Abstract
A cell-free expression platform for making bacterial ribosomes was encapsulated within giant liposomes. The liposomes were prepared using double emulsion template, and compartmentalized in vitro protein synthesis was analysed using spinning disk confocal microscopy. Two different liposome phospholipids formulations were investigated to characterize their effects on the compartmentalized reaction kinetics. This study was performed as a necessary step towards the synthesis of a minimal cell.
Minimal cell construction seeks to assemble the smallest number of cellular components such as DNA, RNA and protein encapsulated in a lipid membrane necessary for life 1–6. The goals are advancing fundamental knowledge, testing our understanding of life and its origins, and facilitating engineering for biotechnology applications. In recent years, key developments in the construction of minimal cells have centred on integrating the subsystems of biology necessary for self-replication. This includes creating compartments (or membrane vesicles) 7, 8, enabling substrates and waste products to flow in and out of compartments 9, and activating complex biochemical reactions and networks inside compartments 10, 11. Despite these advances, the ability to co-activate the processes of ribosome synthesis, ribosome assembly, and translation in liposomes has not been shown. Such an advance is important for minimal cell development because the ribosome, which is encoded by 57 genes, comprises approximately 1/3 of the genes proposed necessary for a protein-based self-replicating system 3.
Up to now, the inability to build ribosomes from in vitro transcribed ribosomal RNA (rRNA) in a way that mimics in vivo ribosome biogenesis has precluded the cell-free construction of ribosomes 3. However, we recently developed an integrated synthesis, assembly, and translation (iSAT) technology to construct Escherichia coli ribosomes in vitro. iSAT enables one-step co-activation of ribosomal RNA (rRNA) transcription, assembly of transcribed rRNA with native ribosomal proteins into functional ribosomes, and synthesis of active protein by these ribosomes in the same reaction 12–14. In this communication, we report the use of double emulsion template to generate giant liposomes 15–17 to compartmentalize iSAT reactions. This demonstration is important towards the systems’ integration and synthesis of a minimal cell 1.
With the goal of encapsulating iSAT reactions in liposomes (Figure 1A), we chose to build-up complexity in three stages. First, we validated iSAT protein synthesis activity in the presence of 2% (w/v) polyvinyl alcohol 13,000–23,000 molecular weight (PVA) in test tubes (Supplementary Figure S1). This was necessary because PVA is used in the generation of liposomes, but not previously used in iSAT. Consistent with our previous studies 12–14, we observed synthesis of the reporter gene superfolder green fluorescent protein (sfGFP) following a lag time of ~0.5–1 h. It is hypothesized that this is the time necessary for rRNA transcription and ribosome assembly into functional particles in the ribosome free crude S150 extract 14. We calculated sfGFP synthesized by converting the fluorescent units into concentrations with a standard curve as previously described18. Approximately 3.3 μM +/− 0.26 μM of sfGFP was produced using the iSAT system with PVA in test tubes.
Figure 1. System illustrations.
(A) Illustration of the iSAT system and its components inside a liposome. (B) Schematic of the microfluidic capillary device to generate ultra-thin double emulsions encapsulating iSAT solution.
Second, we proceeded to compartmentalize iSAT reactions in single emulsions to test the compatibility of the system in emulsion droplets. After single emulsion generation, we monitored sfGFP synthesis with time over a ten-hour reaction using spinning disk confocal microscopy. From Supplementary Figure S2, we can see that the synthesis of sfGFP showed a slight lag during the first ~ 0.5 h and then progressed linearly from approximately 0.5 to 6 h. Between 6 and 8 h, sfGFP synthesis stopped with 0.51 +/− 0.06μM of sfGFP produced. We observed that the protein synthesis duration was longer in emulsions when compared to the test tube (~6–8 h versus 4 h, respectively). These data showed that single emulsions are compatible containers that can be used to host the iSAT system.
With these results at hand, we then generated liposomes with membranes of two different phospholipid compositions (1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC); and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)) to characterize and demonstrate the ability of iSAT to be compartmentalized. We used double emulsion droplet microfluidics 15, 16 to compartmentalize an aqueous phase containing the iSAT reaction mixture with ultra-thin volatile oil containing phospholipids (Figure 1B). Upon evaporation of oil, liposomes encapsulating the iSAT reaction form. We found the encapsulation process to be highly reproducible, with ~ 30% of the initial population generated by double emulsion template not bursting in a 15 h time course (or 30% yield; Supplementary Figure S3). The double emulsion template liposomes include crude ribosome-free S150 E. coli extract containing cytoplasmic translation and assembly factors, and salts, buffers and substrates necessary for rRNA and mRNA transcription, ribosome assembly, and translation. Following liposome generation, we measured overall protein synthesis activity over time by directly quantifying the expression of sfGFP. Initially, we encapsulated the iSAT system in DOPC based liposomes (Figure 2A). Unfortunately, our initial experiments failed and we did not observe sfGFP synthesis. We suspect that this was due to the fact that some of the small molecules needed for protein synthesis, e.g. amino acids, salts, co-factors and phosphoenolpyruvate (PEP) could cross the membrane of liposomes. This hypothesis is consistent with previous results 19, 20, which showed the importance of incubating cell-free translation systems encapsulated in liposomes in a feeding solution.
Figure 2. Cell-free synthesis of bacterial ribosomes in double emulsion template liposomes.
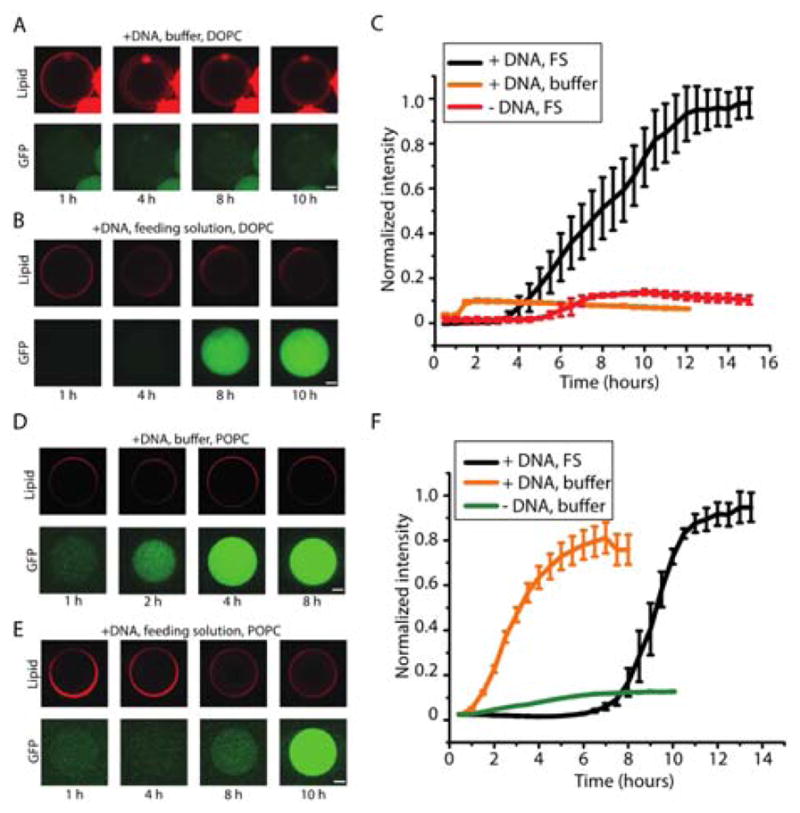
(A) Confocal images of iSAT with sfGFP encapsulated in liposomes over 10 hours using 12 mM lipid consisting DOPC/cholesterol/Rhod-PE (referred to as DOPC liposomes) at 69.5/30/0.5 ratio with buffer solution on the outside (scale = 20 μm) and (B) feeding solution on the outside (scale = 20 μm). (C) Kinetics of DOPC liposomes with and without feeding solution. +DNA and +FS case – average size is 106.8 μm with standard error of 3.4 μm (n=3). +DNA and buffer case – average size is 103 μm with standard error of 1.8 μm (n=5). -DNA and +FS case – average size is 111 μm with standard error of 2.4 μm (n=3). (D) Confocal images of iSAT with sfGFP encapsulated in liposomes over 8 hours using 12 mM lipid consisting POPC/cholesterol/Rhod-PE (referred to as POPC liposomes) at 69.5/30/0.5 ratio lipids with buffer solution on the outside (scale = 20 μm). (E) Confocal images of iSAT with sfGFP encapsulated in liposomes over 10 hours using POPC lipids with feeding solution on the outside (scale = 20 μm). (F) Kinetics of POPC liposomes with and without feeding solution. +DNA and +FS case – average size is 82.8 μm with standard error of 2.5 μm (n=3). +DNA and buffer case – average size is 94.7 μm with standard error of 2.5 μm (n=3). -DNA and buffer case – average size is 97.1 μm with standard error of 0.9 μm (n=3). DNA concentrations of 4 nM pJL1-sfGFP and 4 nM pT7Op WT were used.
We therefore carried out iSAT reactions encapsulated by double emulsion template liposomes in a feeding solution to enable protein synthesis. Figure 2B shows the resulting expression of the reporter gene inside the compartment, when using an outer feeding solution. Expression was quantified using image analysis of micrographs and, notably, all vesicles that do not burst show sfGFP synthesis. Figure 2C displays the different kinetics of compartmentalized gene expression of the iSAT reaction within liposomes without feeding solution and liposomes without DNA. The cell-free reaction within DOPC liposomes began after ~ 4 hours, and reached plateau after ~ 12 hours. Protein synthesis initiates much more slowly in liposomes than in test tube reactions (~4h versus ~1 h). Further, the total protein yield in liposomes (0.2 +/− 0.012 μM of sfGFP) is about an order of magnitude less than that in test tubes, which provides an opportunity for future system improvements. The negative control reaction without DNA did not synthesize protein.
We next investigated the effect of using POPC based liposomes on the rRNA synthesis, ribosome assembly, and sfGFP synthesis iSAT reaction, with and without the feeding solution. In contrast to DOPC liposomes, fluorescence microscopy observations showed that POPC based liposomes allowed protein synthesis without the feeding solution (Figure 2D). This may be due to permeability differences across the different membranes since POPC contains one saturated fatty acid tail compared to two in DOPC. It has been shown, for example, that permeability is different between the two compositions with POPC being less permeable than DOPC membrane 21, 22. Somewhat surprisingly, we observed a significant delay in protein expression (a lag time ~ 7 hours) when combining iSAT in POPC liposomes with a feeding solution (Figures 2E and 2F). One possible model that may explain the delayed sfGFP produced in the presence of the feeding solution is that there is some inhibitory molecule(s) that can pass through both the DOPC and POPC lipid layers at different rates. The reduced permeability of the POPC membrane extends the delayed time compared to DOPC membrane. Looking forward, we plan to carry out a separate comprehensive study to fully understand this phenomenon now that we are able to build ribosomes in liposomes for the first time, which was the goal of our study. Indeed, our data convincingly showed that the cell-free expression platform for making bacterial ribosomes encapsulated within giant liposomes was capable of synthesizing sfGFP. Additional images are shown in Supplementary Figure S4.
In summary, we report the development of experimental conditions necessary to encapsulate iSAT reactions into liposomes. We also measured the kinetics of compartmentalized gene expression when E. coli ribosomes are produced from the transcription of its natural rRNA operon and assembled with purified ribosomal proteins in ribosome free S150 crude extracts. Toward the synthesis of a minimal cell, it is important that DNA encodes biomolecular machines, such as ribosomes. This will be important during self-replication of the information and self-reproduction of the phospholipid bilayer. Liposome division and liposomes fusion are also important processes during evolution 6, 11, 23, 24. Looking forward, our approach may be integrated with more complex designs, such as the expression of cytoskeletal and membrane proteins 9, 25, 26, and even encapsulation of gene circuits for spatial-temporal control 10, 27. To our knowledge, this is the first time that a cell-free transcription and translation system where a DNA molecule encoding the formation of ribosomes has been encapsulated in a model cell-like compartment (i.e. liposome). Thus, our work represents an important step towards the construction of a minimal cell.
Supplementary Material
Acknowledgments
M.C.J. and F.C. gratefully acknowledge the Army Research Office (W911NF-11-1-0445), the David and Lucile Packard Foundation (2011-37152), the Camille Dreyfus Teacher Scholar program, and the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust for support. J.W.L. is supported by the NIH’s Microfluidics in Biomedical Sciences Training Program: NIH NIBIB T32 EB005582. A.P.L. and K.K.Y.H. are supported by the NIH Director’s New Innovator Award: NIH DP2 HL117748-01.
Footnotes
Electronic Supplementary Information (ESI) available: Materials and Methods, FigS1 – Fig S4, See DOI: 10.1039/x0xx00000x
Author Contributions: F.C. designed the research, prepared the iSAT system, made the feeding solution, and wrote the manuscript. J.W.L. performed the experiments, prepared the figures, analysed the data, and wrote the manuscript. K.K.Y.H performed the experiments, and wrote the manuscript. A.P.L. and M.C.J. conceived respectively the double emulsion template for preparing the liposomes and the iSAT system for making ribosomes in vitro. They also wrote the manuscript, aided in research design, and directed the studies.
Notes and references
- 1.Caschera F, Noireaux V. Current opinion in chemical biology. 2014;22C:85–91. doi: 10.1016/j.cbpa.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 2.Noireaux V, Maeda YT, Libchaber A. Proc Natl Acad Sci USA. 2011;108:3473–3480. doi: 10.1073/pnas.1017075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jewett MC, Forster AC. Curr Opin Biotechnol. 2010;21:697–703. doi: 10.1016/j.copbio.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu AP, Fletcher DA. Nat Rev Mol Cell Biol. 2009;10:644–650. doi: 10.1038/nrm2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luisi PL, Ferri F, Stano P. Naturwissenschaften. 2006;93:1–13. doi: 10.1007/s00114-005-0056-z. [DOI] [PubMed] [Google Scholar]
- 6.Szostak JW, Bartel DP, Luisi PL. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 7.Coyne CW, Patel K, Heureaux J, Stachowiak J, Fletcher DA, Liu AP. Journal of visualized experiments : JoVE. 2014:e51510. doi: 10.3791/51510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stachowiak JC, Richmond DL, Li TH, Liu AP, Parekh SH, Fletcher DA. P Natl Acad Sci USA. 2008;105:4697–4702. doi: 10.1073/pnas.0710875105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noireaux V, Libchaber A. Proc Natl Acad Sci U S A. 2004;101:12672–12677. doi: 10.1073/pnas.2135496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elani Y, Law RV, Ces O. Nat Commun. 2014;5:5305. doi: 10.1038/ncomms6305. [DOI] [PubMed] [Google Scholar]
- 11.Caschera F, Sunami T, Matsuura T, Suzuki H, Hanczyc MM, Yomo T. Langmuir : the ACS journal of surfaces and colloids. 2011;27:13082–13090. doi: 10.1021/la202648h. [DOI] [PubMed] [Google Scholar]
- 12.Fritz BR, Jamil OK, Jewett MC. Nucleic acids research. 2015;43:4774–4784. doi: 10.1093/nar/gkv329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritz BR, Jewett MC. Nucleic acids research. 2014;42:6774–6785. doi: 10.1093/nar/gku307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jewett MC, Fritz BR, Timmerman LE, Church GM. Molecular systems biology. 2013;9:678. doi: 10.1038/msb.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho KK, Murray VL, Liu AP. Methods in cell biology. 2015;128:303–318. doi: 10.1016/bs.mcb.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arriaga LR, Datta SS, Kim SH, Amstad E, Kodger TE, Monroy F, Weitz DA. Small. 2014;10:950–956. doi: 10.1002/smll.201301904. [DOI] [PubMed] [Google Scholar]
- 17.Shum HC, Lee D, Yoon I, Kodger T, Weitz DA. Langmuir : the ACS journal of surfaces and colloids. 2008;24:7651–7653. doi: 10.1021/la801833a. [DOI] [PubMed] [Google Scholar]
- 18.Hong SH, Kwon YC, Martin RW, Des Soye BJ, de Paz AM, Swonger KN, Ntai I, Kelleher NL, Jewett MC. Chembiochem. 2015;16:844–853. doi: 10.1002/cbic.201402708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nourian Z, Danelon C. ACS synthetic biology. 2013;2:186–193. doi: 10.1021/sb300125z. [DOI] [PubMed] [Google Scholar]
- 20.Nishimura K, Matsuura T, Nishimura K, Sunami T, Suzuki H, Yomo T. Langmuir : the ACS journal of surfaces and colloids. 2012;28:8426–8432. doi: 10.1021/la3001703. [DOI] [PubMed] [Google Scholar]
- 21.Mathai JC, Tristram-Nagle S, Nagle JF, Zeidel ML. J Gen Physiol. 2008;131:69–76. doi: 10.1085/jgp.200709848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagle JF, Mathai JC, Zeidel ML, Tristram-Nagle S. J Gen Physiol. 2008;131:77–85. doi: 10.1085/jgp.200709849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuji G, Fujii S, Sunami T, Yomo T. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1516893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurihara K, Okura Y, Matsuo M, Toyota T, Suzuki K, Sugawara T. Nat Commun. 2015;6:8352. doi: 10.1038/ncomms9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda Y, Nakadai T, Shin J, Uryu K, Noireaux V, Libchaber A. ACS synthetic biology. 2012;1:53–59. doi: 10.1021/sb200003v. [DOI] [PubMed] [Google Scholar]
- 26.Kuruma Y, Stano P, Ueda T, Luisi PL. Biochimica et biophysica acta. 2009;1788:567–574. doi: 10.1016/j.bbamem.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 27.Shin J, Noireaux V. ACS Synth Biol. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



