Abstract
Summary
Bipolar disorder (BPD) is a highly heritable polygenic disorder. Recent enrichment analyses suggest that there may be true risk variants for BPD among the expression quantitative trait loci (eQTL) in the brain.
Aims
We sought to assess the impact of eQTL variants on BPD risk by combining data from both BPD genome-wide association study (GWAS) and brain eQTL.
Method
To detect single-nucleotide polymorphisms (SNPs) that influence expression levels of genes associated with BPD, we jointly analyzed data from a BPD GWAS (7,481 cases and 9,250 controls) and a genome-wide brain (cortical) eQTL (193 healthy controls) using a Bayesian statistical method, with independent follow-up replications. The identified risk SNP was then further tested for association with hippocampal volume (N=5,775) and cognitive performance (N=342) among healthy subjects.
Results
Integrative analysis revealed a significant association between a brain eQTL rs6088662 in 20q11.22 and BPD (Log Bayes Factor=5.48; BPD p-val=5.85×10−5). Follow-up studies across multiple independent samples confirmed the association of the risk SNP (rs6088662) with gene expression and BPD susceptibility (p-val=3.54×10−8). Further exploratory analysis revealed that rs6088662 is also associated with hippocampal volume and cognitive performance in healthy subjects.
Conclusions
Our findings suggest that 20q11.22 is likely a risk region for BPD, highlighting the informativeness of integrating functional annotation of genetic variants for gene expression in advancing our understanding of the biological basis underlying complex diseases such as BPD.
Bipolar disorder (BPD) is a severe, chronic psychiatric disorder with worldwide lifetime prevalence ranging from 0.5–1.5%.1 BPD is characterized by a variety of profound mood symptoms including episodes of mania, hypomania and depression, and is often accompanied by psychotic features and cognitive deficits. To date, there has been a fair amount of data from family and twin studies to highlight a strong genetic predisposition for BPD.1 That said, BPD is a highly polygenic disorder that can vary substantially from population to population. While linkage analyses and genetic association studies have yielded numerous candidate variants for BPD, only a few of these have been satisfactorily replicated across independent samples.2,3
With the advances in knowledge of human genetic variations—such as data generated by the HapMap and 1000-Human-Genome projects and several subsequent genome-wide association studies (GWASs) by a number of international collaborators—a wealth of novel susceptible variants for BPD have been reported, particularly SNPs in CACNA1C, ANK3, ODZ4, NCAN and TRANK1.4–8 These GWAS-identified risk SNPs unfortunately only account for a small portion of the genetic risk for BPD, which suggests there should be additional loci contributing to the genetic susceptibility. Previous aggregated analyses indicated there may be valid risk loci underlying genetic markers passing only nominal significance in the GWASs,9 a possibility confirmed by several later studies. For example, a number of schizophrenia (SCZ) and BPD susceptibility SNPs did not reach genome-wide significance in initial GWAS samples, but showed consistent replications in subsequent independent samples, thus implying that these loci might reflect weak but true risk signals.10
Genetic loci associated with clinical diagnosis also are expected to be related to so-called intermediate phenotypes implicated in the biology of genetic risk for BP disorder. Previous studies have reported hippocampal dysfunction (e.g., memory impairment) in BPD patients and their unaffected relatives, implying that variation in hippocampal biology is an intermediate phenotype related to the genetic risk of BPD.11 In addition, smaller hippocampal volume has been reported in BPD patients.12,13 Meanwhile, functional neuroimaging studies have revealed that dysfunctions of hippocampus and its closely related regions underpin abnormal affective responses and dysfunctional emotion regulation in BPD.14 Finally, postmortem studies further support the hypothesis that hippocampal abnormalities are relevant to the altered synaptic plasticity and diminished resilience in BPD.15 Therefore, analysis of the BPD-associated SNPs on these hippocampus-related phenotypes may provide a plausible way to uncover their functions in neurodevelopment, and possibly, their involvements in disease susceptibility.
Recent successes in integrating disease GWAS and gene expression data for several other complex diseases have been promising,16–18 and we wondered if such an approach may yield novel results for BPD. Predictably, several lines of evidence have suggested an enrichment of expression quantitative trait loci (eQTL) among BPD susceptibility SNPs in the brain,19 further highlighting the importance of integrating functional annotation of genetic variants for gene expression to advance our understanding of the biological bases of BPD. In light of these findings, we integrated a BPD GWAS data from 16,731 individuals and a genome-wide eQTL data from 193 normal human brains, followed by a set of independent replications on both eQTL and disease associations.
METHOD
Discovery brain eQTL and BPD GWAS datasets
The brain eQTL dataset used in this study was reported previously.20 In brief, after excluding ethnic outliers and samples that were possibly related, a total of 193 independent healthy old (age>65) human cortex samples of European origin were included in the eQTL analysis. Detailed information about genotyping and expression profiling as well as statistical methods can be found in the Supplemental Material or the original publication.20
For the BPD GWAS data, the Psychiatric Genomics Consortium (PGC) BPD working group recently conducted a meta-analysis of large-scale genome-wide data on BPD among populations of European descent (PGC1 GWAS).6 In this prior study, they opted to compare BPD patients that had experienced pathologically relevant episodes of elevated mood (mania or hypomania) and control subjects from the same geographic and ethnic populations. In sum, we utilized 2,117,872 SNPs across the genome from the GWAS samples (7,481 cases and 9,250 controls), and the association significance (P-value) for these SNPs were downloaded from the PGC1 data sharing website (https://pgc.unc.edu/Sharing.php#SharingOpp). Detailed descriptions of the samples, data quality, genotype imputation, genomic controls and statistical analyses can be found in the original GWAS report.6
Integrative analysis of eQTL and BPD GWAS data
We integrated the eQTL and BPD GWAS data using a Bayesian statistical framework. Statistical analyses for the eQTL and BPD GWAS was achieved by using the program Sherlock (http://sherlock.ucsf.edu/), which has been described elsewhere.17 In brief, Sherlock is based on the rationale that a risk gene for the disease may have at least one eQTL, and these eQTL could alter gene expression, which in turn affects disease susceptibility. Given the probability that this is true, there should be a significant overlap of the eQTL of a gene and the loci associated with the disorder, which would imply a likely functional role for the gene in that particular disease. At this juncture, Sherlock aligns the eQTL and BPD GWASs and only considers the shared SNPs in both datasets. Sherlock's scoring rubric both increases the total gene score for overlapping SNPs and provides a penalty in the absence of an overlap, though associations found only in the BPD GWAS do not alter the score. Sherlock computes individual Log Bayes Factors (LBFs) for each SNP pair in the alignment, and the sum of these constitutes the final LBF score for each gene.
Brain eQTL data for replication analysis
Considering that bipolar disorder is a mental disorder that reasonably originates from abnormal brain functions, brain samples are presumably appropriate for replication test of the eQTL results. We first utilized a brain DLPFC (dorsolateral prefrontal cortex) sample (N=320) consisting of healthy controls in Caucasians and African Americans (named as “first replication sample”), in which the sample has been previously used to identify psychiatric risk transcripts.21–23
We also used other well-characterized brain expression databases for replication analysis of the eQTL associations. A brief description of the gene expression resources is provided below; more detailed information can be found in the original studies.18,24–26 (1) BrainCloud: BrainCloud contains genetic information and whole transcriptome expression data from postmortem DLPFC of 261 normal human subjects in Caucasians and African Americans. The data in BrainCloud is aimed at exploring temporal dynamics and genetic control of transcription across lifespan.24 Of note, there is partial overlap between BrainCloud data and our “first replication sample”. (2) Data from Webster et al: This report studied whole-genome transcriptome and genome in a series of neuropathologically normal postmortem samples and a confirmed pathologic diagnosis of late-onset Alzheimer disease (LOAD; final N=188 controls, 176 cases), and suggested that studying the transcriptome as a quantitative endophenotype has greater power for discovering risk SNPs influencing expression than the use of discrete diagnostic categories such as presence or absence of disease.25 It should be noted that the control sample in this study was the same as our discovery brain eQTL sample.20 (3) SNPExpress: The authors analyzed genome-wide SNPs that were associated with gene expression in human primary cells at the exon level, using Affymetrix exon arrays, evaluating 93 autopsy-collected cortical brain tissue samples with no defined neuropsychiatric condition.26 (4) Data from Zou et al: They measured expression levels of 24,526 transcripts in brain samples from the cerebellum and temporal cortex of autopsied subjects with Alzheimer’s disease (AD, cerebellar n = 197, temporal cortex n = 202), and conducted a genome-wide expression association study (eGWAS) using 213,528 cis-SNPs within 6,100 kb of the tested transcripts.18 Their results demonstrated the significant contributions of genetic factors to human brain gene expression, which are reliably detected across different brain regions, and also implicated that combined assessment of expression and disease GWAS may provide complementary information in discovery of human disease variants with functional implications.18
BPD samples for replication analysis
Replication analyses on BPD were conducted in two steps (replication-I and II), examining a total of 6,056 BPD cases and 46,614 controls from ten different geographic locations. Detailed information on each sample—including diagnostic assessment, genotyping method and quality control—are shown in the Supplemental Data and Table S1.
Briefly, the BPD samples used in our replication included: (1) Germany II (181 cases and 527 controls);5 (2) Germany III (490 cases and 880 controls);5 (3) Australia (330 cases and 1,811 controls);5 (4) France (451 cases and 1,631 controls);2 (5) Sweden I (836 cases and 2,093 controls);6 (6) Sweden II sample (1,415 cases and 1,271 controls);6 and (7) Iceland (541 cases and 34,546 controls);6 (8) Romania (244 cases and 174 controls),5 and (9) China (350 cases and 888 controls).27 For replication-II, we used a United Kingdom sample (1,218 cases and 2,913 controls).28 The ten samples from replication-I and II showed no overlap with the PGC1 BPD samples.6 Each of the original studies was conducted under appropriate ethical approvals, and written informed consents were obtained from all subjects.
Samples for analysis of hippocampal volume and cognitive performance
For analysis of hippocampal volume, we utilized the data from a recent GWAS conducted by the Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) consortium.29 The GWAS includes a total of 5,775 young healthy individuals (mean age: 34.8 years). Detailed information on the samples, imaging procedures, genotyping methods and statistical analysis can be found in the original GWAS report.29
For analysis of cognitive performance, we used a Chinese sample that included 342 healthy Chinese college students from Beijing Normal University who had self-reported no known history of any neurological or psychiatric disorders (197 females and 145 males, aged 18–23). Cognitive and behavioral measures included working memory, executive functions (as assessed with the Attention Network Test, the Wisconsin Card Sorting Task, and a reversal learning test), and motivation traits etc., which were shown in Table S2. This experiment was approved by the Institutional Review Board of the State Key Laboratory of Cognitive Neuroscience and Learning at Beijing Normal University, China. Written informed consent was obtained from all participants following a full explanation of the study procedure.
Statistical analysis
For the replication analysis on BPD, genomic control was used to correct for relatedness and population stratification in each sample,30 and association P-values and allele-specific odds ratios (ORs) for each individual sample were calculated by a logistic regression model with an additive effect using a lambda value (genomic control) as a covariate to adjust for potential population stratification. Meta-analyses were then conducted based on Z-scores by combining data from different samples in the R package (meta module) using the Mantel-Haenszel method under the fixed effects model. As described in previous GWAS meta-analysis,6 P-values for replication samples are reported as one-tailed tests, while P-values for all combined samples are shown as two-tailed tests. We used a forest plot to graphically present the individual OR and their 95% confidence interval, i.e., each sample was represented by a square in the forest plot. For the analyses on cognitive performance, two-tailed t-tests were conducted with SPSS 16.0 (SPSS, Chicago, USA).
To explain the logic of the study design, a flow chart about the analytical methods and how variants were taken forward from one stage of analysis to the next was shown in Figure 1. All protocols and methods used in this study were approved by the institutional review board of Kunming Institute of Zoology, Chinese Academy of Sciences and adhere to all relevant national and international regulations.
Figure 1. Flow chart of the present study.
Based on the hypothesis that BPD risk variants are enriched among eQTL, we systematically integrated BPD GWAS and genome-wide brain eQTL data by using Sherlock. The top genes identified by Sherlock were then replicated in independent BPD samples and eQTL datasets. Finally, the successfully replicated SNP (rs6088662) was further tested for the association with BPD biological phenotypes including hippocampal volume and cognitive performance.
RESULTS
Integrative analysis of eQTL and BPD GWAS data
The Sherlock identified a total 20,942 SNPs showing significant eQTL effects, and also having BPD data (e.g., p-value), and these SNPs were included for further analyses. Using a Bayesian statistical method to match the “signature” of genes from the brain eQTL with patterns of association in the BPD GWAS, we ranked the top candidate genes for BPD risk according to their LBF scores and P-values. Only genes with LBF scores higher than 5.00 were shown and included for further analyses.
The integrative analysis yielded four candidate risk genes (Table S3). The first gene was glycosyltransferase 8 domain containing 1 (GLT8D1, LBF=6.78), located on chromosome 3p21.1 that has been repeatedly reported for association with BPD.31,32 Detailed analysis found that the significant association with this gene was mainly driven by a cis-associated SNP rs2251219. This SNP has already been reported in an earlier GWAS of BPD,32 and was replicated in independent BPD samples (in which their samples overlapped with our replication samples).33–35 The second top-ranked gene was chemokine (C-X-C motif) ligand 16 (CXCL16, LBF=6.16) on chromosome 17p13. To the best of our knowledge, this gene has never been reported in genetic association studies on BPD, and we observed two trans- associated SNPs showing moderate associations with BPD. The third top-ranked gene was TRPC4AP (LBF=5.57) on chromosome 20q11.22, with the significance mainly driven by a cis-associated SNP (rs6088662, p-val=5.85×10−5 with BPD). The last top-ranked gene was TAF11 (TAF11 RNA polymerase II, TATA box binding protein-associated factor, 28kDa) on chromosome 6p21.31, with a trans-correlated SNP (rs4482754) showed significant association with BPD.
Replication of eQTL effects in diverse samples
Given the myriad confounders in single eQTL database, it is important and necessary to validate the eQTL associations in independent samples. The above four candidate genes and their cis- or trans- associated SNPs were followed-up in independent eQTL datasets.
For the cis-SNP rs2251219 and GLT8D1, we observed significant association in one replication sample of AD source (Table S4),25 and a marginal significant association in the BrainCloud sample.24 However, as demonstrated by a previous study,32 the association of rs2251219 with GLT8D1 expression in our discovery eQTL sample (Myers et al. study)20 may be an artifact since the probes overlapped with other common SNPs, and it could not be replicated in the original cDNA samples of our discovery eQTL dataset by quantitative PCR using the probes not overlapping with known SNPs. In addition to GLT8D1, we also analyzed the expression of other nearby genes around rs2251219, however, no promising findings were observed (Table S4). For the significant trans- eQTL associations in our discovery sample, neither CXCL16 nor TAF11 could be validated in any of the replication samples (Table S5), implying they might be generated by chance.
For the cis-association between rs6088662 and TRPC4AP expression, in the discovery eQTL brain sample,20 the risk allele [G] of rs6088662 showed significantly decreased gene expression (p<1.0×10−8, Figure 2A). This pattern was validated in one of the replication samples (p<1.0×10−8 in Webster et al. study),25 but it should be noted that this replication data includes our discovery sample. We therefore re-analyzed the result using the non-overlapped AD patients, and it showed nominally significant association (p=0.023, Figure 2B). However, rs6088662 showed an opposite effect on TRPC4AP expression in our “first replication sample” (risk allele [G] of rs6088662 showed increased gene expression), and in other replication samples, no significant association between rs6088662 and TRPC4AP was observed (Table S6).18,26 These inconsistencies may not be surprising, given a prior report of low-to-moderate overlap between eQTL loci across eQTL studies (the percentage of overlapped eQTL is 0~35.4% between pairwise brain studies, shown in Table 4 of McKenzie et al. study).36 In addition, with the use of several non-brain tissue eQTL databases,37–39 we also observed significant and consistent associations between rs6088662 and TRPC4AP expression (the p-values range from 0.047 to 3.60×10−7, Figure S1–S3).
Figure 2. Rs6088662 is significantly associated with TRPC4AP mRNA expression.
(A) Results in 193 neuropathologically normal human brain (cortical) samples of European subjects. (B) Results in 176 Alzheimer’s disease human brain (cortical) samples of European subjects.
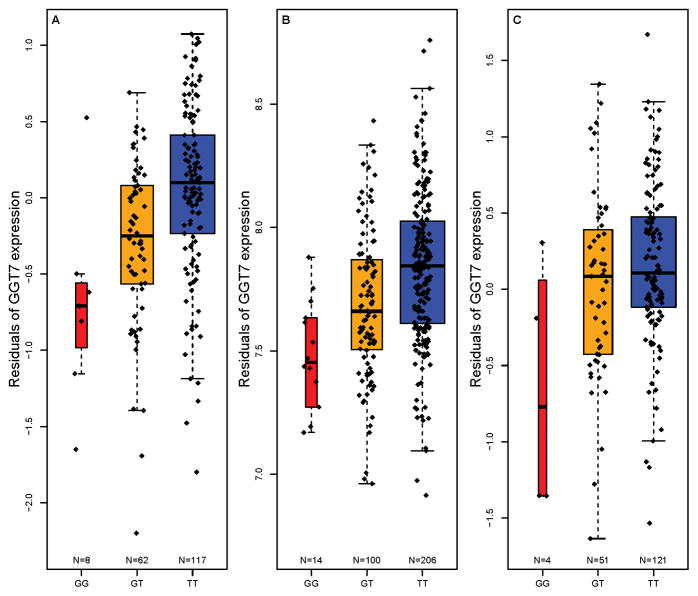
To further dissect if rs6088662 is also associated with the expression of other nearby genes, we screened 14 genes in the 20q11.22 region in both discovery and replication eQTL samples (Table S6). Intriguingly, we observed another gene GGT7 showing significant association in the discovery sample (p<1.0×10−7, Figure 3A), and it remained significant in the “first replication sample” with the same direction of effect (p<1.0×10−8, Figure 3B). In other replication samples, the association is also significant (Webster et al.25 and Zou et al.18 studies, Figure 3C and Table S6) or marginal significant (BrainCloud24), except for Heinzen et al. study (p=0.13),26 however, in the Heinzen et al. sample, rs6088662 still showed one of the strongest associations with GGT7 among the genes in 20q11.22, and the SNP showed significant or marginal significant associations with the expression of several exons in GGT7 (Table S7), which was not observed in most of other nearby genes.
Figure 3. Rs6088662 is significantly associated with GGT7 mRNA expression.
(A) Results in 193 neuropathologically normal human brain (cortical) samples of European subjects. (B) Results in 320 healthy human brain DLPFC samples of Caucasian and African American individuals. (C) Results in 176 Alzheimer’s disease human brain (cortical) samples of European subjects.
For the other genes in 20q11.22, three of them (ACSS2, MYH7B and EDEM2) also showed associations in some of the eQTL samples, but the associations are not consistent, and these genes are unlikely the associated genes (Table S6). To summarize, from the eQTL analyses in both discovery and replication samples, we demonstrated that rs6088662 is likely an authentic eQTL SNP, and we found two potential genes (GGT7 and TRPC4AP) showing association with this risk SNP.
Rs6088662 is associated with bipolar disorder across cohorts
Given the replication of significant associations between rs6088662 and TRPC4AP expression, we opted to pursue further analysis of this SNP on BPD risk. In the stage I replication analysis, rs6088662 was analyzed in nine independent case-control samples. Although the association between rs6088662 and BPD did not achieve even nominal significance (p=0.05) in any single cohort, it does show a trend of association in Germany II and Sweden II samples (p=0.08 and p=0.07, respectively). In a Chinese sample, there is no difference in allele frequencies of this SNP between Han Chinese and Europeans (0.165 versus 0.171 for G allele), and the effect size (OR) in the Chinese sample was even higher than in our discovery sample (1.17 versus 1.12), the non-significant result being likely due to the limited sample size. When all the replication-I samples were combined, the association p-value reached nominal significance level (p=4.95×10−2), with the OR being 1.06 (95% CI=0.99–1.13), consistent with the discovery PGC1 GWAS. There is no significant heterogeneity among the replication-I samples (p=0.77), and detailed results for each individual sample were shown in Table 1. The forest plot of the meta-analysis on all replication-I samples is presented in Figure 4.
Table 1.
Summary of logistic regression results for rs6088662 across cohorts.
| Sample | Ethnicity | Cases | Controls | Effect Allele | Additive P-value | Odds ratio | 95% CI | Data source |
|---|---|---|---|---|---|---|---|---|
| Discovery | ||||||||
| PGC1 | Europeans | 7,481 | 9,250 | G | 5.85×10−5 | 1.12 | 1.06–1.19 | 6 |
| Replication-I | ||||||||
| Germany II | German | 181 | 527 | G | 0.08 | 1.23 | 0.91–1.65 | this study |
| Germany III | German | 490 | 880 | G | 0.16 | 1.09 | 0.90–1.33 | this study |
| Australia | Australian | 330 | 1,811 | G | 0.29 | 1.07 | 0.85–1.33 | this study |
| France | French | 451 | 1,631 | G | 0.42 | 1.02 | 0.85–1.22 | this study |
| Sweden I | Swedish | 836 | 2,093 | G | 0.37 | 1.02 | 0.89–1.18 | this study |
| Sweden II | Swedish | 1,415 | 1,271 | G | 0.07 | 1.12 | 0.97–1.29 | this study |
| Iceland | Icelandic | 541 | 34,426 | G | 0.19 | 0.93 | 0.79–1.10 | this study |
| Romania | Romanian | 244 | 174 | G | 0.42 | 1.04 | 0.74–1.46 | this study |
| China | Han Chinese | 350 | 888 | G | 0.11 | 1.17 | 0.91–1.50 | this study |
| All replication-I samples | 4,838 | 43,701 | G | 4.95×10−2 | 1.06 | 0.99–1.13 | ||
| Replication-II | ||||||||
| UK | British | 1,218 | 2,913 | G | 1.06×10−6 | 1.34 | 1.19–1.51 | 28 |
| Discovery + Replications | 13,537 | 55,864 | G | 3.54×10−8 | 1.12 | 1.07–1.16 | ||
Test of heterogeneity
All replication-I cohorts: p=0.77, I2=0%; meta-analysis was conducted under fixed effect model
Discovery + Replication samples: p=0.07, I2=41.8%; meta-analysis was conducted under fixed effect model
Abbreviations:
CI, confidence interval.
Note:
P-values are two-sided for the discovery cohort and combined analysis; one-sided P-values are listed for the replication-I samples
Figure 4. Forest plot of odds ratios with 95% confidence interval for total replication-I bipolar disorder samples included in meta-analysis of rs6088662.

The G allele of rs6088662 is overrepresented in BPD cases in all of the tested cohorts (except for the Iceland sample).
Notably, a previous study28 has reported a significant association of a proxy SNP of rs6088662 (rs13041792, r2=1.00 with rs6088662 in Europeans) with BPD in an independent UK sample (1,218 cases and 2,913 controls), which is in concordant with our results and was also included in our analysis, denoted as “replication-II” sample. Meta-analysis by combining PGC1 GWAS, replication-I and replication-II samples yielded a genome-wide significant association of rs6088662 with BPD (p=3.54×10−8, OR=1.12, 95% CI=1.07–1.16, Table 1). We used the fixed effect model for meta-analysis because there was no significant heterogeneity among the samples (p>0.05).
Considering the genetic overlap between BPD and other psychiatric disorders 1, we also tested the association of rs6088662 with two other mental disorders, SCZ and major depressive disorder (MDD). It showed nominally significant association with SCZ in the latest PGC2 GWAS (p=0.0037, OR=1.04, 95% CI=1.00–1.08, N=35,476/46,839),40 however, it did not show any significant associations with MDD when using data from the PGC1 MDD GWAS plus Colaus study samples (10,541/11,208) (Table S8),41,42 implying that rs6088662 is likely a psychosis risk SNP rather than a risk SNP for a broader spectrum of mood disorders.
A proxy search for SNPs in high LD with rs6088662 was performed on the SNAP website (http://www.broadinstitute.org/mpg/snap/ldsearch.php) with the European panel from the 1000 Genomes Project (pilot 1) dataset. This identified 43 SNPs in high LD (r2>0.8) with rs6088662, all of which are located within MYH7B and TRPC4AP regions (Figure 5). Among these, there are 1 non-synonymous SNP, 3 synonymous SNPs, 1 SNP in the 5’ untranslated region (UTR), and 1 SNP located in the non-coding RNA (ncRNA) region (Table S9). However, to identify causal variants for BPD, further studies are needed.
Figure 5.
Plot of chromosome region showing a genomic area of high linkage disequilibrium with rs6088662 in European populations.
Rs6088662 is associated with hippocampal volume and cognitive performance
To move beyond statistical association with clinical diagnosis and to obtain convergent evidence for association between rs6088662 and BPD-related biology, we also performed a series of convergent experiments testing risk-associated SNPs on several intermediate biological phenotypes. The hippocampus is a subcortical brain region frequently reported to show dysfunction among BPD patients.18,23 We therefore hypothesized that if the identified risk-associated SNP (e.g., rs6088662) affects the anatomy or function of this brain region, then related cognitive deficits, regardless of illness status should be associated with it. In an exploratory manner, we tested the effects of rs6088662 on the biological phenotypes related to the hippocampus (hippocampal volume and cognitive performance) in healthy subjects.
In the ENIGMA sample, rs6088662 was significantly associated with hippocampal volume across multiple cohorts (p=0.00063, β=27.29 mm3, Table S10), supporting the prior speculation that the BPD-associated SNPs will likely affect hippocampal structure, but detailed analysis found that the risk allele [G] led to larger volume. As a post hoc exploratory test, we then investigated the potential impacts of rs6088662 on cognitive performance, and found that rs6088662 showed nominally significant association with executive functions (the alert attention task) (p=0.0094, Table S11) and language abilities (visual-auditory) (p=0.012, Table S11). Again, the risk allele [G] indicated a better cognitive performance.
Analysis on BPD-related phenotypes further confirmed the role of the risk SNPs in BPD susceptibility and implied it may be functional in the brain. However, as the association results on these intermediate phenotypes (especially for cognitive performance) may not survive multiple correction, further validation in larger samples are needed. In addition, the discrepancy of allelic directionality between clinical diagnosis and intermediate phenotypes suggests that the molecular mechanism at work may be more complicated than what we had initially expected when undertaking this study.
DISCUSSION
Findings relating to 20q11.22 region
In this study, with an integrative analysis on both expression and BPD data, we identified a potential risk region 20q11.22 for BPD, although it is still unclear which SNPs are actually responsible. This genomic region contains an extensive area of high LD spanning ~276 kb, including at least 5 protein coding genes (Figure 5). Of the 43 common SNPs in high LD (r2>0.8) with rs6088662, there is one non-synonymous SNP, three synonymous SNPs, one SNP in the 5’ UTR area of genes, and one SNP located in the ncRNA region, all of which are potentially functional but as of yet unknown roles (Table S9).
We found a nominally significant association of BPD-risk SNPs with hippocampal volume and cognitive performance, which is consistent with the prevalent perspective that many BPD-related genes also affect brain structures and cognitive functions. Rather perplexingly though, the risk allele of rs6088662 actually seemed to be associated with larger hippocampal volume and better cognition, running entirely opposite to the conventional view that risk alleles generally lead to smaller hippocampal volume and worse cognition. One potential speculative explanation is that the risk genes (GGT7 and/or TRPC4AP) may play diverse roles in neural development, and the SNP has pleiotropic effects--some detrimental and some beneficial. Another possible explanation is that gene-behavior association differs by diagnosis status, as previous studies also reported other similar situations: for example, the psychosis risk allele of rs1344706 in ZNF804A is associated with better cognitive performance in SCZ patients seen in two independent samples.43,44 Likewise, another psychosis risk SNP (rs1006737) in CACNA1C was shown to be associated with larger gray matter volume for those with the risk allele.45,46
Additional evidence of GGT7 and TRPC4AP in bipolar disorder
TRPC4AP is known to be a substrate-specific adapter of a DCX (DDB1-CUL4-X-box) E3 ubiquitin-protein ligase complex required for cell cycle control, and GGT7 is a member of a gene family that encodes enzymes involved in metabolism of glutathione and in the transpeptidation of amino acids, however, their roles in susceptibility to BPD are still unclear. Here we studied the spatial expression profiling of GGT7 and TRPC4AP in multiple human tissues to see if they are enriched in brain tissues, as BPD is a mental disorder that mainly originates from abnormal brain function, and if these genes are preferentially expressed in brain, which would make more sense of taking them as potential risk genes for BPD. We used the expression data from Genotype-Tissue Expression project (GTEx),47 in which 3,797 tissues from 150 post-mortem donors have been collected and subsequently analyzed using a RNA sequencing (RNA-seq) based gene expression approach. Notably, we found GGT7 is abundantly expressed in human brain tissues, such as cerebellum (Figure S4-A), while the expression level of GGT7 is generally low in non-neural tissues. However, the expression of TRPC4AP in brains is relatively lower than other tissues (Figure S4-B), but this gene has been previously reported in association with Alzheimer’s disease (AD),48,49 a neurological disorder showing a high comorbidity with affective disorders (such as BPD and MDD) in geriatric populations.50
Implications
Alongside our specific findings for genetic susceptibility to BPD, our results highlight several advantages of convergent analysis using BPD and eQTL GWAS datasets (Figure 1) over conventional analytical strategies aimed at uncovering susceptibility genes. First, analyses such as ours may identify genes that may be missed by traditional univariate analytical approaches, because these genes tend to be authentic risk genes but with small effects. Second, the identification of eQTL effects of the risk SNPs could provide insights for future focused studies, since conventional analyses often observed a large LD region containing numerous genes showing association with the illness, but actually determining which one is the susceptibility gene is difficult at best. Third, significant association between eQTL and illness has been consistently replicated across independent datasets, providing convergent validity for findings and suggesting potentially higher reproducibility for this kind of system-level analysis. Given these advantages, it is likely that further studies using similar methods will strengthen the case for such studies in trying to uncover genetic risk factors for psychiatric diseases.
Study limitations
While this study offers some interesting observations, it should be noted that the present evidence is limited, and we are cautious in interpreting these results. (1) In the integrative analysis on BPD and eQTL GWAS data, we arbitrarily selected genes that were scored higher than 5.0 (LBF scores). As such, it is possible that some genes that may contribute to BPD risk but did not meet our selection criteria could have been missed. (2) Similarly, while we used GWAS data in our analysis, the SNP coverage is still relatively low and other true risk SNPs may have been missed. Due to the dearth of functional data, it is difficult to identify the causative variant(s). (3) Likewise, we cannot exclude the possibility that the positive association signal was actually caused by the hitch-hiking effect of rare missense mutations, copy number variations or variants in a distant region. Further focused studies may provide a more complete survey. (4) The SNPs in the discovery eQTL sample were not imputed, thus reducing the overlap between eQTL and GWAS data sets and the power of our method, although we believe the obtained results are valuable. (5) The gene expression coverage in the discovery eQTL dataset is relative low, and we cannot exclude the possibility of other missing risk genes during the integrative analyses, although we conducted a comprehensive replication and fine-mapping analyses to localize the actual risk genes. Further studies using a high-coverage array or RNA-sequencing are warranted. (6) It also should be acknowledged that the eQTL databases that we used are highly variable, in terms of expression platforms and tissue quality, age, and diagnoses. It is highly likely that biological factors mediating eQTL associations, such as epigenetic regulation, transcription factor binding, and microRNA dynamics will vary across age and diagnosis. (7) We also would note that our results reached genome wide significance in the final meta-analysis of our ten new samples added to the public BPD dataset. Our understanding of the association of rs6088662 with BPD and with gene expression and hippocampal biology might have started first with the combined GWAS result, but this was not our strategy.
Conclusions
In conclusion, our data from large-scale samples support that SNPs in a region on chromosome 20q11.22 are significantly associated with BPD. We observed associations with GGT7 and TRPC4AP mRNA expression, hippocampal volume and cognitive performance. Although the actual risk gene(s) for BPD in this genomic region are yet to be determined, future studies may give a more compelling picture on the association between these potential risk factors and genetic susceptibility to BPD.
Supplementary Material
Acknowledgments
We would like to acknowledge the efforts of the Bipolar Disorder Working Group of Psychiatric GWAS Consortium in their contribution to this study. We are deeply grateful for Stacy Steinberg, Hreinn Stefansson, Kari Stefansson, Thorgeir Thorgeirsson (deCODE genetics, Reykjavik, Iceland) and Engilbert Sigurdsson (Landspitali University Hospital, Reykjavík, Iceland) for their results in Iceland samples. We are also grateful to Andrew Willden (Kunming Institute of Zoology) for language editing of the manuscript.
Funding
This work was supported by grants from the National 973 project of China (2011CBA00401), the National Natural Science Foundation of China (U1202225, 31130051, 31071101 and 31221003), the German Federal Ministry of Education and Research (BMBF), the National Genome Research Network (NGFN), the Integrated Genome Research Network (IG) MooDS (grant 01GS08144 to SC and MMR, grant 01GS08147 to MR and TGS), 111 Project (B07008) of the Ministry of Education of China, the Strategic Priority Research Program (B) of the Chinese Academy of Sciences (XDB02020000), the National Authority for Scientific Research, Bucharest, Romania (UEFISCDI - PN-II-89/2012) and Personal Genetics SRL, Bucharest, Romania.
Footnotes
Declaration of interest: None
Supplementary material cited in this article is available online.
References
- 1.Craddock N, Jones I. Genetics of bipolar disorder. J Med Genet. 1999;36:585–94. doi: 10.1136/jmg.36.8.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etain B, Dumaine A, Mathieu F, Chevalier F, Henry C, Kahn JP, et al. A SNAP25 promoter variant is associated with early-onset bipolar disorder and a high expression level in brain. Mol Psychiatry. 2009;15:748–55. doi: 10.1038/mp.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M, Luo XJ, Rietschel M, Lewis CM, Mattheisen M, Muller-Myhsok B, et al. Allelic differences between Europeans and Chinese for CREB1 SNPs and their implications in gene expression regulation, hippocampal structure and function, and bipolar disorder susceptibility. Mol Psychiatry. 2014;19:452–61. doi: 10.1038/mp.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJ, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. 2011;18:195–205. doi: 10.1038/mp.2011.157. [DOI] [PubMed] [Google Scholar]
- 5.Cichon S, Muhleisen TW, Degenhardt FA, Mattheisen M, Miro X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88:372–81. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–8. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muhleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. 2014;5:3339. doi: 10.1038/ncomms4339. [DOI] [PubMed] [Google Scholar]
- 9.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinberg S, de Jong S, Andreassen OA, Werge T, Borglum AD, Mors O, et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet. 2011;20:4076–81. doi: 10.1093/hmg/ddr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quraishi S, Walshe M, McDonald C, Schulze K, Kravariti E, Bramon E, et al. Memory functioning in familial bipolar I disorder patients and their relatives. Bipolar Disord. 2009;11:209–14. doi: 10.1111/j.1399-5618.2008.00661.x. [DOI] [PubMed] [Google Scholar]
- 12.Rimol LM, Hartberg CB, Nesvag R, Fennema-Notestine C, Hagler DJ, Jr, Pung CJ, et al. Cortical thickness and subcortical volumes in schizophrenia and bipolar disorder. Biol Psychiatry. 2010;68:41–50. doi: 10.1016/j.biopsych.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 13.Haukvik UKWLT, Mørch-Johnsen L, Jørgensen KN, Lange EH, Dale AM, Melle I, Andreassen OA, Agartz I. In vivo hippocampal subfield volumes in schizophrenia and bipolar disorder. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Phillips ML, Ladouceur CD, Drevets WC. A neural model of voluntary and automatic emotion regulation: implications for understanding the pathophysiology and neurodevelopment of bipolar disorder. Mol Psychiatry. 2008;13:829, 33–57. doi: 10.1038/mp.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey BN, Andreazza AC, Nery FG, Martins MR, Quevedo J, Soares JC, et al. The role of hippocampus in the pathophysiology of bipolar disorder. Behav Pharmacol. 2007;18:419–30. doi: 10.1097/FBP.0b013e3282df3cde. [DOI] [PubMed] [Google Scholar]
- 16.Conde L, Bracci PM, Richardson R, Montgomery SB, Skibola CF. Integrating GWAS and expression data for functional characterization of disease-associated SNPs: an application to follicular lymphoma. Am J Hum Genet. 2013;92:126–30. doi: 10.1016/j.ajhg.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92:667–80. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamazon ER, Badner JA, Cheng L, Zhang C, Zhang D, Cox NJ, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry. 2012;18:340–6. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 21.Morita Y, Callicott JH, Testa LR, Mighdoll MI, Dickinson D, Chen Q, et al. Characteristics of the cation cotransporter NKCC1 in human brain: alternate transcripts, expression in development, and potential relationships to brain function and schizophrenia. J Neurosci. 2014;34:4929–40. doi: 10.1523/JNEUROSCI.1423-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci U S A. 2009;106:15873–8. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao R, Li C, Newburn EN, Ye T, Lipska BK, Herman MM, et al. Transcript-specific associations of SLC12A5 (KCC2) in human prefrontal cortex with development, schizophrenia, and affective disorders. J Neurosci. 2012;32:5216–22. doi: 10.1523/JNEUROSCI.4626-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–23. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–58. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Zhang C, Wu Z, Wang Z, Peng D, Chen J, et al. Association of genetic variation in CACNA1C with bipolar disorder in Han Chinese. J Affect Disord. 2013;150:261–5. doi: 10.1016/j.jad.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E, et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry. 2013;18:1302–7. doi: 10.1038/mp.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–61. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 31.Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A. 2009;106:7501–6. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet. 2010;42:128–31. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breen G, Lewis CM, Vassos E, Pergadia ML, Blackwood DH, Boomsma DI, et al. Replication of association of 3p21.1 with susceptibility to bipolar disorder but not major depression. Nat Genet. 2011;43:3–5. doi: 10.1038/ng0111-3. author reply. [DOI] [PubMed] [Google Scholar]
- 34.Kondo K, Ikeda M, Kajio Y, Saito T, Iwayama Y, Aleksic B, et al. Genetic variants on 3q21 and in the Sp8 transcription factor gene (SP8) as susceptibility loci for psychotic disorders: a genetic association study. PLoS One. 2013;8:e70964. doi: 10.1371/journal.pone.0070964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassos E, Steinberg S, Cichon S, Breen G, Sigurdsson E, Andreassen OA, et al. Replication study and meta-analysis in European samples supports association of the 3p21.1 locus with bipolar disorder. Biol Psychiatry. 2012;72:645–50. doi: 10.1016/j.biopsych.2012.02.040. [DOI] [PubMed] [Google Scholar]
- 36.McKenzie M, Henders AK, Caracella A, Wray NR, Powell JE. Overlap of expression quantitative trait loci (eQTL) in human brain and blood. BMC Med Genomics. 2014;7:31. doi: 10.1186/1755-8794-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nica AC, Parts L, Glass D, Nisbet J, Barrett A, Sekowska M, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2012;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preisig M, Waeber G, Vollenweider P, Bovet P, Rothen S, Vandeleur C, et al. The PsyCoLaus study: methodology and characteristics of the sample of a population-based survey on psychiatric disorders and their association with genetic and cardiovascular risk factors. BMC Psychiatry. 2009;9:9. doi: 10.1186/1471-244X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walters JT, Corvin A, Owen MJ, Williams H, Dragovic M, Quinn EM, et al. Psychosis susceptibility gene ZNF804A and cognitive performance in schizophrenia. Arch Gen Psychiatry. 2010;67:692–700. doi: 10.1001/archgenpsychiatry.2010.81. [DOI] [PubMed] [Google Scholar]
- 44.Chen M, Xu Z, Zhai J, Bao X, Zhang Q, Gu H, et al. Evidence of IQ-modulated association between ZNF804A gene polymorphism and cognitive function in schizophrenia patients. Neuropsychopharmacology. 2012;37:1572–8. doi: 10.1038/npp.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang F, McIntosh AM, He Y, Gelernter J, Blumberg HP. The association of genetic variation in CACNA1C with structure and function of a frontotemporal system. Bipolar Disord. 2011;13:696–700. doi: 10.1111/j.1399-5618.2011.00963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perrier E, Pompei F, Ruberto G, Vassos E, Collier D, Frangou S. Initial evidence for the role of CACNA1C on subcortical brain morphology in patients with bipolar disorder. Eur Psychiatry. 2011;26:135–7. doi: 10.1016/j.eurpsy.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poduslo SE, Huang R, Huang J. The frequency of the TRPC4AP haplotype in Alzheimer’s patients. Neurosci Lett. 2009;450:344–6. doi: 10.1016/j.neulet.2008.11.050. [DOI] [PubMed] [Google Scholar]
- 49.Poduslo SE, Huang R, Huang J, Smith S. Genome screen of late-onset Alzheimer’s extended pedigrees identifies TRPC4AP by haplotype analysis. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:50–5. doi: 10.1002/ajmg.b.30767. [DOI] [PubMed] [Google Scholar]
- 50.Teipel SJ, Walter M, Likitjaroen Y, Schonknecht P, Gruber O. Diffusion tensor imaging in Alzheimer’s disease and affective disorders. Eur Arch Psychiatry Clin Neurosci. 2014 doi: 10.1007/s00406-014-0496-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.