Abstract
The naked mole-rat maintains robust proteostasis and high levels of proteasome-mediated proteolysis for most of its exceptional (~31y) life span. Here, we report that the highly active proteasome from the naked mole-rat liver resists attenuation by a diverse suite of proteasome-specific small molecule inhibitors. Moreover, mouse, human, and yeast proteasomes exposed to the proteasome-depleted, naked mole-rat cytosolic fractions, recapitulate the observed inhibition resistance, and mammalian proteasomes also show increased activity. Gel filtration coupled with mass spectrometry and atomic force microscopy indicates that these traits are supported by a protein factor that resides in the cytosol. This factor interacts with the proteasome and modulates its activity. Although HSP72 and HSP40 (Hdj1) are among the constituents of this factor, the observed phenomenon, such as increasing peptidase activity and protecting against inhibition cannot be reconciled with any known chaperone functions. This novel function may contribute to the exceptional protein homeostasis in the naked mole-rat and allow it to successfully defy aging.
Keywords: Proteasome, Heat Shock Protein, Naked mole-rat, Protease Inhibitor, Protein degradation, Aging
1.0 INTRODUCTION
The longest-lived rodent, the naked mole-rat, Heterocephalus glaber, lives nearly an order of magnitude longer than similar-sized mice (1). Despite high levels of oxidative stress evident even at young ages (2), naked mole-rats maintain cancer-free good health and reproductive potential well into their third decade of life (3). Furthermore, these rodents show pronounced in vivo and in vitro resistance to a wide spectrum of toxins including oxidative stressors, heavy metals, and chemotherapeutics (4,5). This is also evident at the macromolecular level with mole-rat proteins markedly resistant to both oxidative damage and unfolding stressors (6). This generalized resilience against stress is likely due to efficient maintenance of protein quality control, involving both proteolytic machinery to remove damaged proteins and molecular chaperones [HSPs] that assist in protein repair or elimination.
HSPs bind to exposed hydrophobic regions of proteins preventing their aggregation and promoting their correct folding (7,8). If the process is unsuccessful, HSPs direct protein removal via either the ubiquitin proteasome system [UPS] or autophagy. The UPS degrades the majority of intracellular proteins and is considered pivotal for the digest of oxidatively damaged substrates (7,9). Proteolysis of damaged proteins occurs primarily in the cytosol (10,11). Here, ubiquitinylated, misfolded, oxidized, or otherwise damaged proteins are recognized by the proteasome (12) and cleaved into peptides by active centers located in the proteasome 20S catalytic core (10). The active proteolytic centers display three major specificities designated chymotrypsin-like [ChT-L], trypsin-like [T-L], and post-glutamyl, peptide-hydrolyzing [PGPH], reflecting the divergent chemical properties of the amino acid residues on the carboxyl side of the scissile bond (10).
Stress resulting from protein damage challenges both HSPs and the UPS by firstly increasing the load of substrates destined for degradation, and secondly by directly damaging the proteasome and thereby impairing its function (9). Indeed, the reported decline in mouse proteolytic degradative capacity with age is attributed to the stress induced increase in misfolded protein load and accompanying reduction in proteasome efficiency (9,13,14). In contrast, proteasome activity in aged naked mole-rats, like that in the cells of supercentenarians (15), remains at high levels even though these rodents from an early age bear a greater burden of proteotoxic stress from oxidatively-damaged proteins (6). We postulate that as this species evolved mechanisms to prevent damage from both the barrage of endogenous and environmental stressors, they developed better maintenance of somatic integrity and proteostasis and thereby longer lives.
RNA sequence analysis [RNA-Seq] reveals that many of the genes involved in the regulation of UPS as well as those of HSPs are detected at much higher levels in the naked mole-rat relative to mice (16,17). However, the particular expression pattern of UPS components as well as HSPs can only partially explain the high and sustained levels of proteasome activity in the naked mole-rat, such that young mole-rats exhibit five-fold higher specific peptidase activities compared to physiologically aged mice (18). Moreover, RNA-Seq data do not explain the resilience of naked mole-rat proteasomes to competitive inhibitors (6,19). Although published studies have documented that proteasome activity may be elevated in response to mild oxidative challenge (9,20), to date there has been no report of resistance of proteasomes to inhibition (20). We hypothesize that the naked mole-rat employs novel molecular mechanisms to protect proteasome function and achieve sufficiently high levels of catalytic activity necessary to effectively maintain proteostasis.
2. MATERIALS AND METHODS
2.1 Animals
This study used two similarly sized physiologically age-matched rodent species namely Mus musculus (C57BL/6 mice; 4–6 months) and Heterocephalus glaber (naked mole-rats; 2–3 years). The mice were fed a standard NIH-31 chow ad libitum and maintained in cohorts of four animals in microisolator mouse cages at 25°C, on a 12-h dark/light cycle. Naked mole-rats were from the well-characterized colonies of Dr. Rochelle Buffenstein housed at the University of Texas Health Science Center, San Antonio (3). Naked mole-rats were housed in simulated, multi-chambered burrow systems under constant climatic conditions that aimed to approximate their native habitat (30°C; 30–50% RH). Naked mole-rats were given an ad libitum supply of fresh fruits and vegetables supplemented weekly with a high protein and vitamin enriched cereal (Pronutro, South Africa). In this study, female mice were used to correspond with past studies undertaken in this field by our lab (13). As the subordinate naked mole-rats are sexually monomorphic (21), and we found no sex specific differences in any of our measurements we used tissues from both male and females (18).
Animals were anesthetized with isofluorane, euthanized by cardiac exsanguination and the liver tissue immediately excised and flash frozen in liquid nitrogen. All procedures involving animals were approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center (San Antonio, TX)
2.2 Whole Tissue Lysates and Subcellular Fractionation
Mouse and naked mole-rat liver lysates were separated into cytosolic, microsomal, and nuclear fractions using a modified Millipore Corp. procedure as previously described (13,18). These various fractions were then used in peptidolytic assays.
2.3 Peptidolytic Assay
Proteasome activity was measured using fluorogenic model peptide substrates (Boston Biochem, Boston, MA) specific for the ChT-L and T-L active centers of the proteasome as previously described (18). Parallel activity assays were performed with varying concentrations of proteasome inhibitors that represented four different classes of compounds with distinct modes of inhibitory action. These compounds modify active centers using a boronate group (bortezomib [BZ] (Sigma-Aldrich, St. Louis, MO)); an aldehyde (N-(benzyl-oxycarbonyl) leucinyl-leucinyl-leucinal [MG132]); a vinyl sulfone, (adamantane-acetyl-(6-aminohexanoyl)3-(leucinyl)3-vinyl-(methyl)-sulfone [Adh (VS)]); or a lactone (lactacystin [LC] (Calbio-chem/EMD Millipore, Billerica, MA)). Concentrations ranged from 5nM to 10μM for BZ, 0.2μM to 8μM for LC, and 10μM to 250μM for MG132 and Adh(VS) based on determinations from previous studies (13,18,22,23). Specific peptidolytic activity was presented as pmol of released AMC in 1 min per 1 μg of total protein in the test sample. This was determined after generating a standard curve using serial dilutions of 1mM AMC and measuring the fluorescence using a Spectra-Max Multi-mode microplate reader (Molecular Devices, Sunnyvale, CA) as previously described (18).
The IC50 concentration was determined as the concentration of inhibitor that reduced proteasome activity by 50%. For that purpose exponential decay or sigmoidal functions were fitted to the titration data and the corresponding IC50 extracted (OriginPro, OriginLab Corp or CompuSyn).
2.4 Crossover Assays
A partial proteasome purification of five separate naked mole-rat and five mouse samples was performed using high-speed centrifugation (27,28). Briefly, the samples were centrifuged at 100,000xg in an S120 AT-2 (Sorvall) rotor for 5 hours. The supernatant was decanted and pooled, while the pellet was re-constituted in 50mM Tris, pH 7.4 containing 20% glycerol and 1mM DTT. The pellet was left on ice for 30 minutes and then centrifuged again for 5 minutes at 16,000xg. Protein content was measured using the BCA Protein Assay Kit (Pierce, Thermo Scientific, Rockford, IL, USA) and the peptidolytic assay for ChT-L activity described above was performed starting with a 1:1 ratio of mouse or naked mole-rat purified proteasome to naked mole-rat or mouse pooled supernatant. We also performed these experiments using purified human 26S proteasome and yeast 20S proteasome (Enzo Life Sciences, Plymouth Meeting, PA, USA). Parallel with these samples, supernatants alone, and original mouse and naked mole-rat cytosolic fractions were tested on the same 96-well plate used for the assay.
2.5 Size-exclusion spin filtration
The size-exclusion filtration procedure was performed according to the manufacturer’s instructions (Amicon Microcon ultracel YM-3 cellulose filter, 3000Da) (EMD Millipore, Billerica, MD, USA). Briefly, 500 μL of supernatant prepared as described under “Crossover Assay” was placed on the filter and spun at 14,000xg for 30 minutes (3 x 10 minute washes). The filtrate was collected and then the sample reservoir was upended, and spun for 3 minutes at 14,000xg to collect the retentate. Protein content was measured using the BCA Protein Assay Kit (Pierce, Thermo Scientific, Rockford, IL, USA) and ChT-L activity was measured as described previously using a 1:1 mouse or naked mole-rat purified proteasome to naked mole-rat pooled supernatant, retentate, or filtrate.
2.6 Atomic Force Microscopy (AFM) Imaging
Chromatographic fractions showing inhibition resistance (see below), purified human 20S proteasome, and their mixture were subjected to AFM imaging with tapping (oscillating) mode in liquid using the Multimode Nanoscope IIIa AFM (Bruker) as previously described (24). Briefly, 3 μL of sample containing 2 ng of h20S or undiluted fr. 23, or a mixture of both, were deposited on freshly cleaved muscovite mica and left for 2–3 min. at room temperature to allow for electrostatic binding to mica. Then, the sample was overlaid with 30 μL of imaging buffer (5 mM Tris/HCl, pH 7.0), and subjected to AFM imaging. Oxide-sharpened silicon nitride tips on cantilevers with a nominal spring constant 0.32 N/m mounted in the wet chamber (Bruker Corp.) were used for imaging. The resonant frequency was tuned to 9–10 kHz, with a drive voltage of 200–500 mV. To minimize “tapping” force interference with the imaging molecules, relatively high values of the set point, 1.5 V to 1.9 V, were applied. Fields of 0.56 μm2 to 1 μm2 were scanned at a rate of 3.05 Hz with trace and retrace images collected with a resolution of 512 x 512 pixels. The images were processed only with the standard plane-fit and flattening; therefore they should be considered as “raw”. For display purposes the brightness and contrast of the images was adjusted and the occasional scan lines were removed with the Nanoscope software. Morphometric analysis of particles was performed with the grain analysis function in SPIP software (Image Metrology). To detect distinct classes of particles, the Peak Analyzer function was applied to the footprint area of all particles followed by hierarchical cluster analysis executed on footprint area, height, aspect, perimeter, and fiber length as unique and independent morphometric parameters (OriginPro). A new class of objects characterized by the set of identified morphometric parameters representing complexes of resistasome with 20S proteasome was then sourced to the specific objects in the original AFM images. As a self-test, the method correctly identified the top view and side view proteasomes in samples containing only 20S particles and in the mixture with fr. 23.
2.7 Multiplex Western Blot Analysis of Heat Shock Proteins
Tissue lysates or sub-cellular fractions were separated in 12% SDS-PAGE (Biorad Life Sciences, Hercules, CA) and transferred to nitrocellulose membranes (Biorad Life Sciences, Hercules, CA). The membranes were probed with antibodies against the following proteins: HSP110 (rabbit, SPA-1101, 1:5k), HSP90 (mouse, SPA831, 1:20K), HSP70/72 (mouse, SPA810, 1:10K), HSP40 (HDJ1) (rabbit, SPA400, 1:2.5K), HSP25/HSPB1 (rabbit, SPA801, 1:10K), (Enzo Life Sciences, Plymouth Meeting, PA, USA). We also used antibodies against HSC70 (mouse, sc-7298, 1:10K) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Blots were stained with Ponceau-S to measure total protein load. Primary antibodies were detected using anti-mouse IRDye 680LT, or anti-rabbit IR Dye800 CW (Li-Cor) conjugated antibodies. Secondary antibodies were incubated at 1:10K (anti-rabbit) or 1:20K (anti-mouse) for 2 hours at room temperature and images were captured using the Odyssey Imaging System (Li-Cor, Lincoln) for IRDye 680LT, IR Dye800. Immunoblots were quantified using the ImageJ public domain Java image processing program (http://rsbweb.nih.gov/ij/).
2.8 Gel Filtration Chromatography
The 5-hr supernatant was fractionated with gel filtration chromatography on a Superose 6 GL 10/30 column (GE Healthcare) fitted into a BioCad Sprint (Perseptive Biosystems) HPLC. A 100μL sample of approximately 2μg/μL protein concentration was loaded. Chromatograms were developed with a column buffer (50mM Tris/HCl pH 7, 20% glycerol) using 0.4mL/min flow rate. The volume of collected fractions was 500μL. About 48 fractions per a single chromatogram were collected. Fractionation progress was monitored with absorption readings at 260 and 280nm. The apparent molecular weight of separated proteins was determined based on elution volumes of a set of gel filtration markers (Bio-Rad) ranging from 1,350 (vitamin B12) to 670,000 Da (bovine thyroglobulin).
Concentration of protein in fractions was determined with a BCA assay (Pierce, Thermo Scientific, Rockford, IL, USA). Fractions were concentrated as necessary using a Centricon 3000Da cutoff filter (Amicon Microcon ultracel YM-3 cellulose filter; see Size Exclusion Column above; EMD Millipore, Billerica, MA, USA) and the peptidolytic assay for ChT-L activity was performed as described above (see Peptidolytic Assay, main and supplementary text) with 0 and 20μM of MG132.
2.9 In vitro Depletion Assays
Anti-HSP72 (Enzo, SPA-810), anti-HSP40 (Hdj1) (Enzo, SPA-400), anti-HSP25 (Enzo, SPA-801), or anti-HSP90 (Enzo, SPA831) specific anti-bodies were added in increasing concentrations ranging from 10pg to 400pg (made as serial dilutions from the original stock using PBS with 50% glycerol) to either naked mole-rat cytosolic lysates or naked mole-rat supernatants separated from these lysates (see Crossover Assays above). Because of the sequence similarity between mice and naked mole-rats of the key chaperones measured in this study, it was assumed that the antibodies could recognize the targeted protein in each species. For chemical inhibition of HSP72, increasing concentrations of VER155008 (25nM to 1000nM, Tocris Bioscience, UK) and pifithrin-μ (10μM to 1000μM; both Calbiochem/EMD Millipore, Billerica, MA USA) were added to lysates or supernatants in this manner. Next, the mixture was incubated at 30°C for 1hr in the presence of 1mM ATP. Immediately, the antibodies or small-molecule inhibitors and purified human 26S proteasome or 5hr-naked mole-rat partially purified proteasome as necessary were added to the sample mixture. Then ChT-L activity was tested as described above using Suc-LLVY-AMC in the presence or absence of 20μM of MG132.
2.10 Immunoprecipitation Assay
Immuoprecipitation was performed with the Protein A/G PLUS-Agarose Immunoprecipitation Reagent (Santa Cruz Biotechnology, Santa Cruz, CA) using the protocols suggested by Enzo (www.enzolifesciences.com/support/antbodies/protocols/immunoprecipitation-protocol/) with 1μg of anti-HSP70/72 antibodies (Enzo, SPA-801) per 100μL of mouse or naked mole-rat liver lysates (final concentration 1mg/mL).
After an overnight incubation at 4°C of the immune complex, the protein A/G beads were added and incubated for 2hrs at 4°C. The beads were collected by microcentrifugation, washed 5X with PBS, resuspended in 2X Laemmli reducing running buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue, 0.125M Tris HCL; 95°C for 5 min) and then subjected to SDS-PAGE separation under de-naturing conditions. The gels were transferred to PVDF membrane and then tested with the same antibodies against HSP90, HSP72, HSP40, and HSP25 used in previous Western blots and antibodies for proteasome subunits, RPT5 (Enzo, mouse PW8770, 1:2K), and α7 (Enzo, mouse PW8110, 1:5K). HRP-conjugated secondary antibodies for goat, rat, or rabbit (Santa Cruz) were used to visualize the immuno-reaction using the ECL Prime Western Blotting Detection Reagent, a chemiluminescent substrate (Amersham, Buckinghamshire, UK). Immunoblots were visualized using the Typhoon 9410 variable mode imager or on x-ray film (GE Healthcare).
2.11 Protein Identification with Mass Spectrometry
Proteins were separated by 1-D SDS-PAGE and proteins in each gel lane were digested in situ with trypsin (Promega). The digests were analyzed by capillary HPLC electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) on a Thermo Fisher Orbitrap Velos mass spectrometer. The MS data were searched against the rodent subset of the NCIBnr protein database (NCBInr_20130102; 316,972 sequences) by Mascot (Matrix Science). The Mascot results were subjected to a subset search by X! Tandem followed by determination of probability assessments of the peptide assignments and protein identifications by Scaffold (Proteome Software).
2.12 Statistical Analysis
A two-tailed Student’s t-test on two different statistical platforms (Microsoft Excel 2010; SigmaPlot) was used to determine significant differences in the means for the peptidolytic assays and Western blot quantitation. One-way ANOVA was used in the inhibition resistance experiments to analyze the variances between species and fractions while two and three-way ANOVAs were used to test variance when comparisons between treatments, species, and concentration were necessary (SigmaPlot). Statistical significance was set at the p < 0.05 level. All pairwise multiple comparison procedures used the Bonferroni and Holm-Sidak corrections to counteract the probability of false positives.
3. RESULTS AND DISCUSSION
3.1 Naked mole-rat proteasomes are resistant to proteasome inhibition
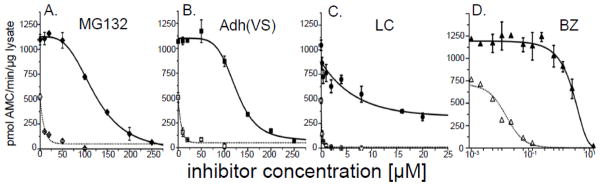
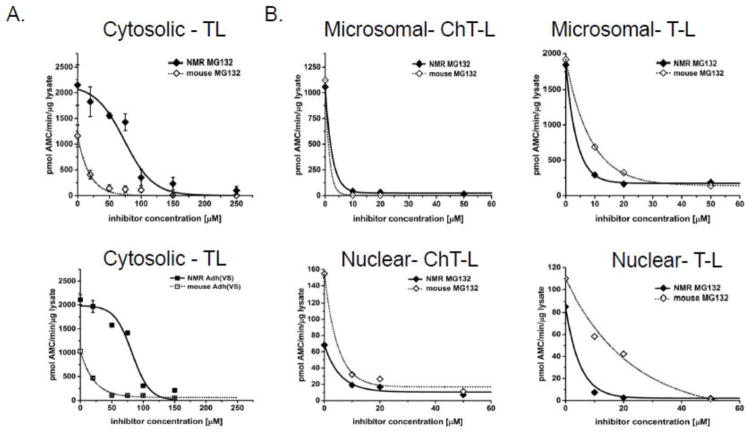
To test this hypothesis, we measured proteasome activity in the various subcellular fractions of liver lysates of mice and mole-rats when treated with several proteasome-specific small molecule inhibitors, namely MG132, [Adh(VS)] (25), [BZ] (22), and LC (23). These well-characterized competitive inhibitors bind to the proteasome catalytic centers using distinct chemical mechanisms. ChT-L activity is the primary target for all four inhibitors. BZ, an approved anti-cancer drug (26,27), is regarded as the most specific for this peptidase activity. T-L activity is known as a secondary target for both MG132 and LC (23), and Adh(VS) inhibits all three peptidases with relatively similar efficiency. We compared these peptidase activities in untreated and inhibitor-treated samples. In sharp contrast to data acquired with mouse proteasomes, we found that naked mole-rat proteasomes from cytosolic extracts maintained ChTL activity when treated with specific competitive proteasome inhibitors, (Figure 1). Remarkably, naked mole-rats required 10-fold higher concentrations to ablate 50% of ChT-L activity (the IC50) for both MG132 and LC while the IC50 for Adh(VS) was 22-fold greater (Figure 1; Table 1). Strikingly, the ChT-L IC50 concentration for BZ was more than two orders of magnitude (163-fold) higher in the naked mole-rat than in mouse cytosolic extracts (Figure 1; Table 1). T-L activity also showed inhibition resistance to MG132 and Adh(VS) with the T-L IC50 5-fold greater in naked mole-rats compared to mice for both agents (Figure 2A; Table 1).
FIGURE 1. Proteasomes from naked mole-rat [NMR] cytosol show resistance to proteasome specific competitive inhibitors.
Dramatically different ChT-L activity was evident in response to increasing concentrations of inhibitors representing four different classes of the compounds: (A) an aldehyde, MG132; (B) a vinyl sulfone, Adh(VS); (C), a lactone, LC and (D) a boronate, BZ. To compare the inhibition resistance between NMR (solid symbols) and mouse (open symbols), we calculated the IC50 value, which corresponds to the concentration of inhibitor required to ablate 50% of activity. The IC50 for (A) MG132 was 15X, (B) Adh(VS) 18x, (C) LC 40x, and (D) BZ 163x greater in NMR samples compared to those of mice. See also Table 1 for IC50 values. Calculations are based on ChT-L activity assessments from lysates of at least 3 mice or 6 NMRs.
TABLE 1.
NMR proteasome ChT-L and T-L activity only in the cytosolic fraction showed remarkable resistance to competitive inhibition as indicated by significant differences (*, p > 0.05) when compared to mouse values. IC50 values are indicated in μM ± S.E.
| Fraction/Inhibitor | ChT-L IC50 (μM) | T-L IC50 (μM) | ||
|---|---|---|---|---|
| NMR | Ms | NMR | Ms | |
| Cytosolic/ Adh (VS) | 125.0±5.6* | 5.56±1.57* | 80.6±2.5* | 16.2±1.9* |
| Cytosolic/ MG132 | 122.0±5.5* | 8.22±2.75* | 71.4±8.7* | 14.6±4.4* |
| Cytosolic/LC | 6.15±1.4* | 0.156±0.005* | -- | -- |
| Cytosolic/BZ | 2.45±0.74* | 0.015±0.004* | -- | -- |
| Microsomal/MG132 | 1.79 | 0.99 | 3.01 | 6.63 |
| Nuclear/ MG132 | 6.49±2.70 | 4.6±1.62 | 2.70±1.4 | 12.8±4.7 |
-- = no data; Adh (VS) = adamantane-acetyl-(6-aminohexanoyl)3-(leucinyl)3- vinyl-(methyl)-sulfone ; MG132 = N-(benzyl-oxycarbonyl) leucinyl-leucinyl- leucinal; LC = lactacystin; BZ = bortezomib
FIGURE 2. Trypsin-like activity of proteasome in NMR cytosolic fractions was more resistant to competitive inhibitors than mouse proteasome, although less profoundly than the ChT-L activity. However, resistance was not observed in the microsomal or nuclear fractions of either species.
(A) Based on IC50 values, the proteasome in NMR cytosol is about 5 times less susceptible to both MG132 (top) and Adh(VS) (bottom) than was the mouse cytosol. See also Table 1. (B) In contrast, no significant differences in inhibitor resistance between NMR and Ms were found in the microsomal or nuclear fractions as tested with MG132. See also Table 1.
Inhibition resistance was restricted to the cytosolic fraction since it was not observed in the microsomal or nuclear fractions of either species (Figure 2B; Table 1). Since the cytosolic fraction is most likely to encounter cellular stressors, we concluded that cytosol-specific resistance to proteasome inhibition observed in the naked mole-rat could be an important component of the cytoprotective arsenal that underlies naked mole-rat resilience against potentially harmful conditions (5).
3.2 Proteasome resistance is transferrable among species
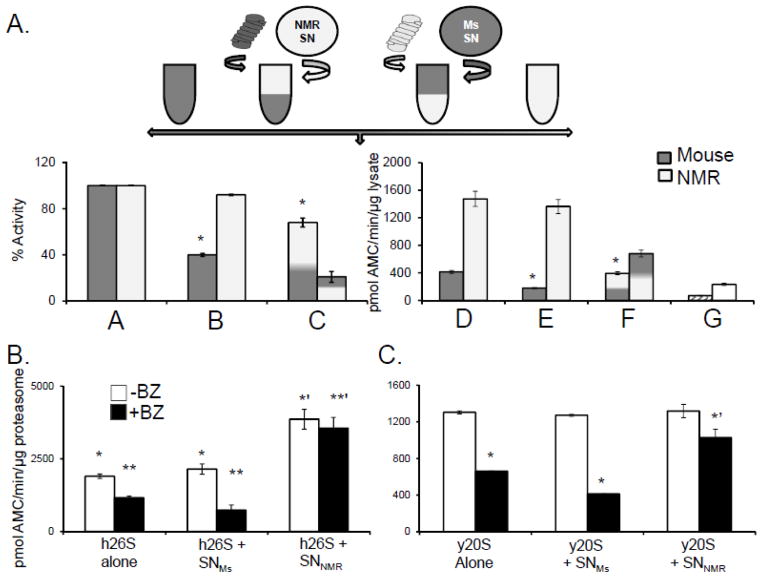
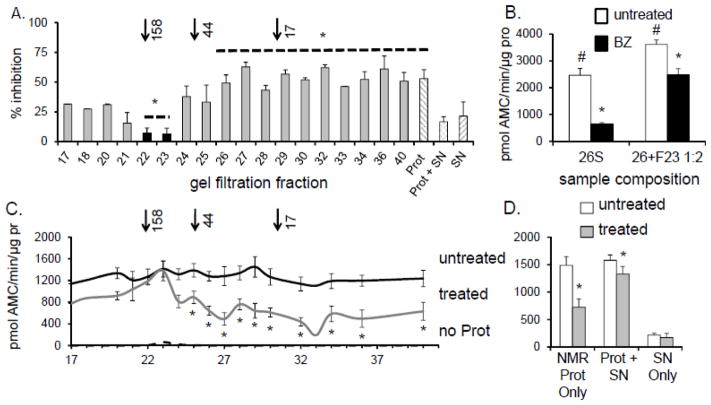
To evaluate whether the observed resistance to inhibition was an intrinsic property of the naked mole-rat proteasome or is rather mediated by specific factors within the intracellular milieu, we used a simple “cross-over” experimental design (Figure 3A). Mole-rats and mice proteasomes were partially purified from the cytosolic fractions using differential centrifugation (28,29) and the proteasome-enriched pellet and the proteasome-depleted supernatant were separately retained. The supernatant of both species had very low ChT-L activity (Figure 3A, bar G) confirming efficient removal of proteasomes from the cytosol. As a next step, ChT-L activity was measured in the “reconstituted cytosol” containing proteasome-depleted (SN) plus proteasome-enriched (Prot) preparations from either species in a 1:1 ratio of the original protein contents. When the ProtNMR was resuspended in SNMs, it no longer showed resistance to MG132, and also exhibited similar proteasome specific activity to that observed in mouse lysates (Figure 3A, bars C, F). Conversely, ProtMs activity more than doubled upon mixing with the SNNMR (Figure 3A, bar F). Furthermore, the ProtMs, when incubated in SNNMR, became resistant to inhibition thus mirroring the cytosolic proteasome activity profile of naked mole-rat (Figure 3A, bar C). These data reveal that both elevated proteasome activity of naked mole-rats and inhibition resistance are not due to intrinsic proteasome properties conferring a more stable and more efficient proteasome, but rather, are due to protective modulators of proteasome activity within the cytosolic milieu.
FIGURE 3. An NMR cytosolic factor confers resistance to mammalian and yeast proteasomes from competitive inhibition.
(A)Top panel: Schematic showing the design of the cross over experiment set up to evaluate if species differences in proteasome resistance to inhibition reflect an intrinsic property of the proteasome or the presence of a protective cytosolic factor. See Crossover Assays in Methods below for details. Bottom panel: When ProtMs were resuspended in SNNMR they showed elevated activity and inhibition resistance, whereas ProtNMR exposed to SNMS displayed both lower activity and greater sensitivity to inhibition. Bars A and D represents the activity of reconstituted MS and NMR cytosols (A = percentage set to 100% activity and D= ChT-L activity per mg lysate) in the absence of any inhibitor. B and E reveal the change in activity following treatment of the reconstituted cytosols with 20 μM MG132 and indicate that in comparison with mouse, the proteasome in NMR cytosol is resistant to inhibition. Bars C and F demonstrate that when ProtMs were resuspended in SNNMR they exhibited elevated activity and acquired inhibition resistance (p<0.05), whereas ProtNMR resuspended in SNMs showed both lower activity and greater sensitivity to inhibition (p<0.05) (means ± S.E.M.; n =5). Bar G reveals that the SN alone had very low peptidolytic activity. (B) Human 26S proteasomes [h26S] treated with SNNMR, but not SNMs or buffer, showed increased ChT-L activity (solid bars) and pronounced resistance to 10nM BZ (hatched bars) (* to *’, p<0.003; ** to **’, p<0.0004; means ± S.E.M; n = 6). (C) SNNMR conveyed inhibition resistance to the yeast proteasome [y20S]. SNMS did not show this effect. No significant increase in activity in the presence of SNNMR was detected. (* and ** indicated p < 0.01; means ± S.E.M.; n = 3)
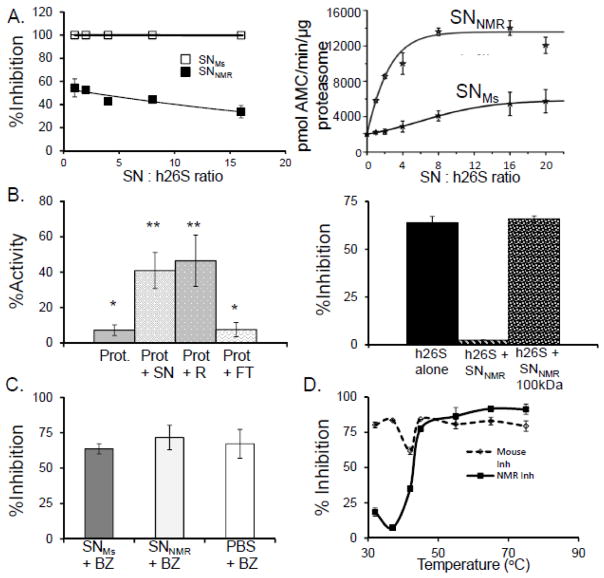
We found that the cytosolic milieu not only modulated mouse proteasome activity but that the SNNMR also conferred these same effects on both human and yeast proteasomes. When we exposed purified human 26S or yeast 20S proteasome to the SNNMR we noticed an enhanced resistance to BZ inhibition (Figure 3B,C). Indeed levels of proteasome activity and inhibition resistance, when treated with SNNMR in these evolutionarily divergent species, converged to that observed in naked mole-rats. Moreover, exposure of human 26S to increasing concentrations of SNNMR resulted in a systematic increase of ChT-L activity and inhibition resistance (Figure 4A). In contrast, the treatment with SNMS resulted in trivial increases in ChT-L activity at only the highest ratio of SNMS to human 26S proteasome [h26S], and no inhibition resistance at any concentration of SNMs (Figure 4A). These data provide strong evidence that components within the naked mole-rat cytosol protect its proteasome from agents that commonly impair proteasome function. This cytosolic factor is transferrable to other species and capable of inducing similar protective and modulatory effects.
FIGURE 4. NMR cytosol contains a protein factor conveying resistance to inhibition and leading to activation of the proteasome.
(A) Titration of human 26S proteasome [h26S] with increasing protein concentration of mouse supernatant [SNMS] (open squares) did not result in acquiring resistance to inhibition with 20 μM MG132 even at 16-fold excess of SNMS. In contrast, resistance was evident even at lowest used SNNMR: h26S ratio (1:1, 250ng of each component), and was slightly stronger with increasing SNNMR: h26S ratio (left panel; closed squares). Chymotrypsin-like [ChT-L] activity of h26S was rapidly and profoundly enhanced by the addition of SNNMR, reaching a plateau with more than 6-fold activation at about 8:1 (SNNMR: h26S) ratio (right panel). However, treatment with the 8-fold excess of SNMS resulted in only 2-fold activation of h26S, and the activation never exceeded 3-fold under the conditions used (n =5). (B) SNNMR was fractionated with a spin filter with 3,000 Da pore cutoff membrane (top panel) or with a 100,000 Da pore cutoff membrane (bottom panel). Top panel demonstrates ChT-L activities of ProtNMR after treatment with 20 μM MG132, alone or with addition of the whole SNNMR, the 3,000 Da retentate (RNMR; molecules with apparent molecular weights higher than 3,000 Da) or the 3,000 Da filtrate [FTNMR; molecules with apparent molecular weights lower than 3,000 Da]. The activity of inhibitor-challenged ProtNMR was markedly higher after treatment with the whole SNNMR (compare with Fig. 2A, bottom panel) or with the RNMR, but not with FTNMR. Spin-filtering the SNNMR through a 100,000 Da pore cutoff membrane, and performing the ChT-L activity assay with h26S treated with 20μM of MG132 (left bar) the inhibition resistance was conveyed by the SNNMR (middle bar; compare with Fig. 2B) but not by the filtrate (right bar; MW>100,000 Da, n =5 each treatment). (C) The ChT-L activity of h26S proteasome was markedly inhibited by all three filtrates, indicating that BZ was not significantly sequestered by SNNMR, as compared with SNMs or buffer (n = 6). (D) To test if the factor is heat liable we subjected SNNMR and SNMs to heat stress at temperatures ranging from 32°C to 75°C. The heat-treated SNs were then added to h26S and ChT-L proteasome activity and inhibition resistance were assessed with 10nM BZ. Heat-treatment above 45°C ablated the inhibition resistance conveyed by SNNMR strongly suggesting a protein nature of the proteasome-affecting factor (n = 5).
Both partially purified mouse proteasomes and purified human 26S also showed marked increases in specific proteasome activity even in the absence of chemical inhibition (Figure 3A,B). The mechanism(s) facilitating this increase in activity are unknown and possibly modulation of proteasome activity in the absence of inhibitory agents may be a mammal-specific property for this was not evident when yeast 20S proteasomes where incubated with SNNMR (Figure 3C). It is intriguing that at least the inhibition resistance conveyed by the naked mole-rat cytosolic factor is universal from yeast to mammalian proteasomes.
3.3 Identification of the naked mole-rat activation/resistance factor
We tested if the observed changes in proteasome function in the presence of the SNNMR were conveyed by small molecules or specific macromolecule(s) that directly interact with the proteasome and alter its properties. The involvement of cytosolic “small molecules” was ruled out by evaluating proteasome activity and inhibition resistance in the presence of the flow-through (i.e., metabolites, small peptides; Figure 4B) or retentate (proteins and other macro-molecules) after the 3-kDa cutoff spin filtration of cytosols. When the mouse or naked mole-rat proteasome was exposed to the retentate, it also exhibited a similar degree of inhibition resistance to that observed when it was exposed to the SNNMR in the cross-over experiment (Figure 4B; Table 2). However, when proteasomes were treated with filtrate alone, the resistance was not observed (Figure 4B; Table 2). Interestingly, when we fractionated the supernatant with a 100-kDa cut-off filter and then measured the peptidolytic activity in the presence of the flow-through, the inhibition resistance capability was removed suggesting that this feature of the cytosolic factor was facilitated by a molecule or a group of molecules larger than 100 kDa (Figure 4B). To rule out sequestration of the inhibitors or their degradation in the supernatant, we pre-incubated BZ with SNNMR, SNMs and buffer alone. Then, we recovered the BZ by spin filtering each sample through a 3-kDa cutoff membrane. Next, we tested ChT-L activity of human 26S proteasome treated with the obtained filtrates. All three samples apparently contained highly potent BZ (Figure 4C), therefore we concluded that the inhibitor is not substantially degraded, modified or sequestrated by components of the supernatant.
TABLE 2. The inhibition resistance factor was associated with high molecular weight molecule(s) that were preserved in a retentate after SN fractionation through a Centricon 3000.
Naked mole-rat supernatant [SN], retentate [R], or flow-through [FT] was added to proteasomes [Prot] of both species [ProtNMR or ProtMS]. Neither the partially purified proteasomes alone nor the proteasomes mixed with FT showed resistance to inhibition when treated with a range of MG132 concentrations. In contrast, addition of either the SN or R did convey inhibition resistance. These data complement the results presented in Figure 4B, showing expanded concentrations of MG132 tested as well as percent inhibition. Results are presented as pmol AMC/min/μg protein.
| conc. MG132 (μM)→ | 0 | 10 | 20 | 50 | |||
|---|---|---|---|---|---|---|---|
| Sample | Mean ± SE | Mean ± SE | % Inhibition | Mean ± SE | % Inhibition | Mean ± SE | % Inhibition |
| ProtNMR | 514 ± 9 | 72 ± 13 | 86.0 | 43 ± 0.3 | 91.6 | 23 ± 1.0 | 95.5 |
| ProtNMR + SN | 1011 ± 15*# | 488 ± 58*# | 51.7 | 499 ± 71*# | 50.6 | 327 ± 104*# | 67.7 |
| ProtNMR + R | 785 ± 106*# | 542 ± 181*# | 31.0 | 479 ± 146*# | 39.0 | 378 ± 104*# | 51.8 |
| ProtNMR + FT | 237 ± 45 | 40 ± 18 | 83.1 | 27 ± 18 | 88.6 | 16 ± 13 | 93.2 |
| ProtMS | 201 ± 1.0 | 25 ± 0.1 | 87.6 | 17 ± 0.2 | 91.5 | 11 ± 0.1 | 94.5 |
| ProtMS + SN | 263 ± 2.0*# | 126 ± 22*# | 52.1 | 116 ± 20*# | 55.9 | 84 ± 34*# | 68.1 |
| ProtMS + R | 282 ± 28*# | 178 ± 72*# | 36.9 | 172 ± 70*# | 39.0 | 89 ± 12*# | 68.4 |
| ProtMS + FT | 117 ± 13 | 17 ± 0.3 | 85.5 | 15 ± 3.0 | 87.2 | 3.0 ± 1.9 | 97.4 |
Statistical significance is indicated on the table below (ANOVA,
p < 0.05 to Prot;
p < 0.05 to FT).
Based upon these results we hypothesized that both proteasome activation and inhibition resistance are conferred by cytosolic proteins. To assess this we subjected both mouse and naked mole-rat lysates to heat stress for 1hr at temperatures ranging from 32°C to 75°C. Human 26S was not any longer protected from inhibition when treated with NMR lysate exposed to temperatures higher than 45°C (Figure 4D). We envisioned two possible scenarios: a) that multiple proteins present in high levels in naked mole-rat but not in mouse cytosols create an environment generally supporting activation and resistance, or that b) the naked mole-rat cytosol contains a specific protein or a protein complex that interacts with the proteasome, commanding its activation and resistance.
In an attempt to identify the components of the SNNMR that confer inhibition resistance, the 5-hr supernatant was fractionated by gel filtration chromatography. ProtNMR was added to each chromatographic fraction and ChT-L activity was measured in the presence or absence of the MG132 proteasome inhibitor. The presence of a distinct macromolecular resistance factor particle would be evident if only one or a few fractions convey the resistance. The total loss of protective capabilities after the cytosol fractionation would suggest that macromolecular resistance factor is not stable to gel filtration conditions or that the cytosolic environment in general is responsible for the protection. The potential factor(s) conveying inhibition resistance were localized to only two adjacent chromatographic fractions (fr.22 and fr.23) corresponding to relative molecular mass of about 100kDa – 160 kDa (Figure 5A,C). These two fractions were pooled, collectively called fr.23, and used in subsequent experiments. Purified human 26S proteasomes treated with fr.23 were clearly resistant to inhibition when challenged with 10nM BZ (Figure 5B). Therefore, we concluded that the resistance factor present in fr.23 is a stable macromolecule, capable of withstanding the purification procedure, and whose actions on the proteasome can be transferred to other species.
FIGURE 5. NMR proteasome resistance to inhibition was found in two gel filtration fractions.
Proteasome-depleted SNNMR was fractionated by gel filtration. (A) ProtNMR was added to the gel filtration fractions and proteasome-specific inhibition resistance was measured after treatment of each fraction with 20 μM MG132. Only fractions 22 and 23 consistently protected the ProtNMR ChT-L activity from MG132 inhibition (*p<0.05; mean ± S.E.M., n = 5). Although fr.21 contained HSP72 and HSP 40 it did not confer significant inhibition resistance. Arrows above A indicate elution of the molecular weight standards. (B) ChT-L activity (expressed per mg protein, [pro]) of human 26S proteasome [h26S] was significantly increased and protected from inhibition by 10 nM BZ when suspended in the resistance factor-enriched fr.23 (#p<0.01, *p<0.0004). Treatment with 10 nM BZ ablated activity of h26S by 75%. In the presence of fr. 23 ChT-L activity of h26S exposed to BZ was reduced only by 30%. (C) Naked mole-rat [NMR] proteasome-enriched pellet [ProtNMR] was added to the gel filtration fractions and chymotrypsin-like [ChT-L] activity was measured after treatment with 20μM, MG132 (“treated”) or DMSO vehicle (“untreated”). Essentially no ChT-L activity was detected in the fractions in the absence of ProtNMR (“no Prot”). (*p<0.05; mean ± S.E.M.; n = 5). Only ProtNMR added to fractions 22 and 23 was refractory to MG132. (D) ProtNMR alone was sensitive to MG132 inhibition, unlike ProtNMR re-suspended in SNNMR (*p < 0.02). Very low peptidolytic activity was detected in SNNMR (see also C, hashed columns).
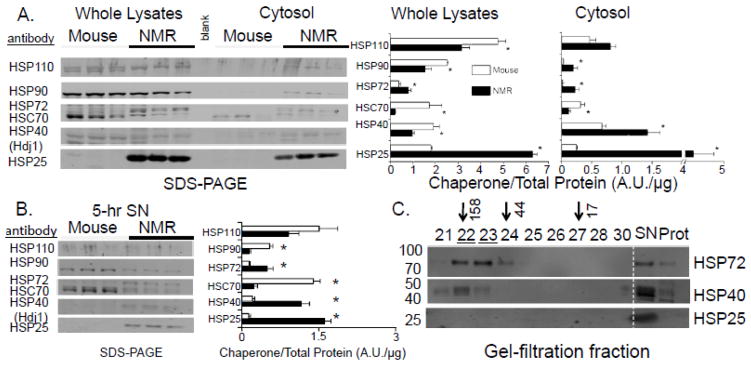
Interestingly, studies examining changes in gene expression in response to MG132 treatment, reveal overlapping features with the heat shock response signaling pathway (30). As such, compounds or pathways that stimulate or maintain chaperone response could also aid in the preservation of proteasome function and recognition of damaged substrates. Western blot analyses revealed that expression levels of three key chaperones, HSP72, HSP40, and HSP25 were significantly higher in naked mole-rat liver cytosolic fractions and 5-hr supernatants than in respective mouse samples (Figure 6A,B; Figure S1). The high levels of these HSPs in naked mole-rats concur with the previously described strong correlation between levels of molecular chaperones and species longevity in reptiles, birds, and mammals (31,32). Cytosolic abundance of these key HSPs in naked mole-rat tissues may contribute to enhanced protection at the molecular, cellular, and whole animal level against the many potential stressors these subterranean-dwelling rodents encounter over the course of their lifespan, and may be responsible for providing a protective intracellular resistant environment. Mass spectrometry analysis of the fr.22 and fr.23 content, the chromatographic fractions conferring inhibition resistance, revealed the presence of naked mole-rat HSP72 (inducible heat shock protein 70, Table 3; Figure S2; See also Supplementary Material Table S1). Consistently, Western blot analyses detected a high level of HSP72 in fr.21, fr.22, and fr.23. HSP40 was also present (Figure 6C) in these fractions. Although HSP25 was the most abundant HSP in the cytosol and 5-hr supernatants (Figure 6A,B), this particular HSP was not found in fr.21–23 but only in the supernatant fraction (SN) (Figure 6C).
FIGURE 6. NMR showed higher levels of majority of cytosolic HSPs than did mice (A, B). On the other hand, gel filtration fractions influencing PRS resistance contained only high levels of HSP72 and HSP40 (C).
(A) Multiplex Western blots of whole lysates and cytosolic preparations and their quantification (bar graphs) after standardization to total protein (Ponceau-S, see Figure S1) showed dramatic species differences in relative content of HSP90, HSP72, HSC70, HSP40, and HSP25. Mean values and standard error bars are shown (*p<0.005). (B) 5-hr SNs showed similar differences in HSPs content to that of the cytosolic preparations. (C) As demonstrated by Western blotting (left, molecular weights), SNNMR, fr.22, and fr.23 (underlined) contained high levels of HSP72, some HSP40, and virtually no HSP25. Arrows indicate elution of the molecular weight standards and numbers on the left of the figure represent molecular weights.
TABLE 3.
Mass spectrometry of fractions 22 and 23 revealed the presence of several molecular chaperones. For a complete list please see also Table S1.
| Identified Proteins (8/223) | Accession Number | Molecular Weight | fr.22 | fr.23 |
|---|---|---|---|---|
| inducible heat shock protein 70 (HSP72) [Heterocephalus glaber] | gi|13242237 (+26) | 71 kDa | 28 | |
| Ubiquitin-like modifier-activating enzyme 1 [Heterocephalus glaber] | gi|351699501 | 119 kDa | 20 | 12 |
| 78 kDa glucose-regulated protein [Heterocephalus glaber] | gi|351702099 | 72 kDa | 10 | 5 |
| inducible heat shock protein 70 [Mus musculus] | gi|118490060 (+7) | 70 kDa | 8 | 6 |
| Hsp90aa1 protein [Mus musculus] | gi|118142832 (+23) | 66 kDa | 8 | 7 |
| heat shock protein 90 beta [Equus caballus] | gi|12082134 (+17) | 82 kDa | 7 | 8 |
| Protein disulfide-isomerase [Heterocephalus glaber] | gi|351706419 | 57 kDa | 7 | 5 |
| stress-70 protein, mitochondrial [Mus musculus] | gi|162461907 (+9) | 73 kDa | 7 | 6 |
Quantitative value as calculated by the Scaffold v3 program are shown in the table under fr.22 and fr.23 columns.
3.4 Functional role for HSP72 and HSP40 in the naked mole-rat activation/resistance factor
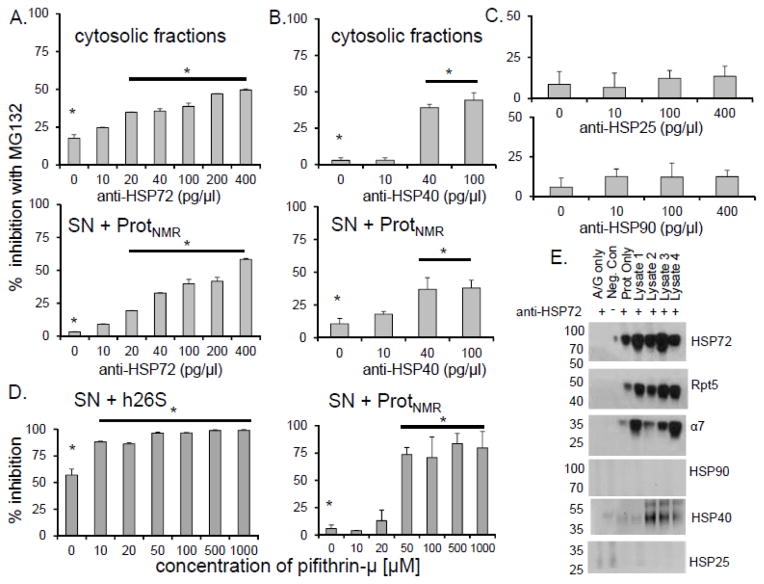
We next set to test which HSPs contributed to the observed cytosolic protective and regulatory effects on proteasomes by using neutralizing antibodies for HSP40, HSP72, or HSP90. Only anti-HSP70/72 and anti-HSP40 altered sensitivity to inhibition (Figure 7). With increasing concentration of anti-HSP72, activity of proteasome was also reduced (Figure 8A,B). Since mass spectrometry revealed the presence of HSP90 in fr.23 (Table 3), we also tested the influence of specific anti-HSP90 antibodies on proteasomes. This treatment had no effect on inhibition resistance, nor did HSP90 co-precipitate with HSP72 in naked mole-rat lysates (Figure 7C,E). Collectively, these results confirm a key role for the canonical chaperone HSP72, and its co-chaperone HSP40 in the protection of naked mole-rat proteasome function.
FIGURE 7. HSP72 and HSP40 played a critical role in inhibition resistance of naked mole-rat proteasomes.
(A, B) Titration of the naked mole-rat cytosolic fraction and SNNMR with increasing amounts of neutralizing antibodies against HSP72 and HSP40 significantly decreased the level of inhibition resistance (*p<0.001) (mean ± S.E.M., n = 6). (C) Anti-HSP25 or anti-HSP90 did not affect sensitivity to inhibition. (D) Treatment with increasing concentrations of the HSP72-specific inhibitor pifithrin-μ attenuated resistance to inhibition (*p<0.01) (mean ± S.E.M., n = 5). (E) Immunoprecipitation with anti-HSP70/72 antibody showed interaction between HSP72 and the NMR 26S (α7, Rpt5) as well as with HSP40, but not with HSP25 or HSP90.
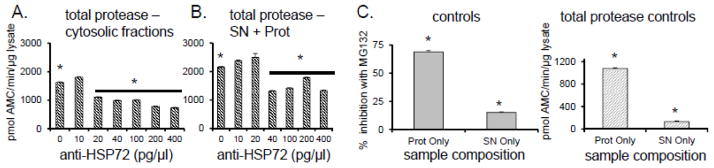
FIGURE 8. HSP72 depletion affects not only inhibition resistance but also peptidase activity of proteasome.
(A) Up to 40% decline in ChT-L activity was observed when cytosolic lysates were mixed with at least anti-HSP70/72 antibodies (*p<0.001). (B) A similar decrease of activity with reconstituted naked-mole rat supernatant [SN] and partially purified naked mole-rat proteasomes [Prot] was observed (*p<0.001). (C) NMR proteasome [Prot] in the absence of SN does not exhibit inhibitor resistance (left). Moreover, NMR supernatant [SN] does not present any appreciable ChT-L peptidase activity (right).
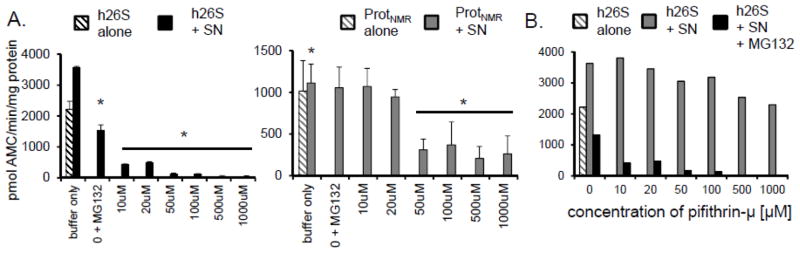
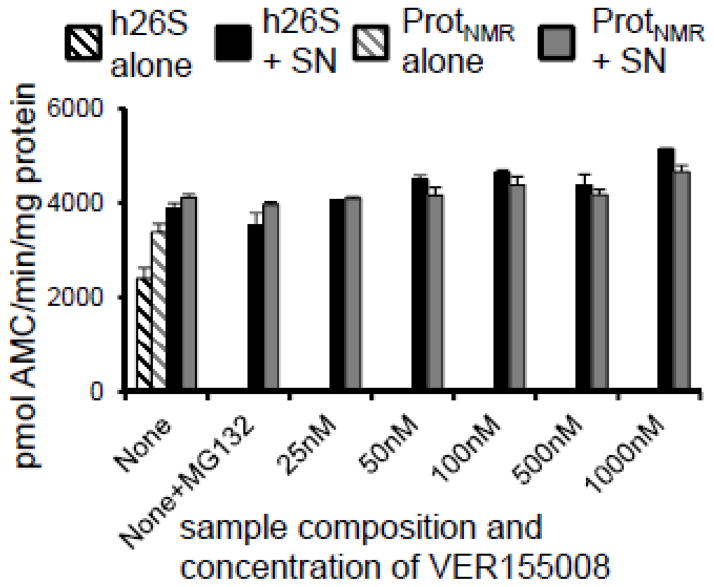
To gain insight into the mode of action of the chaperones we challenged HSP72 with two distinct inhibitors: pifithrin-μ (33) and VER155008 (34). Pifithrin-μ, like the neutralizing antibody, binds to the substrate-binding domain of HSP72 (33,35) whereas VER155008 binds to the ATPase domain of HSP72 and prevents ATP hydrolysis (34). Intriguingly, only pifithrin-μ lessened both the inhibition resistance and the increased proteasome activity associated with the SNNMR (Figure 7D; Figure 9). To the contrary, VER155008 had no effect upon sensitivity to inhibition or activity (Figure 10). Divergent responses to pifithrin-μ and VER155008 suggest that the resistance/activation factor function is independent on ATP hydrolysis, but possibly relies on the substrate-binding domain of HSP72. Immunoprecipitation experiments revealed that HSP72 was associated with both the 26S proteasome and HSP40 (Figure 7E), indicating direct interactions between this factor and proteasome in naked mole-rats, and suggesting a critical role of the HSP72/HSP40 co-chaperone relationship for the observed functions of increased proteasome activity and inhibition resistance for this cytosolic factor.
FIGURE 9. Inhibition of Hsp72 in naked mole-rat supernatants [SNNMR] with a specific small-molecule inhibitor pifithrin-μ attenuates the ability to convey resistance in a dose-dependent manner.
(A) In both reconstituted naked mole-rat samples (Prot + SN; left panel) and human proteasomes [h26S] resuspended in SNNMR (right panel), and then treated with 20μM MG132, the well-preserved chymotrypsin-like [ChT-L] proteasome activity declined with the addition of increasing concentrations of pifithrin-μ. Significant interference with the inhibition resistance was apparent at the 10μ – 50 μM range of pifithrin-μ concentrations. (B) However, pifithrin-μ did not significantly affect activity of the uninhibited proteasome until much higher concentrations (500 μM). Even then, the activity of pifithrin-μ - treated proteasomes was not lower than activity of the h26S in buffer alone.
FIGURE 10. The HSP72-specific inhibitor, VER155008, did not affect inhibition resistance.
Titration of SNNMR with VER155008 did not disrupt the inhibition resistance of the human or naked mole-rat proteasome challenged with MG132.
The potential actions of HSP72 and HSP40, as described above, differ markedly from the well-established roles of these molecular chaperones in proteostasis. HSP72 and HSP40 are known to participate in chaperone mediated autophagy [CMA], protein refolding, the prevention of protein aggregation as well as the unfolding and transport of damaged proteins for proteasome-mediated degradation (36). Moreover, it is well known that HSP40 commonly co-localizes with HSP72, and that HSP40 regulates ATP-dependent HSP72 activity (37). However, no HSP has been previously shown to stimulate proteasome activity or for that matter any other protease. A previous study has shown that increased expression of HSP40 (Hdj1) can confer proteasome resistance to inhibition after exposure to oxidative stress in an in vitro cell system (38), which supports our in vivo findings. Nevertheless, such findings, that HSPs protect the proteasome (or any other protease) from endogenous or environmental stressors or, even more surprisingly, from the various well-documented proteasome-specific competitive inhibitors that induce their inhibition using different mechanisms of action, in a natural animal has not been documented. Moreover, the role of these HSPs in proteasome modulation and protection from inhibition is independent of ATP, further alluding to a previously undocumented novel mechanism for HSP72 action. The C-terminus sequence of NMR and mouse HSP72 show several areas of weaker homology including deletion of a 16 residue long C-terminal peptide in NMR (Figure S3). Since C-terminal part of HSp 72 is responsible for interaction with substrates and Hsp40, such differences may point at alternate binding partners or different efficiency of substrate binding. Interestingly, a strong presence of retinal dehydrogenase was detected in fractions 22 and 23 (Table S1). Although its detection may be simply result of an abundance of the enzyme, it might also be possible that changes in redox status of NADH may play a key role in activating this chaperone response. This possibility is supported by recent finding that NADH binds to the 26S proteasome without ATP (39) and so could be a part of a larger chaperone-proteasome complex.
Despite the fact that mouse cytosolic lysates contain many of the homologous HSPs, albeit at lower concentrations than those found in the naked mole-rat supernatant, they do not appear to convey any of the proteasome protective properties observed in the naked mole-rat cytosol. This is apparent even when the cytosolic lysates are concentrated, with the exception of a modest activation at the highest concentrations of SNMS used (Figure 4A). It is possible that other macromolecules also contribute to the unique properties of this naked mole-rat cytosolic factor and that this likely forms a complex with the proteasome.
3.5 Morphometric characterization of a complex between 20S proteasome and the resistance factor
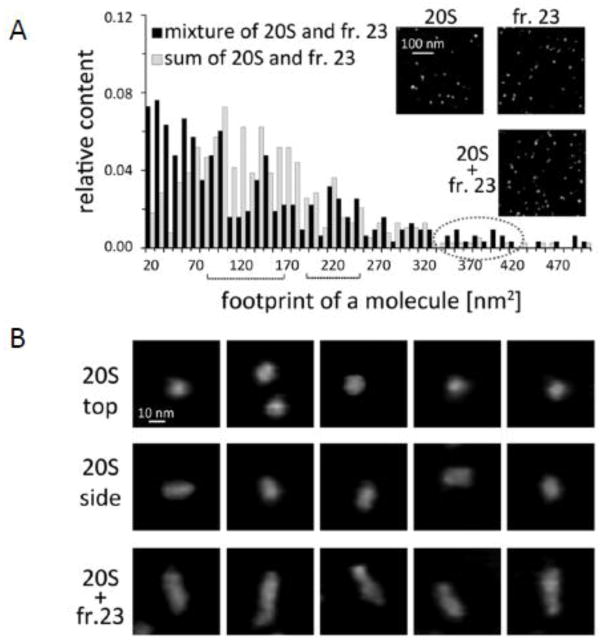
Finally, we used atomic force microscopy [AFM], a nondestructive imaging technology capable of detecting the topography of single native biomacromolecule to see if we could identify distinct complexes of purified human 20S proteasome with the resistance factor present in fr. 23 (Figure 11). As previously found with AFM, the tube-shaped human 20S proteasomes bound to a mica surface in two orientations, “standing” (the majority) and “lying” (24). AFM imaging rendered the standing particles as round cone shaped objects (Figure 11B top row). A small fraction of the 20S proteasomes lay on their side and they were observed as rectangular or slightly oval particles (Figure 11B middle row). AFM images of fr. 23 presented as expected a complex mixture of particles of different sizes that were smaller than proteasomes and devoid of large complexes or aggregates (Figure 11A insert). Following the mixing of purified human 20S proteasomes with the naked mole-rat supernatant fr. 23, AFM produced images of a heterogenous mixture of particles. To determine if the mixture contains a new class of particles besides those found separately in fr. 23 and in the purified 20S proteasome preparation, we performed morphometric analysis of images collected for each investigated case followed by hierarchical cluster analysis. We found that about 5% of the particles in the fr. 23 + 20S proteasome mixture was classified as a distinct new population of elongated (33nm long and 10nm wide) objects (Figure 11A; Figure 11B bottom row). At the same time slightly lower abundance of free proteasomes was also observed. Since we could not consistently find any other larger or uniquely shaped molecules in the mixture or changes in object abundance, the identified particles most likely correspond to complexes of the resistance factor with the core proteasome. The dimensions of the complex may imply that the resistance factor binds to the α ring of 20S proteasome since the length of side view proteasome alone is 15–18 nm (31). Furthermore, the topography of identified complexes indicated that both α faces were saturated with the resistance factor. Likely, half saturated complexes also existed but could have been obscured in the morphometric analyses of AFM images by the presence of other similarly sized protein particles in fr. 23. These exciting data support our premise that the cytosolic factor complexes with proteasomes and thereby both modulates their activity and protects them from inhibiting agents.
FIGURE 11. Atomic force microscopy [AFM] imaging identified putative complexes between proteasome and resistance factor.
(A) Frequency histogram of footprint area of the AFM detected particles in a mixture of purified h20S proteasome with a gel filtration fr. 23 containing resistance factor (black columns) and algebraic sum of the particle counts obtained separately with purified h20S proteasome and fr. 23 (gray columns). Inserts show AFM images of particle fields representing h20S, fr. 23 and their mixture (B) A gallery of representative images of particles zoomed-in from the fields of h20S (top-view and side-view) and h20S – fr. 23 mixture. The elongated molecules representing presumed proteasome-resistance factor complexes form the new class of particles, which appeared only when h20S was mixed with fr. 23. Tapping mode in liquid was used to collect height images (see Methods). The grey scale in all AFM images represents height of the particles, with black color corresponding to the background (0 nm) and white color corresponding to 20 nm. A gray dot line oval marks bins comprising a new class of particles present only in the mixture and centered around 360–380 nm2 (3B, bottom row). Black braces indicate ranges of area sizes characteristic for upright (A, left brace; B top row) and lying side view (A, right brace B middle row) h20S. Between 200 and 300 particles were analyzed for each case. Relative abundance of the new complexes in the mixture was 4.7% and average size of particles was 33x10nm (length x width). Inserts show fragments of AFM images of fields with respective particles.
4. CONCLUSIONS
Clearly, the preternaturally long-lived naked mole-rats have evolved certain molecular mechanisms that contribute to their ability to prolong good health and attenuate the aging process (1). The high proteasome content coupled with its distinctive composition in naked mole-rats, that we previously described may play an integral role in this regard (18). However, we describe here another important, complementary mechanism. We report here for the first time that naked mole-rats express high levels of key chaperones, HSP72, HSP40, and HSP25 even in untreated tissues when compared to those of the mouse. Further we present evidence suggesting the presence of a novel cytosolic factor that contains two of these chaperones, and that not only protects proteasome function against cell stressors but also enhances proteasome performance. This factor may be a common constitutive feature of long-lived species, or possibly may be induced under specific stress conditions in both naked mole-rats and other organisms. Our finding of a transferable stable factor that protects a critical intracellular proteolytic system may have profound therapeutic significance. We envision that this may guard against the many age-related diseases linked to a dysfunction in proteostasis and the concomitant accrual of protein aggregates, such as occurs in Alzheimer’s and Parkinson’s diseases. This cytosolic factor may also ameliorate the well-documented decrease in proteasome activity with age (13,14,40,41) and if used therapeutically may thus promote prolonged healthspan and longevity in our own aging population.
Supplementary Material
Highlights.
The naked mole-rat proteasome is protected from inhibition
This is accomplished by a transferable chaperone-containing cytosolic factor.
The factor interacts with naked mole-rat proteasome enhancing its activity.
This factor protects and increases proteasome function of widely divergent species.
The factor participates in a novel mechanism that may help to defy cellular aging.
Acknowledgments
We thank Drs. Steve Austad, James Nelson and Shane Rea for their constructive criticism of the earlier versions of this manuscript; Kaitlyn Lewis, Megan Smith and the LAR at UTHSCSA who were responsible for the care and maintenance of the naked mole-rat colony. Mass spectrometry analyses were conducted in the UTHSCSA Institutional Mass Spectrometry Laboratory by Kevin W. Hakala and Sam Pardo. KAR was supported by a Training Grant through the NIH/NIA (T32 AG021890) and an American Federation for Aging Research (AFAR)/Ellison Postdoctoral Fellowship to continue this work under the mentorship of RB. This work was also funded by awards to RB from the NIH/NIA (1R21AG043912), Glenn Foundation, the Ellison Medical Foundation and Breakthroughs in Gerontology from the AFAR.
Abbreviations
- Adh(VS)
adamantane-acetyl-(6-aminohexanoyl)3-(leucinyl)3-vinyl-(methyl)-sulfone
- BZ
bortezomib
- ChTL
chymotrypsin-like
- FT
filtrate
- HSP
heat shock protein
- IC50
concentration required to ablate 50% of activity
- LC
lactacystin
- PRSMS
mouse partially-purified proteasome 5h pellet
- SNMs
mouse supernate MG132, N-(benzyl-oxycarbonyl) leucinyl-leucinal
- NMR
naked mole-rat
- PRSNMR
NMR partially-purified proteasome 5h pellet
- SNNMR
NMR supernate
- PGPH
post-glutamyl peptide hydrolyzing
- PRS
proteasome
- RT
retentate
- SN
supernate
- TL
trypsin-like
- Ub
ubiquitin
- UPS
ubiquitin proteasome system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B. 2008;178:439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- 2.Andziak B, Buffenstein R. Disparate patterns of age-related changes in lipid peroxidation in long-lived naked mole-rats and shorter-lived mice. Aging Cell. 2006;5:525–532. doi: 10.1111/j.1474-9726.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- 3.Buffenstein R. The naked mole-rat; a new long-living model for human aging research. J Gerontol Biol Sci. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- 4.Salmon AB, Akha AAS, Buffenstein R, Miller RA. Fibroblasts from naked mole-rats are resistant to multiple forms of cell injury, but sensitive to peroxide, ultraviolet light, and endoplasmic reticulum stress. J Ger Biol Sci Med Sci. 2008;63:232–241. doi: 10.1093/gerona/63.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis KN, Mele J, Hornsby PJ, Buffenstein R. Stress Resistance in the Naked Mole-Rat: The Bare Essentials - A Mini-Review. Gerontology. 2012;58:453–462. doi: 10.1159/000335966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey YH, Friguet B, Ward W, Richardson A, Chaudhuri A. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci USA. 2009;106:3059–3064. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morimoto RI, Cuervo AM. Protein Homeostasis and Aging: Taking Care of Proteins From the Cradle to the Grave. J Ger Biol Sci Med Sci. 2009;64:167–170. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 9.Grimm S, Hohn A, Grune T. Oxidative protein damage and the proteasome. Amino acids. 2012;42:23–38. doi: 10.1007/s00726-010-0646-8. [DOI] [PubMed] [Google Scholar]
- 10.Hanna J, Finley D. A proteasome for all occasions. FEBS Lett. 2007;581:2854–2861. doi: 10.1016/j.febslet.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwickl P, Seemuller E, Kapelari B, Baumeister W. The proteasome: a supramolecular assembly designed for controlled proteolysis. Adv Protein Chem. 2001;59:187–222. doi: 10.1016/s0065-3233(01)59006-3. [DOI] [PubMed] [Google Scholar]
- 12.Jung T, Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 2008;60:743–752. doi: 10.1002/iub.114. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez KA, Gaczynska M, Osmulski PA. Molecular mechanisms of proteasome plasticity in aging. Mech Ageing Dev. 2010;131:144–155. doi: 10.1016/j.mad.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrington DA, Husom AD, Thompson LV. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005;19:644–646. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- 15.Chondrogianni N, Petropoulos I, Franceschi C, Friguet B, Gonos ES. Fibroblast cultures from healthy centenarians have an active proteasome. Exp Gerontol. 2000;35:721–728. doi: 10.1016/s0531-5565(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 16.Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, Yim SH, Zhao X, Kasaikina MV, Stoletzki N, Peng C, Polak PXZ, Kiezun A, Zhu Y, Chen Y, Kryukov GV, Zhang Q, Peshkin L, Yang L, Bronson RT, Buffenstein R, Wang B, Han C, Li Q, Chen L, Zhao W, Sunyaev SR, Park TJ, Zhang G, Wang J, Gladyshev VN. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu C, Li Y, Holmes A, Szafranski K, Faulkes CG, Coen CW, Buffenstein R, Platzer M, de Magalhaes JP, Church GM. RNA sequencing reveals differential expression of mitochondrial and oxidation reduction genes in the long-lived naked mole-rat when compared to mice. PLoS One. 2011;6:e26729. doi: 10.1371/journal.pone.0026729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez KA, Edrey YH, Osmulski P, Gaczynska M, Buffenstein R. Altered composition of liver proteasome assemblies contributes to enhanced proteasome activity in the exceptionally long-lived naked mole-rat. PLoS One. 2012;7:e35890. doi: 10.1371/journal.pone.0035890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis KN, Andziak B, Yang T, Buffenstein R. The Naked Mole-rat Response to Oxidative Stress: Just Deal With it. Antioxid Redox Signal. 2012;19:1388–1399. doi: 10.1089/ars.2012.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickering AM, Davies KJ. Differential roles of proteasome and immunoproteasome regulators Pa28alphabeta, Pa28gamma and Pa200 in the degradation of oxidized proteins. Arch Biochem Biophys. 2012;523:181–190. doi: 10.1016/j.abb.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto M, Jepsen KJ, Terranova CJ, Buffenstein R. Lack of sexual dimorphism in femora of the eusocial and hypogonadic naked mole-rat: a novel animal model for the study of delayed puberty on the skeletal system. Bone. 2010;46:112–120. doi: 10.1016/j.bone.2009.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cvek B, Dvorak Z. The ubiquitin-proteasome system (UPS) and the mechanism of action of bortezomib. Current pharmaceutical design. 2011;17:1483–1499. doi: 10.2174/138161211796197124. [DOI] [PubMed] [Google Scholar]
- 23.Kisselev AF, Callard A, Goldberg AL. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J Biol Chem. 2006;281:8582–8590. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- 24.Gaczynska M, Osmulski PA. Atomic force microscopy of proteasome assemblies. Methods Mol Biol. 2011;736:117–132. doi: 10.1007/978-1-61779-105-5_9. [DOI] [PubMed] [Google Scholar]
- 25.Kessler BM, Tortorella D, Altun M, Kisselev AF, Fiebiger E, Hekking BG, Ploegh HL, Overkleeft HS. Extended peptide-based inhibitors efficiently target the proteasome and reveal overlapping specificities of the catalytic beta-subunits. Chemistry & biology. 2001;8:913–929. doi: 10.1016/s1074-5521(01)00069-2. [DOI] [PubMed] [Google Scholar]
- 26.de Bettignies G, Coux O. Proteasome inhibitors: Dozens of molecules and still counting. Biochimie. 2010;92:1530–1545. doi: 10.1016/j.biochi.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- 28.Gaczynska M, Rock KL, Goldberg AL. Gamma-Interferon and Expression of MHC Genes Regulate Peptide Hydrolysis by Proteasomes. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 29.Cascio P, Goldberg AL. Preparation of hybrid (19S-20S-PA28) proteasome complexes and analysis of peptides generated during protein degradation. Methods Enzymol. 2005;398:336–352. doi: 10.1016/S0076-6879(05)98028-2. [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, Joo HJ, Kim YH, Ahn S, Chang J, Hwang KB, Lee DH, Lee KJ. Systemic analysis of heat shock response induced by heat shock and a proteasome inhibitor MG132. PLoS One. 2011;6:e20252. doi: 10.1371/journal.pone.0020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salway KD, Gallagher EJ, Page MM, Stuart JA. Higher levels of heat shock proteins in longer-lived mammals and birds. Mech Ageing Dev. 2011;132:287–297. doi: 10.1016/j.mad.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Krivoruchko A, Storey KB. Forever young: mechanisms of natural anoxia tolerance and potential links to longevity. Oxidative medicine and cellular longevity. 2010;3:186–198. doi: 10.4161/oxim.3.3.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey AJ, Williamson DS, Browne H, Murray JB, Dokurno P, Shaw T, Macias AT, Daniels Z, Geoffroy S, Dopson M, Lavan P, Matassova N, Francis GL, Graham CJ, Parsons R, Wang Y, Padfield A, Comer M, Drysdale MJ, Wood M. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer chemotherapy and pharmacology. 2010;66:535–545. doi: 10.1007/s00280-009-1194-3. [DOI] [PubMed] [Google Scholar]
- 35.Dehghani M, Xiao CF, Money TGA, Shoemaker KL, Robertson RM. Protein expression following heat shock in the nervous system of Locusta migratoria. Journal of Insect Physiology. 2011;57:1480–1488. doi: 10.1016/j.jinsphys.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 36.Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Q, Keller JN. Proteasome inhibition in oxidative stress neurotoxicity: implications for heat shock proteins. J Neurochem. 2001;77:1010–1017. doi: 10.1046/j.1471-4159.2001.00302.x. [DOI] [PubMed] [Google Scholar]
- 39.Tsvetkov P, Myers N, Eliav R, Adamovich Y, Hagai T, Adler J, Navon A, Shaul Y. NADH Binds and Stabilizes the 26S Proteasomes Independent of ATP. J Biol Chem. 2014;289:11272–11281. doi: 10.1074/jbc.M113.537175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dasuri K, Nguyen A, Zhang L, Fernandez-Kim SO, Bruce-Keller AJ, Blaloc BA, deCabo R, Keller JN. Comparison of Liver and Brain Proteasomes for Oxidative Stress Induced Inactivation: Influence of Aging and Dietary Restriction. Free Rad Res. 2009;43:28–36. doi: 10.1080/10715760802534812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dasuri K, Zhang L, Ebenezer P, Fernandez-Kim SO, Bruce-Keller AJ, Szweda LI, Keller JN. Proteasome alterations during adipose differentiation and aging: links to impaired adipocyte differentiation and development of oxidative stress. Free Radic Biol Med. 2011;51:1727–1735. doi: 10.1016/j.freeradbiomed.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.