deficit in context dependent fear conditioning, indicative of intact spatial memory. However, Jakmip1 KO mice displayed a diminished noise-cued fear response, which suggests a potential disruption of auditory-amygdala circuits (Phillips and LeDoux, 1992), which we interpret as consistent with their potentially reduced anxiety/increased impulsivity.
Marble burying is a naturalistic behavior in mice that has been related to obsessive compulsive and repetitive behaviors and anxiety in mice (Deacon, 2006b; Thomas et al., 2009). Decreased digging and decreased marble burying as observed here in conjunction with increased stereotypies has been observed in other mouse models including Shank2 and Synapsin knockouts (Greco et al., 2013; Won et al., 2012). The reduced digging and marble burying in parallel with increases in other forms of repetitive behaviors is likely due to a combination of factors, including decreased anxiety (Deacon, 2006b; Njung'e and Handley, 1991) which is consistent with their light-dark box behavior. Alternatively, the decreased digging and marble burying may be caused by the increased time spent in other stereotyped activities (Greco et al., 2013). The reduction in digging and marble burying in the presence of increases in several other repetitive behaviors is consistent with the presence of independent forms of repetitive behaviors in ASD patients (Bodfish, 2011). Although work over several decades has clearly implicated striatal-cortical pathways in repetitive behaviors (Langen et al., 2011), specific circuits underlying the many different forms of repetitive behavior are not yet well elucidated in mice. Thus, the Jakmip1 KO model and others with similar phenotypes provide important opportunities to understand how specific pathways may lead to different forms of ASD-related restrictive and repetitive behaviors.
Here we describe a biochemical interaction between JAKMIP1 and FMRP, which is likely indirect, involving association with members of a translational complex including FMRP, but not FMRP directly, as we only observed co-immunoprecipitation with lower salt concentrations. The precise nature of this interaction will require further investigation. Intriguingly, both proteins promote the expression of PSD95 protein at the synapse (Muddashetty et al., 2007; Todd et al., 2003; Zalfa et al., 2007) and, therefore, may act in parallel. Irrespective of JAKMIP1’s relationship with FMRP, we provide clear evidence that JAKMIP1 regulates protein synthesis in neurons during a specific postnatal developmental window coincident with peak synaptogenesis, regulates the protein expression of key targets involved in glutamatergic transmission at the synapse, and affects the membership of proteins and RNA in monosome and polyribosome fractions, which has significant behavioral consequences.
JAKMIP1’s involvement in translation during the peak of synaptogenesis is intriguing as translational regulation has been proposed as a potential convergent mechanism in ASD (Ebert and Greenberg, 2013; Kelleher and Bear, 2008; Waung and Huber, 2009), and the consequences of Jakmip1 deletion in mouse mirror ASD-like behaviors observed in other mouse models of ASD where specific components of the neural translational machinery are dysregulated in the CNS (Gkogkas et al., 2013; Santini et al., 2013). The mechanisms by which JAKMIP1 reduction leads to these behaviors and the relative contribution of its roles in mRNA transport versus translational regulation to behavior warrants further detailed investigation. Careful cellular anatomical examination to complement the gross analysis performed here with MRI will be important for a complete examination of this question. This work identifying JAKMIP1 as a new molecule regulating neural translation during the peak period of postnatal synaptogenesis opens up many new avenues of future research and further highlights the need to gain a better understanding of neural translational regulation during synaptic development as a potential convergent pathway in ASD.
EXPERIMENTAL PROCEDURES
Mass Spectrometry and Immunoprecipitation (IP)
IPs were conducted using Dynabeads according to manufacturer’s instructions with slight modifications (Invitrogen, Carlsbad, CA). Mass spectrometry analysis was conducted as previously described (Wohlschlegel, 2009). Further details regarding IPs, interactor selection criteria, and immunoblotting are listed in Supplemental Experimental Procedures.
Generation of a Jakmip1 knockout mouse
Jakmip1 knockout mice were generated in collaboration with the UC Davis KOMP Repository Knockout Mouse Project. Chimeras were produced from Velocigene embryonic stem cells (Jakmip1tm1(KOMP)Vlcg ) in which Jakmip1 coding exons 2–8 were replaced with a LacZ-Neo cassette by homologous recombination. Methods related to confirmation of JAKMIP1 loss are detailed in Supplemental Experimental Procedures.
Gene ontology analysis
GO analysis was conducted using Ingenuity Pathway Analysis software (http://www.ingenuity.com). MudPIT-identified JAKMIP1 protein interactors listed in Table 1 were analyzed after removing JAKMIP1 and two proteins not recognized by Ingenuity, 1810049H19Rik and Igkv1-117. Permutation analysis details are listed in Supplemental Experimental Procedures.
Polyribosome fractionation
Polyribosome fractionation was conducted as previously described (del Prete et al., 2007) with modifications described in Supplementary Experimental Procedures.
Nascent synthesis of proteins
FUNCAT analysis was performed on mouse neural progenitors according to manufacturer’s instructions (ClickIT, Invitrogen, Carlsbad, CA). Custom software using the R environment (http://www.r-project.org/) and the R packages rimage and pixmap was developed to quantify the amount of new translation (average 555 signal intensity) in the space defined by TUJ1 positive pixels. Images were subsequently manually curated to remove overlapping glial signal and to divide the neuron into cell body and neurite compartments (See Supplemental Experimental Procedures).
Electrophysiology
Electrophysiology experiments were conducted from coronal slices of postnatal and adult male mice. Whole-cell patch-clamp recordings in voltage clamp mode were obtained from neurons. For evoked synaptic currents, the stimulating electrode was placed in the corpus callosum 300μm from the recorded cell. For additional methods, see Supplemental Experimental Procedures.
Neuroimaging
Adult male mice were imaged in a 7T small bore scanner using Rapid Acquisition with Refocused Echoes (RARE) pulse sequence and analyzed for brain volumetric changes. See Supplemental Experimental Procedures for further details.
Behavioral Tests
Experiments were conducted using C57BL/6 Jakmip1 WT and KO mice generated from matings of heterozygous mice. Mice had ad-lib access to food and water and were housed in a 12 hour light/12 hour dark cycle. Mice were ear tagged or, in the case of pups, given a paw tattoo. Assessment of general health and gross neurologic function was conducted according to previously published methods (Crawley and Paylor, 1997). Experiments were carried out in accordance with UCLA animal research committee. Home cage behavior was conducted by placing mice in juxtaposed cages separated by opaque panels that contained fresh bedding. Behavior was recorded by the automated system, Top Scan (Clever Sys, Inc., Reston, VA) for 20 minutes. The last 10 minutes of the recording was scored for repetitive hindlimb jumping, digging, and grooming. The T maze spontaneous alternation test was performed as previously described (Penagarikano et al., 2011). The Three-Chamber Social Interaction Test was conducted as previously described (Silverman et al., 2010). The Rotarod Test was conducted by placing mice on a rod that rotated for 5 rpm and then 20 rpm (constant test) or 5 rpm and then 60 rpm (accelerating test) for a maximum time of 180 seconds. Latency to fall was measured. In the Wire Hang Test, mice were scored for their latency to fall from a swiftly inverted wire lid. The Light Dark Exploration Test was conducted as previously described (Penagarikano et al., 2011). Ultrasonic Vocalizations (USVs) were recorded from pups separated from their mother at three postnatal time points. USVs were processed and characterized according to previous methods (Burkett et al., 2015; Scattoni et al., 2011)). Return to Huddle was conducted as previously described (Wang et al., 2011) with modifications. The Hot plate Startle Test was conducted by placing mice on a 55 degrees Celsius hot plate and time was recorded from time of placement to the first sign of pain, which included licking or kicking of the paws. Auditory Fear Conditioning was performed as previously described (Kim and Fanselow, 1992) with modifications. The Nesting Behavior Test was performed by exposing mice to a tightly packed unit of nest material overnight. Nests were scored according to previously published criteria (Deacon, 2006a). The Marble Burying Test was conducted as previously described (Deacon, 2006b) with modifications. The Open Field Test was conducted as previously described (Penagarikano et al., 2011). Olfaction was measured by testing latency to find a buried food item after 24 hour food deprivation according to previously published methods (Yang and Crawley, 2009). Please see Supplemental Experimental Procedures for additional details.
Statistical analysis
Significance was determined using a Student’s t-test, 2way repeated measures ANOVA with Sidak multiple comparisons test, a mixed effects model, or the non-parametric Mann-Whitney test where noted. For FUNCAT analysis, a mixed effects regression was used to statistically control for littermate set and technical variation across day of imaging. Significance was estimated using a mixed effects regression model with a fixed effect covariate by date and a random intercept for mouse set. Hypergeometric probability was calculated using R code 1-phyper (k-1, j, m-j, n) with the following definitions: ‘m’ is the universe of proteins defined as all brain expressed genes (15,132) (Kang et al., 2011), k is number of overlapping proteins, j is number of FMRP complex members, and ‘n’ is JAKMIP1 interactors. For additional details related to Hypergeometric calculations (Table 2), please see Supplemental Experimental Procedures. *p <= 0.05, ** p <= 0.01, *** p <= 0.001, and data are presented as mean +/− SEM.
Supplementary Material
Acknowledgments
We thank Dr. Sandra Pellegrini for J1269-286 rabbit polyclonal antibody, rabbit preimmune serum, and human JAKMIP1 expression plasmid; Dr. Joseph Dougherty for N2A BacTrap cell lines; Dr. Claudia Bagni for protocols related to IP/RT-PCR experiments; Dr. Larry Simpson for use of an Isco Fractionator; Dr. Andrew Poulos and Dr. Jesse Cushman and the UCLA behavioral testing core for help with mouse behavioral experiments; Dr. Eunjoon Kim for the anti SHANK2 rabbit polyclonal antibody; Hongmei Dong for technical assistance with C57BL/6 mice for developmental profiling; Grace Shin for technical assistance with genotyping; Michelle Bailhe for technical assistance with protein preparation and RNA extraction; Dr. Lars Ittner for pLVU/RED plasmid ID 24178; Elizabeth Fraley for consultation regarding USV tests; Lauren Kawaguchi for laboratory management. The vector, ES cell(s), and/or mouse strain used for this research project was generated by the trans-NIH Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository (https://www.komp.org). NIH grants to Velocigene at Regeneron Inc (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ES cells for 8500 genes in the KOMP Program, which are archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244). This work was supported by grants NIH/NIMH F31MH088083 (J.M.B.), NIH/NIMH T32MH073526 (fellow: J.M.B.; P.I.: D.H.G.), UCLA Chancellor’s Award (J.M.B.), Achievement Rewards for College Scientists (J.M.B.), NIH/NIMH K99MH102357 (J.L.S.), Autism Speaks Translational Postdoctoral Fellowship (J.L.S.), NIH/NIMH R01MH070712 (S.A.W.), NIH/NIMH K08MH086786 (S.C.F.), NIH/NICHD P30HD004612 (J. de Vellis), NIH/NIGMS R01GM089778 (J.A.W.), the Jonsson Cancer Center at UCLA (J.A.W.), Autism Center of Excellence Network grant NIH/NIMH R01MH081754 and 5R01MH100027 (D.H.G.), and CART/Autism Center of Excellence NIH/NICHD P50HD055784 (S. Bookheimer). The content described is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
J.M.B and D.H.G designed the experiments and conducted the data analysis; J.M.B., C.L., and D.H.G. wrote the manuscript; Most experiments were performed or overseen by J.M.B; C.L. performed polyribosome fractionation experiments, including corresponding immunoblotting and qRTPCR. L.C. performed IPs, immunoblotting, FUNCAT, qRTPCR, and immunocytochemistry; L.G., C.C, J.Y.C. and M.S.L. designed, conducted, and analyzed electrophyisological experiments; O.P. helped design mouse behavioral experiments and helped guide initial polyribosome fractionation experiments; J.L.S. developed the custom computer analysis program for FUNCAT experiments and performed statistical analysis for this experiment; A.L. helped perform mouse behavioral experiments and helped analyze the corresponding data; A.O. provided a subset of tissue used for developmental expression profiling and gave critical insights into earlier phases of the study; J.A.M. cross-referenced MudPIT interactors with HPRD and conducted statistical testing of PPI enrichment; A.A.V. and J.A.W. performed mass spectrometry; M.E.S. helped perform mouse behavioral experiments, conducted genotyping, and conducted some qRTPCR experiments; E.P.K. provided technical assistance including cell culture, IPs, immunocytochemistry, and helped with behavioral experiments; E.T. provided technical assistance with image curation, gross morphological analysis, and E.T and A.G helped conduct spine density experiments and analysis; N.B.A and S.C.F. conducted brain imaging; Z.D.B and S.A.W. provided USV equipment and helped with USV analysis. All authors provided comments on the manuscript.
The content described is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health.
Supplementary Information includes eight figures, one movie, supporting images for Table 3 and Supplementary Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autism Genome Project C. Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Geschwind DH. Autism genetics: searching for specificity and convergence. Genome Biol. 2012;13:247. doi: 10.1186/gb-2012-13-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodfish J. Repetitive Behaviors in Individuals with Autism Spectrum Disorders. New York, New York: Oxford University Press; 2011. [Google Scholar]
- Burkett ZD, Day NF, Peñagarikano O, Geschwind DH, White SA. VoICE: A semiautomated pipeline for standardizing vocal analysis across models. Scientific Reports. 2015;5 doi: 10.1038/srep10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couve A, Restituito S, Brandon JM, Charles KJ, Bawagan H, Freeman KB, Pangalos MN, Calver AR, Moss SJ. Marlin-1, a novel RNA-binding protein associates with GABA receptors. J Biol Chem. 2004;279:13934–13943. doi: 10.1074/jbc.M311737200. [DOI] [PubMed] [Google Scholar]
- Crawley JN, Paylor R. A proposed test battery and constellations of specific behavioral paradigms to investigate the behavioral phenotypes of transgenic and knockout mice. Hormones and behavior. 1997;31:197–211. doi: 10.1006/hbeh.1997.1382. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Bagni C. Regulation of molecular pathways in the Fragile X Syndrome: insights into Autism Spectrum Disorders. J Neurodev Disord. 2011;3:257–269. doi: 10.1007/s11689-011-9087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon RM. Assessing nest building in mice. Nature protocols. 2006a;1:1117–1119. doi: 10.1038/nprot.2006.170. [DOI] [PubMed] [Google Scholar]
- Deacon RM. Digging and marble burying in mice: simple methods for in vivo identification of biological impacts. Nature protocols. 2006b;1:122–124. doi: 10.1038/nprot.2006.20. [DOI] [PubMed] [Google Scholar]
- del Prete MJ, Vernal R, Dolznig H, Mullner EW, Garcia-Sanz JA. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA. 2007;13:414–421. doi: 10.1261/rna.79407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DSM-V. American Psychiatric Association: Diagnostic and statistical manual of mental disorders. 5. 2013. text rev. [Google Scholar]
- Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegood J, Anagnostou E, Babineau BA, Crawley JN, Lin L, Genestine M, DiCicco-Bloom E, Lai JK, Foster JA, Penagarikano O, et al. Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol Psychiatry. 2015;20:118–125. doi: 10.1038/mp.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez E, Rajan N, Bagni C. The FMRP regulon: from targets to disease convergence. Frontiers in neuroscience. 2013;7:191. doi: 10.3389/fnins.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Parikshak N, Konopka G, Geschwind DH. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21:4171–4186. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco B, Manago F, Tucci V, Kao HT, Valtorta F, Benfenati F. Autism-related behavioral abnormalities in synapsin knockout mice. Behavioural brain research. 2013;251:65–74. doi: 10.1016/j.bbr.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78:8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor L, et al. Proteomic, functional, and domain-based analysis of in vivo 14–3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedia S, Chattarji S. Marble burying as a test of the delayed anxiogenic effects of acute immobilisation stress in mice. Journal of neuroscience methods. 2014;233:150–154. doi: 10.1016/j.jneumeth.2014.06.012. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nature reviews Neuroscience. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Fanselow MS. Modality-specific retrograde amnesia of fear. Science. 1992;256:675–677. doi: 10.1126/science.1585183. [DOI] [PubMed] [Google Scholar]
- Lai JK, Sobala-Drozdowski M, Zhou L, Doering LC, Faure PA, Foster JA. Temporal and spectral differences in the ultrasonic vocalizations of fragile X knock out mice during postnatal development. Behavioural brain research. 2014;259:119–130. doi: 10.1016/j.bbr.2013.10.049. [DOI] [PubMed] [Google Scholar]
- Langen M, Durston S, Kas MJ, van Engeland H, Staal WG. The neurobiology of repetitive behavior: ...and men. Neuroscience and biobehavioral reviews. 2011;35:356–365. doi: 10.1016/j.neubiorev.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Beaudin AE, Verosky S, Driscoll LL, Weiskopf M, Levitsky DA, Crnic LS, Strupp BJ. Attentional dysfunction, impulsivity, and resistance to change in a mouse model of fragile X syndrome. Behavioral neuroscience. 2006;120:1367–1379. doi: 10.1037/0735-7044.120.6.1367. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, Tomonaga S, Watanabe Y, Chung YJ, Banerjee R, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11–13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Martin CL, Vazquez-Lopez A, Spence SJ, Alvarez-Retuerto AI, Sigman M, Steindler C, Pellegrini S, Schanen NC, Warren ST, et al. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum Mol Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- Njung'e K, Handley SL. Effects of 5-HT uptake inhibitors, agonists and antagonists on the burying of harmless objects by mice; a putative test for anxiolytic agents. British journal of pharmacology. 1991;104:105–112. doi: 10.1111/j.1476-5381.1991.tb12392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RG, LeDoux JE. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behavioral neuroscience. 1992;106:274–285. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- Poultney CS, Goldberg AP, Drapeau E, Kou Y, Harony-Nicolas H, Kajiwara Y, De Rubeis S, Durand S, Stevens C, Rehnstrom K, et al. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am J Hum Genet. 2013;93:607–619. doi: 10.1016/j.ajhg.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TS, Kandasamy K, Pandey A. Human Protein Reference Database and Human Proteinpedia as discovery tools for systems biology. Methods Mol Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- Presti MF, Watson CJ, Kennedy RT, Yang M, Lewis MH. Behavior-related alterations of striatal neurochemistry in a mouse model of stereotyped movement disorder. Pharmacology, biochemistry, and behavior. 2004;77:501–507. doi: 10.1016/j.pbb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Sawicka K, Zukin RS. Dysregulation of mTOR signaling in neuropsychiatric disorders: therapeutic implications. Neuropsychopharmacology. 2012;37:305–306. doi: 10.1038/npp.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav. 2011;10:44–56. doi: 10.1111/j.1601-183X.2010.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nature reviews Neuroscience. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg J, Webber C. The roles of FMRP-regulated genes in autism spectrum disorder: single- and multiple-hit genetic etiologies. Am J Hum Genet. 2013;93:825–839. doi: 10.1016/j.ajhg.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C, Li Z, Algarte M, Alcover A, Libri V, Ragimbeau J, Pellegrini S. Jamip1 (marlin-1) defines a family of proteins interacting with janus kinases and microtubules. J Biol Chem. 2004;279:43168–43177. doi: 10.1074/jbc.M401915200. [DOI] [PubMed] [Google Scholar]
- Szczypka MS, Kwok K, Brot MD, Marck BT, Matsumoto AM, Donahue BA, Palmiter RD. Dopamine production in the caudate putamen restores feeding in dopamine-deficient mice. Neuron. 2001;30:819–828. doi: 10.1016/s0896-6273(01)00319-1. [DOI] [PubMed] [Google Scholar]
- Thomas A, Burant A, Bui N, Graham D, Yuva-Paylor LA, Paylor R. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology. 2009;204:361–373. doi: 10.1007/s00213-009-1466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci U S A. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzetis M, Kitsiou-Tzeli S, Frysira H, Xaidara A, Kanavakis E. The clinical utility of molecular karyotyping using high-resolution array-comparative genomic hybridization. Expert review of molecular diagnostics. 2012;12:449–457. doi: 10.1586/erm.12.40. [DOI] [PubMed] [Google Scholar]
- Vidal RL, Valenzuela JI, Lujan R, Couve A. Cellular and subcellular localization of Marlin-1 in the brain. BMC Neurosci. 2009;10:37. doi: 10.1186/1471-2202-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villace P, Marion RM, Ortin J. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 2004;32:2411–2420. doi: 10.1093/nar/gkh552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CC, Held RG, Chang SC, Yang L, Delpire E, Ghosh A, Hall BJ. A critical role for GluN2B-containing NMDA receptors in cortical development and function. Neuron. 2011;72:789–805. doi: 10.1016/j.neuron.2011.09.023. [DOI] [PubMed] [Google Scholar]
- Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr Opin Neurobiol. 2009;19:319–326. doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol. 2009;497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- Yang M, Crawley JN. Simple behavioral assessment of mouse olfaction. Curr Protoc Neurosci. 2009;Chapter 8(Unit 8):24. doi: 10.1002/0471142301.ns0824s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaichenko MI, Vanetsian GL, Merzhanova G. Differences in behavior of impulsive and self-controlled rats in the open-field and light-dark tests. Zhurnal vysshei nervnoi deiatelnosti imeni I P Pavlova. 2011;61:340–350. [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.