Abstract
CD4+Foxp3+ regulatory T cells (Tregs) play a non-redundant role in control of excessive immune responses, and defects in Tregs have been shown both in patients and murine models of primary biliary cirrhosis (PBC), a progressive autoimmune biliary disease. Herein, we took advantage of a murine model of PBC, the dominant negative transforming growth factor β receptor II (dnTGFβRII) mice, to assess Treg genetic defects and their functional effects in PBC. By using high-resolution microarrays with verification by PCR and protein expression, we found profound and wide-ranging differences between dnTGFβRII and normal, wild type Tregs. Critical transcription factors were down-regulated including Eos, Ahr, Klf2, Foxp1 in dnTGFβRII Tregs. Functionally, dnTGFβRII Tregs expressed an activated, pro-inflammatory phenotype with upregulation of Ccl5, Granzyme B and IFN-γ. Genetic pathway analysis suggested that the primary effect of loss of TGFβ pathway signaling was to down regulate immune regulatory processes, with a secondary upregulation of inflammatory processes. These findings provide new insights into T regulatory genetic defects; aberrations of the identified genes or genetic pathways should be investigated in human PBC Tregs. This approach which takes advantage of biologic pathway analysis illustrates the ability to identify genes/pathways that are affected both independently and dependent on abnormalities in TGFβ signaling. Such approaches will become increasingly useful in human autoimmunity.
Keywords: Primary biliary cirrhosis, cholangitis, regulatory T cells, transcription profile and pathway analysis
Introduction
Although there have been significant improvements in our understanding of the immunological events that occur in patients with primary biliary cirrhosis (PBC), there remains a paucity of data on underlying molecular mechanisms that facilitate breach of tolerance[1–6]. This problem is compounded by the fact that patients develop clinical symptomatology long after the earliest causal events that initiate disease [7]. Antibodies to mitochondrial antigens, the serologic hallmark of PBC, are found many years before diagnosis [8]. Hence, an approach to understanding of the earliest events that lead to cholangitis are critical. In this respect our laboratory has studied a murine model of PBC, dnTGFβRII mice [9]. These animals develop high titer autoantibodies to mitochondrial antigens of the same specificity as humans with PBC and also exhibit portal infiltrates, ductopenia, granulomas, and a number of immunological features that are shared by patients with PBC [10].
In both patients with PBC and human autoimmune diseases, CD4+Foxp3+ regulatory T cells (Tregs) play a pivotal role in controlling excessive immune responses [11]. Tregs defects have been reported in both PBC patients and murine models [12, 13]. Recent data suggest that dnTGFβRII CD4+Foxp3+ Tregs possess weaker suppressive function than wild type Tregs [14]. In fact, the immunopathology of dnTGFβRII mice requires defects in both Tregs and pathogenic cytotoxic CD8+ T cells [15]. To better define the dnTGFβRII genetic Treg abnormalities, we have performed comprehensive transcriptome analysis, quantitative PCR and protein expression of various Treg populations.
Materials and methods
Mice
dnTGFβRII mice were derived from the vivarium at the University of California at Davis. Foxp3GFP mice (Foxp3tm2Ayr) were kindly provided by Dr. A.Y. Rudensky [16]. dnTGFβRII; Foxp3GFP mice were generated by selective breeding of our dnTGFβRII colony with female Foxp3GFP/GFP mice. All mice were housed under specific pathogen-free and controlled environmental conditions in the animal facility of the School of Life Sciences of the University of Science and Technology of China. All experiments were performed following approval from the USTC Animal Care and Use Committee.
Flow cytometry, immuno-phenotype detection and intracellular staining
All mice were studied at 10–13 weeks of age. Liver, spleen or mesenteric lymph nodes mononuclear cells were isolated and prepared as described [17]. Single cell suspensions were incubated with anti-mouse CD16/32 (BioLegend, San Diego, CA). All additional flow antibodies, unless otherwise noted, were purchased from BioLegend. To identify subpopulations of CD4+ T cells, cells were stained with Pacific Blue-CD3 (17A2), PE-Cy7-NK1.1 (PK136), APC-Cy7-CD4 (GK1.5) [9]. To detect Tregs related surface markers and confirm the data from gene expression profile, cells were labeled with PE-CD25 (PC61), APC-GITR (YGITR765), PE-CXCR3 (CXCR3-173), Alexa 647-CCR6 (29-2L17), PerCP/Cy5.5-ICOS (C398.4A), or PE-CD62L (MEL-14). In some cases, intracellular staining was performed using a FOXP3 Fix/Perm Buffer Set (BioLegend) to detect CTLA-4 using PE-CTLA-4 (UC10-4B9) [18]. To detect the level of intracellular cytokine and confirm the data from microarray, cells were resuspended in RPMI-1640 with 10% fetal bovine serum and stimulated with Cell Stimulation Cocktail (plus protein transport inhibitors) at 37°C 5% CO2 for 3 hours [19]. Thence, cells were stained with surface markers by CD3, CD4, CD8β, and NK1.1 as described [17], fixed with Fixation Buffer (BioLegend), and permeabilized with Permeabilization Wash Buffer (BioLegend), and stained for intracellular PE-IFN-γ (XMG1.2), Alexa Fluro 647-Granzyme B (GB11), APC-IL-10 (JES5-16E3). For purposes of control, normal IgG isotypes were used (BioLegend). Stained cells were analyzed using a flow cytometer FACS verse (BD) and data analyzed using Flowjo software (Tree Star, Inc., Ashland, OR).
Gene-expression profiling analysis of regulatory T cells
CD4+ T cells from spleens of 10-week old female dnTGFβRII; Foxp3GFP or WT;Foxp3GFP mice were first enriched by MACS using anti-CD4 microbeads (Miltenyi, Bergisch Gladbach, German) and regulatory T cells (CD4+Foxp3+) were isolated by FACS Aria (BD) to attain a purity greater than 95%. RNA was extracted with RNAiso Plus (Takara, Dalian, China) and hybridized to Affymetrix MOE 430 2.0 chips. Fluorescence was detected using an Affymetrix GeneChip Scanner 3000 and images were analyzed using Affymetrix GeneChip Operating Software (GCOS). Transcription profile chip service was provided by Shanghai Biotechnology Cooperation (Shanghai). Expression fluorescence values were log2-transformed, and subsequent analyses conducted using SAS statistical software online (http://sas.ebioservice.com/). Differentially expressed genes were defined as equal to or more than 2 fold differences between the two groups, signal values were confirmed beyond background signals, and the genes classified using the annotation of the Gene Ontology (GO) project [20]. Heat map and blue-pink scale schemes were designed by using Multiple Experiment Viewer 4.9 software and Microsoft Excel. A scatter plot was performed using Graph Pad Prism and color labeled to facilitate identification.
Biological pathway analysis
The open source Bioinformatics software Cytoscape 3.2.0 [21–23] (http://www.cytoscape.org/) was used to visualize gene-gene interactions. We loaded a BioGRID [24] interaction network for Mus musculus, then imported the gene expression data we acquired before by setting Entrez Gene identifiers (IDs) as the node primary identifiers. The up-regulated genes or down-regulated genes in dnTGFβRII Tregs compared with WT Tregs (more than 2-fold change) were selected and formed two sub-networks, and their first interaction neighbor genes were shown in the interaction sub-networks. Gene Ontology (GO) annotations and P-values for different biological processes, in which the target genes were involved, were assigned and analyzed using BiNGO 3.0.2, an application for Cytoscape [25]. BiNGO was also applied to generate the GO biological process network [25]. The hypergeometric test was used as the statistical testing of BiNGO, the Bonferroni correction was used to control the family-wise error rate (FWER) [26], and the Benjamini and Hochberg correction [27] was used in multiple testing corrections to control False Discovery Rate (FDR). The P-Value significance level, the EASE score, was set as 0.01, then statistical biological process pathway analysis was performed [28].
Quantitative PCR
Regulatory T cells (CD4+Foxp3+) or non-regulatory conventional CD4+ T cells (CD4+Foxp3−) from spleens of dnTGFβRII mice and B6 wild type control mice were sorted using FACS Aria (BD); the purity was confirmed to be greater than 95%. Total RNA from different sorted cells was extracted with RNAiso Plus (Takara) separately, and cDNA synthesized with the PrimeScript® RT reagent Kit (Takara). Quantitative PCR was performed using SYBR® Premix Ex TaqTM II (Takara) as previously described [17]. Data were collected by an ABI StepOne real-time PCR system (Applied Biosystems, Carlsbad, CA). The primers used in this study are listed in Table 1 and were based on PrimerBank (http://pga.mgh.harvard.edu/primerbank), then blasted to confirm the target genes using NCBI primer blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) [29]. Amplification specificity was further analysis using melt curve [30]. The expression levels of target genes were thence normalized to the housekeeping gene Gapdh (ΔCt), and the results were calculated by 2−ΔΔCt method.
Table 1.
Primers used in q-PCR
| Gene | Forward Primer5′-3′ | Reverse Primer5′-3′ |
|---|---|---|
| Foxp3 | CCCATCCCCAGGAGTCTTG | ACCATGACTAGGGGCACTGTA |
| Ikzf2 | GAGCCGTGAGGATGAGATCAG | CTCCCTCGCCTTGAAGGTC |
| Ikzf4 | TCTGGACCACGTCATGTTCAC | ACGATGTGGGAAGAGAACTCATA |
| Ccl5 | GCTGCTTTGCCTACCTCTCC | TCGAGTGACAAACACGACTGC |
| GzmB | CCACTCTCGACCCTACATGG | GGCCCCCAAAGTGACATTTATT |
| Cxcr3 | TACCTTGAGGTTAGTGAACGTCA | CGCTCTCGTTTTCCCCATAATC |
| Ahr | AGCCGGTGCAGAAAACAGTAA | AGGCGGTCTAACTCTGTGTTC |
| Cxcr4 | GAAGTGGGGTCTGGAGACTAT | TTGCCGACTATGCCAGTCAAG |
| Foxp1 | GGTCTGAGACAAAAAGTAACGGA | CGCACTCTAGTAAGTGGTTGC |
| Itgae | CCTGTGCAGCATGTAAAAGAATG | CAAGGATCGGCAGTTCAGATAC |
| Klf2 | CTCAGCGAGCCTATCTTGCC | CACGTTGTTTAGGTCCTCATCC |
| Vcam1 | AGTTGGGGATTCGGTTGTTCT | CCCCTCATTCCTTACCACCC |
| Gp49a | GCAGTACAGGCAGATTCATTCT | AGTAGCATGGGTTGGCTGATT |
| Igfbp4 | AGAAGCCCCTGCGTACATTG | TGTCCCCACGATCTTCATCTT |
Statistical Analysis
Data are presented as the mean ± standard deviation (SD). The significance of differences was determined using a two-tailed unpaired t test or the Mann-Whitney U test in Graph Pad Prism.
Results
Frequency and number of Tregs
The frequency of Tregs in dnTGFβRII; Foxp3GFP mice compared to controls was decreased in liver (P=0.032) but was unchanged in spleen (P=0.724) or mesenteric lymph nodes (mLN) (P=0.118). Due to splenomegaly and lymphadenopathy, the total Treg number was increased in liver (P=0.0007), spleen (P=0.0002) and mLN (P=0.0002) in dnTGFβRII; Foxp3GFP mice compared to control (Figure 1A, B). Therefore the frequency of Tregs in dnTGFβRII mice was similar to wild type controls, suggesting that qualitative, not quantitative defects of Tregs predominate in this model.
Figure 1.
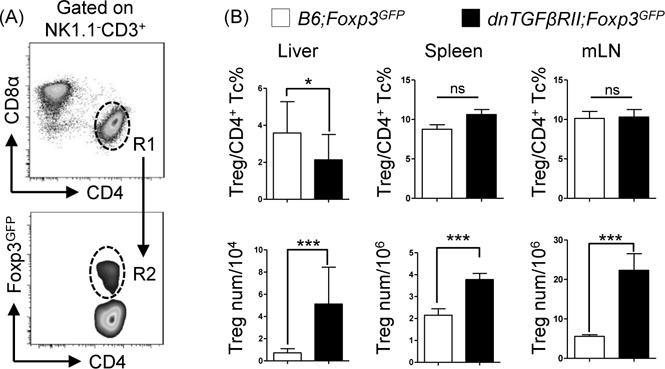
Frequency and total cell numbers of regulatory T cells in dnTGFβRII;Foxp3GFP mice did not demonstrate quantitative defects compared with WT;Foxp3GFP mice. (A) Gate strategy defining NK1.1−CD3+CD4+ total classic CD4+ T cells (R1), NK1.1−CD3+CD4+Foxp3+ regulatory T cells (R2) in liver, spleen and mesenteric lymph nodes. (B) Frequency of regulatory T cells in CD4+ T cells (upper panel) and total cells numbers (lower panel) in liver, spleen and mesenteric lymph nodes. Graphs present mean ± SD of 11–13 week-old 4–8 mice per group. *P <0.05 and ***P <0.001 as determined by Student Test.
Transcriptional features of regulatory T cells
A total of 40,000 genes were analyzed using CD4+Foxp3+ Treg cells derived from the spleens of dnTGFβRII;Foxp3GFP compared to wild type Foxp3GFP mice (GEO serial number GSE62964). Of these 40,000 tested genes, there were positive signal values in 20,332. Of these 20,332, 17,182 did not reveal any changes in dnTGFβRII mice compared to wild-type Tregs. In contrast, a greater than two-fold change was found in 3150 expressed genes (Figure 2A). Amongst these 3150 differentially expressed genes, 832 genes in both dnTGFβRII Tregs and WT Tregs gave a signal value distinct from background; 722 genes had a known gene symbol and title, and 609 were annotated. These 609 genes fell into specific categories (Figure 2B), including T cell-related intracellular proteins, T cell-related membrane proteins, immune response-related membrane proteins, cytokines and cytokine receptors, chemotaxis related, transcription factors, cytoskeleton or structural related, apoptosis related, metabolic process related, cell cycle and proliferation related genes.
Figure 2.
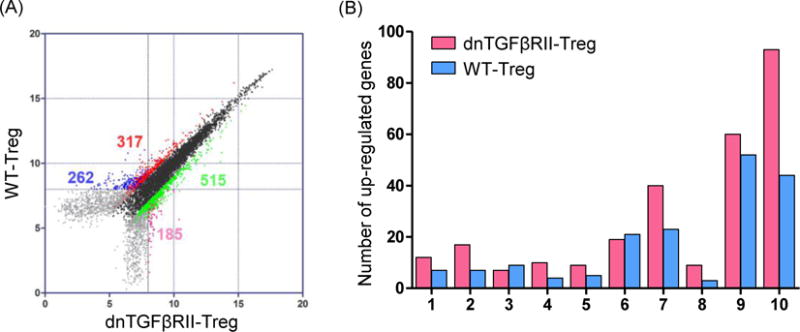
Comparison of transcription profile between dnTGFβRII Tregs and WT Tregs.
(A) Comparative transcriptome analysis between CD4+Foxp3+ Tregs from 10 week-old dnTGFβRII;Foxp3GFP mice (dnTGFβRII) and WT;Foxp3GFP mice (WT). Regulatory T cells were sorted from splenocytes pooled from 5 mice. Analysis was performed by Affymetrix GeneChip Mouse Genome 430 2.0 arrays. Genes of dnTGFβRII Tregs that were expressed greater than 2-fold higher or lower than WT Tregs were highlighted. Differential expression genes were divided into 4 groups by sample calibration. Dots were highlighted brilliant green and pink represented the 2-fold unregulated genes in dnTGFβRII Tregs, red and blue dots represented the 2-fold down-regulated genes in dnTGFβRII Tregs. Signal pairs from brilliant green and red groups were both distinct from chip background signal. Signal pairs from soft pink and blue groups had one probe signal cannot distinct from background, the other probe signal was higher than 8, this design was used to exclude the noise signals and still keep some positive signals. The number of genes for each comparison in the 4 groups mentioned above were indicated. (B) Distribution by functional category of up-regulated genes in dnTGFβRII Tregs and WT Tregs. Genes with greater than 2-fold differences are included. 1, T cell-related intracellular protein, 2, T cell-related membrane protein, 3, immune response-related membrane protein, 4, cytokines and cytokine receptors, 5, chemotaxis related, 6, transcription factors, 7, cytoskeleton or structural related, 8, apoptosis related, 9, metabolic process related, 10, cell cycle and proliferation related.
The quantitative analysis of gene expression differences revealed multiple differences between dnTGFβRII and WT Tregs. (Figure 3). A specific regulatory function, activation related molecule CD274 (Pd-l1) was over-expressed in dnTGFβRII Tregs. Molecules associated with T cell responses, including cell adhesion and chemokine receptors such as CCR5 (Ccr5) or CXCR3 (Cxcr3), were up-regulated in dnTGFβRII Tregs at the transcription level. However, CXCR4 (Cxcr4), CD103 (Itgae), CCR7 (Ccr7), CD24 (CD24a), vascular cell adhesion protein 1, Vcam1 (Vcam1) were down-regulated in dnTGFβRII Tregs (Figure 3B). These data suggest that dnTGFβRII Tregs are differentially activated, with different migratory tendencies, compared to wild type control Tregs. Some transcription factors, such as T-bet (Tbx21) were up-regulated in dnTGFβRII Tregs, while the aryl hydrocarbon receptor, Ahr (Ahr), FoxP1 (Foxp1), KLF2 (Klf2) were expressed at lower levels in dnTGFβRII Tregs than WT Tregs. Moreover, some secreting factors, such as CCL5 (Ccl5), Granzyme B (Gzmb), IL-10 (Il10) and IFN-γ (Ifng) were unregulated in dnTGFβRII Tregs, but TGF-β1 (Tgfb1) was down-regulated in dnTGFβRII Tregs.
Figure 3.
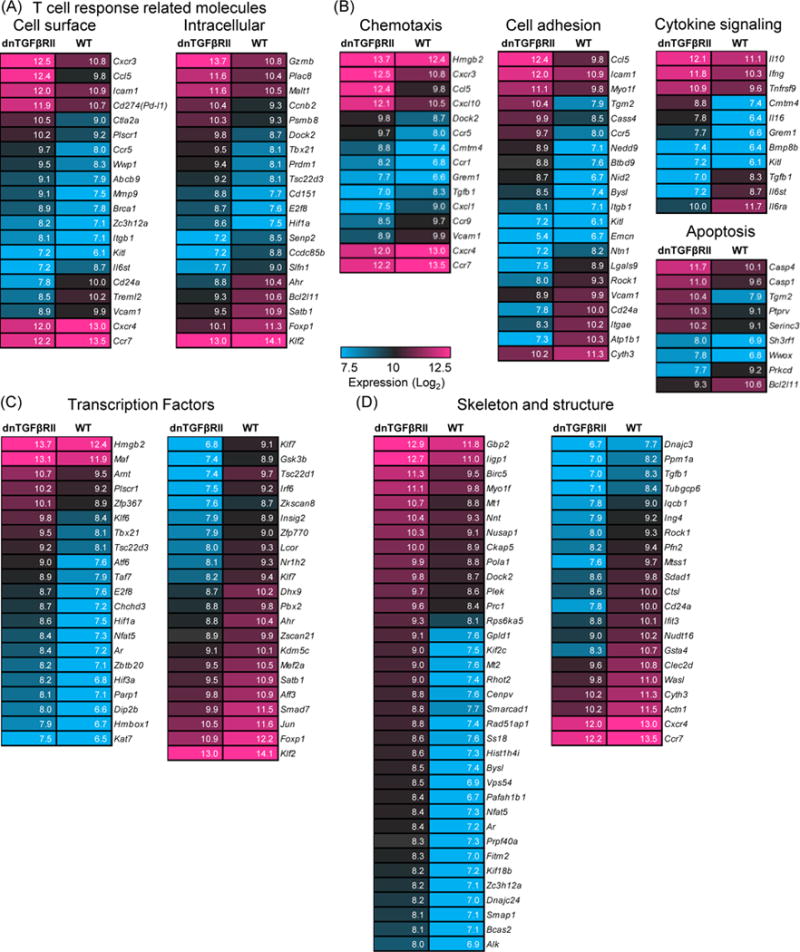
Characterization of different gene categories of dnTGFβRII and WT Tregs.
Differentially expressed genes in dnTGFβRII Tregs and WT Tregs were chosen and classified into different groups by functional category. Heat maps showing signal values of the listed genes had greater than 2-fold differences. Genes are ranked by their signal values. Numbers in each box indicated probe signal value. Some T cell response related molecules were shown in panel (A), chemotaxis, cell adhesion, cytokine signaling, and apoptosis related genes were displayed in panel (B), some transcriptional factors and cell skeleton related genes were exhibited in panel (C) and (D), respectively.
To confirm this data, we performed quantitative PCR (Figure 4A, B). We note that Ikaros family zinc finger transcription factors, Helios (Ikzf2), was significantly up-regulated, but a related transcription factor, Eos (Ikzf4) was down-regulated. Several other transcription factors were down-regulated in dnTGFβRII Tregs compared to WT Tregs, including Ahr (Ahr), KLF2 (Klf2), Foxp1 (Foxp1). Soluble factors such as CCL5 (Ccl5) and Granzyme B (Gzmb) were preferentially expressed in dnTGFβRII Tregs, and TGF-β1 was down-regulated in dnTGFβRII Tregs. Surface molecules, such as CD103 (Itgae), Vcam-1 (Vcam1), CXCR4 (Cxcr4), Insulin-like growth factor-binding protein 4, IGFBP4 (Igfbp4) were also down-regulated in dnTGFβRII Tregs compared to WT Tregs. Interestingly, these differentially expressed genes were largely regulatory T cell specific, as shown by low or absent expression in control WT and dnTGFβRII conventional T cells (Fig 4).
Figure 4.
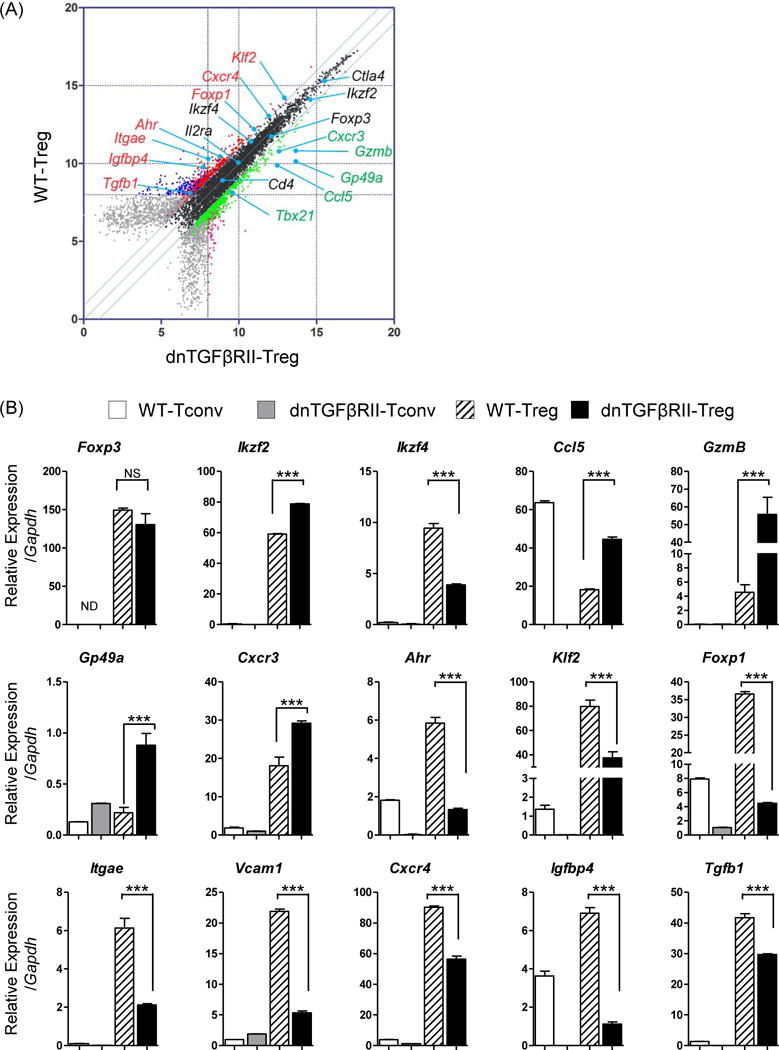
Quantitative PCR analysis to confirm differentially expressed genes.
(A) Selected genes from the transcriptional array were studied by qPCR as indicated in black (references with less than two fold change), green (upregulated in dnTGFβRII Tregs) or in red (upregulated in WT Tregs). (B) Quantitative PCR analysis comparing mRNA level expression of selected genes among WT CD4+ non-Treg cells (WT-Tconv, white bar), dnTGFβRII CD4+ non-Treg cells (dnTGFβRII-Tconv, gray bar), WT Tregs (slash white bar) and dnTGFβRII Tregs (black bar), which were listed in panel (A). P values were determined using Nonparametric Mann-Whitney U test, and *P <0.05 **P<0.01, and ***P <0.001. Graph contains results from RNA sample analyzed in microarray in two independent experiments.
Regulatory T cells in dnTGFβRII mice are skewed to a Th1 activation state
We next examined established Treg markers including interleukin 2 receptor α (CD25), glucocorticoid-induced TNFR-related protein (GITR), inducible co-stimulator (ICOS), cytotoxic T lymphocyte-associated antigen-4 (CTLA-4). Compared with WT;Foxp3GFP mice derived Tregs, intrahepatic dnTGFβRII Tregs over expressed CD25 (P=0.0059), GITR (P=0.0001), ICOS (P=0.0035) (Figure 5 A, B). However, in the secondary lymphoid organs, CD25 demonstrated no significant difference in expression. Although there was no change in CTLA-4 (P=0.375) in intrahepatic Tregs, dnTGFβRII mice derived splenic Tregs (P=0.0008) and mLN derived Tregs (P=0.0004) expressed relatively higher levels. In addition to activation marker up regulation, dnTGFβRII liver derived CD4+Foxp3+ Tregs up regulated Th1 response-related chemokine receptor CXCR3 (P=0.018), but down regulated Th17 response related chemokine receptor CCR6 (P=0.0002), and with a similar result in second lymphoid organs (Figure 5 A, B). Finally, we demonstrate a functional difference in dnTGFβRII murine spleen derived Tregs at the protein level, as they secreted increased Granzyme B and IFN-γ compared with WT Tregs, while the capacity of secreting IL-10 was not different (Figure 6). This protein expression data confirmed our transcriptional microarray analysis.
Figure 5.
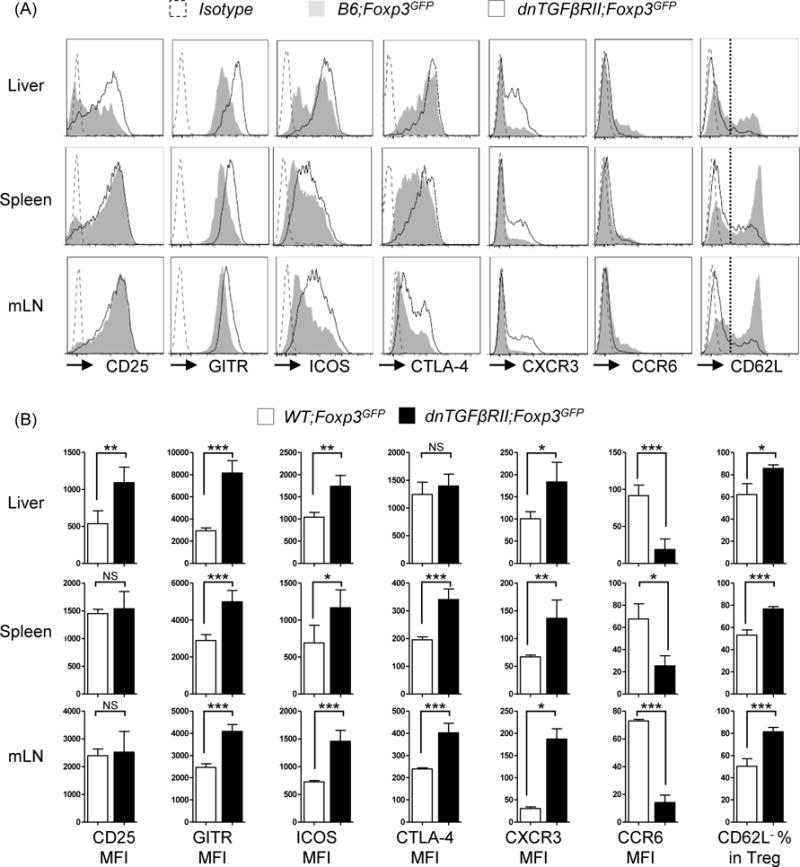
CD4+Foxp3+ cells from dnTGFβRII;Foxp3GFP mice display a highly activated Th1- like phenotype. (A) Expression of activation markers (CD25, GITR, ICOS, CTLA-4) and adhesion molecule (CXCR3, CCR6, CD62L) on CD4+Foxp3+ T cells from liver, spleen and mesenteric lymph nodes of dnTGFβRII;Foxp3GFP mice and WT;Foxp3GFP mice. (B) Statistical analysis of mean fluorescence intensity (MFI) of the markers presented in (A). Graphs present mean ± SD of 11–13 week-old 4–8 mice per group. *P <0.05, **P <0.01 and *** P <0.001 as determined by Student Test.
Figure 6.
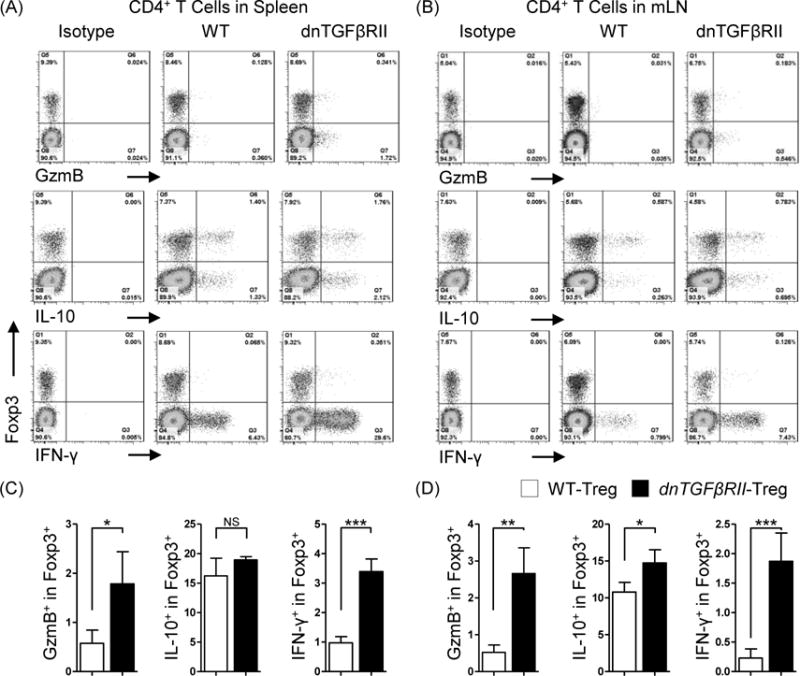
Comparative analysis of cytokine secreting capacity between dnTGFβRII;Foxp3GFP mice Tregs and WT;Foxp3GFP mice derived Tregs.
Total mononuclear cells from spleen (A) and mesenteric lymph nodes (B) of dnTGFβRII;Foxp3GFP and WT;Foxp3GFP mice were stimulated with PMA and ionomycin for 3 hours in the presence of Golgi stop reagent. Secreted IFN-γ, IL-10 and Granzyme B (GzmB) of Tregs were assessed by flow cytometry. Graphs present 10 week-old mice, 4 mice per group. (C) and (D) present the statistics analysis of the data indicated in (A) and (B), respectively. Graphs present mean ± SD. Data is representative of two independent experiments with similar results. *P <0.05, **P <0.01 and ***P <0.001 as determined by Student Test.
Biological pathway analysis of differentially expressed genes
To further determine the gene interaction networks of the differentially expressed genes between dnTGFβRII Tregs and WT Tregs controls, these genes were analyzed in Cytoscape to create an interaction network. We first loaded the gene expression data (along with the unique Gene IDs), distinguished from the array background signal, to an established Mus musculus gene interaction network based on the BioGRID database, and built a sub-network consisting of 3092 nodes and 5631 edges. The nodes of up-regulated genes with their first interaction neighbors were selected to create a sub-network constituted with 251 nodes and 577 edges. Similarly the nodes of down-regulated genes with their first interaction neighbors were used to build a sub-network constituted with 265 nodes and 662 edges (Figure 7A). Of note, there was one Foxp3 hub in the up-regulated network and one Iqcb1 (IQ calmodulin-binding motif containing 1) hub in the down-regulated network, respectively.
Figure 7.
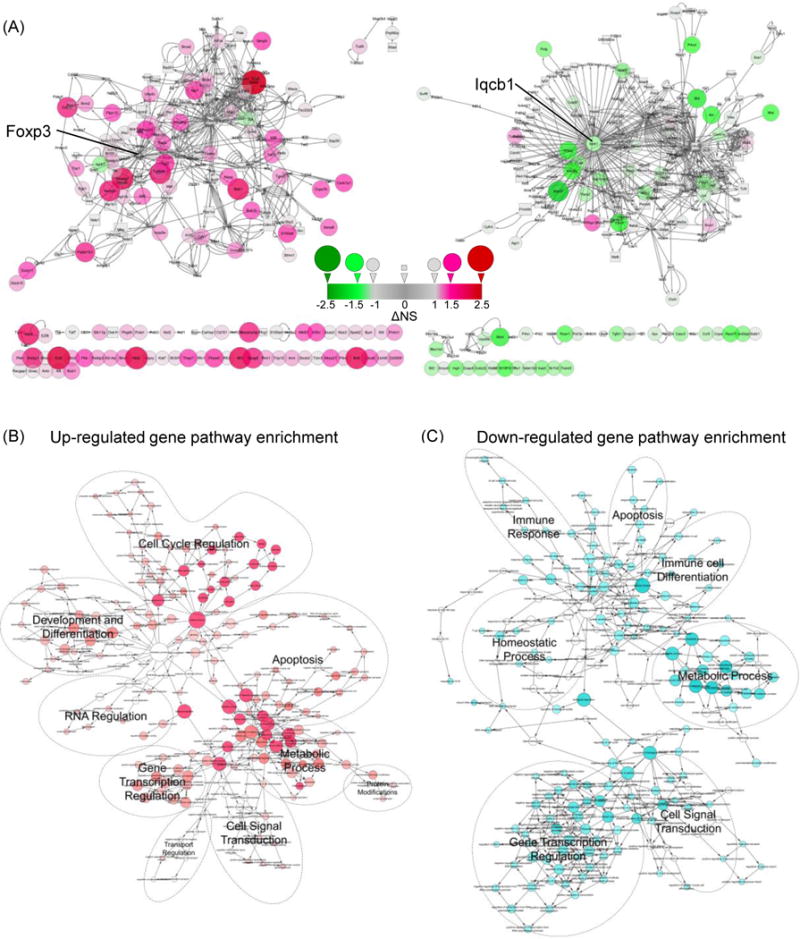
Visualization of up-regulated and down-regulated gene-gene interaction pathways.
(A) Interaction network of up-regulated genes (left panel) and down-regulated genes (right panel) and their first neighbor nodes. Fold changes between dnTGFβRII Tregs and WT Tregs were depicted by the size and color of the nodes. More than 2-fold change genes were depicted as a circular node, while less than 2-fold change neighbor nodes were depicted as squares. Up-regulated genes are indicated in red, while down-regulated genes are indicated in green. ΔNS represents the difference of dnTGFβRII Treg normalized signal minus WT Treg normalized signal. A number greater than 1.0 indicates that the gene expression was up-regulated more than 2-fold, while a number was less than −1.0 means the gene expression was down-regulated more than 2-fold. The cycles represented below the network figure show genes with auto-interaction or lacking interaction information. (B and C) Overview of biological pathway analysis. Enrichment of biological process pathways defined by Gene Ontology were generated with the BiNGO 3.0.2 plugin in Cytoscape 3.2.0. Red nodes depict processes that were targeted by up-regulated genes, while cyan nodes represent processes targeted by down-regulated genes. Number of the genes involved in the biological pathways is depicted by the size of the nodes.
To assess the possible biological pathways in which the differentially expressed genes were involved, BiNGO was applied to categorize GO biological processes and generate a pathway enrichment network. Figure 7B and 7C illustrate the overall representation of biological pathways relatively dominated by up-regulated (red) or down-regulated (cyan) genes, respectively. Both up-regulated and down-regulated genes were involved in: 1) gene transcription regulation, 2) cell signal transduction, 3) apoptosis and 4) metabolic processes. Specifically upregulated processes included: 1) cell cycle regulation, 2) development and differentiation, and 3) RNA regulation; Protein modification and transporter regulation pathways were modestly upregulated. The uniquely down-regulated pathways included 1) immune cell differentiation, 2) immune response and 3) homeostasis processes (Table 2,3). Notably, almost all of the down-regulated pathways include Tgfb1 signaling components whereas the upregulated pathways do not (Table 2,3). This raises the possibility that the primary effect of the dominant negative TGFβ receptor is impaired regulation of the immune system, with the upregulated genetic effects occurring as a secondary response.
Table 2.
GO pathways identified in uniquely up-regulated genes using BiNGO.
| RANK | GO_Term | P-value | Nr. Genes | Associated gene symbol |
|---|---|---|---|---|
| 1 | cell cycle process | 4.2E-23 | 27 | Spag5| Kif2c| Bub1b| Ckap5| Bub1| Ncapg2| Skp2| Anln| Ccna2| Mki67| Nusap1| Hells| Aurkb| Nedd9| Kif11| Pafah1b1| Brca1| Phgdh| Plk1| Cdc20| Cenpv| Cdca8| Csnk1a1| Stmn1| Ube2c| Cdk1 |
| 2 | mitotic cell cycle | 9.7E-23 | 23 | Spag5| Hells| Kif2c| Ckap5| Bub1b| Aurkb| Nedd9| Kif11| Pafah1b1| Bub1| Ncapg2| Plk1| Skp2| Cdc20| Anln| Cenpv| Cdca8| Ccna2| Csnk1a1| Ube2c| Stmn1| Cdk1 |
| 3 | cell cycle phase | 1.9E-22 | 25 | Nusap1| Spag5| Hells| Kif2c| Ckap5| Bub1b| Aurkb| Nedd9| Kif11| Pafah1b1| Bub1| Ncapg2| Plk1| Skp2| Cdc20| Anln| Cenpv| Cdca8| Ccna2| Csnk1a1| Mki67| Ube2c| Stmn1| Cdk1| |
| 4 | M phase | 2.1E-22 | 24 | Nusap1| Spag5| Hells| Kif2c| Ckap5| Bub1b| Aurkb| Nedd9| Kif11| Pafah1b1| Bub1| Ncapg2| Plk1| Cdc20| Anln| Cenpv| Cdca8| Ccna2| Csnk1a1| Mki67| Ube2c| Stmn1| Cdk1 |
| 5 | cell division | 2.9E-22 | 24 | Nusap1| Spag5| Hells| Kif2c| Ckap5| Bub1b| Aurkb| Nedd9| Kif11| Pafah1b1| Bub1| Ncapg2| Top1| Plk1| Cdc20| Anln| Cenpv| Cdca8| Ccna2| Csnk1a1| Ube2c| Cdk1| Pdcd6ip |
| 6 | nuclear division | 4.3E-22 | 21 | Spag5| Hells| Kif2c| Ckap5| Bub1b| Aurkb| Nedd9| Kif11| Pafah1b1| Bub1| Ncapg2| Plk1| Cdc20| Anln| Cenpv| Cdca8| Ccna2| Csnk1a1| Ube2c| Cdk1 |
| 7 | mitosis | 4.3E-22 | 21 | Spag5| Hells| Kif2c| Ckap5| Bub1b| Aurkb| Nedd9| Kif11| Pafah1b1| Bub1| Ncapg2| Plk1| Cdc20| Anln| Cenpv| Cdca8| Ccna2| Csnk1a1| Ube2c| Cdk1 |
| 8 | M phase of mitotic cell cycle | 4.3E-22 | 21 | Spag5| Hells| Kif2c| Ckap5| Bub1b| Aurkb| Nedd9| Kif11| Pafah1b1| Bub1| Ncapg2| Plk1| Cdc20| Anln| Cenpv| Cdca8| Ccna2| Csnk1a1| Ube2c| Cdk1 |
| 9 | cell cycle | 4.9E-22 | 30 | Spag5| Kif2c| Bub1b| Ckap5| Bub1| Ncapg2| Skp2| Anln| Ccna2| Mki67| Pdcd6ip| Racgap1| Nusap1| Hells| Aurkb| Nedd9| Kif11| Pafah1b1| Brca1| Phgdh| Plk1| Cdc20| Cenpv| Cdca8| Csnk1a1| Stmn1| Ube2c| Cdk1 |
| 10 | organelle fission | 5.0E-22 | 21 | Spag5| Hells| Kif2c| Ckap5| Bub1b| Aurkb| Nedd9| Kif11| Pafah1b1| Bub1| Ncapg2| Plk1| Cdc20| Anln| Cenpv| Cdca8| Ccna2| Csnk1a1| Ube2c| Cdk1 |
| 11 | DNA replication | 2.9E-10 | 11 | Brca1| Top1| Pola1| Rfc5| Kat7| Rmi1| Fen1| Rfc1| Rrm2| Pole |
| 12 | microtubule-based process | 7.6E-08 | 10 | Brca1| Nusap1| Kif2c| Ckap5| Ss18| Kif11| Pafah1b1| Stmn1 |
| 13 | DNA metabolic process | 2.7E-07 | 13 | Parp1| Rfc5| Hells| Rmi1| Fen1| Pole| Top1| Brca1| Pola1| Kat7| Rfc1| Rrm2| |
| 14 | regulation of apoptosis | 2.9E-07 | 16 | | Hells| Sh3rf1| Birc6| Mmp9| Brca1| Skp2| Tgm2| Tsc22d3| Bak1| Wwox| Gnaq| Traf4| Pdcd6ip |
| 15 | regulation of programmed cell death | 3.3E-07 | 16 | | Hells| Sh3rf1| Birc6| Mmp9| Brca1| Skp2| Tgm2| Tsc22d3| Bak1| Wwox| Gnaq| Traf4| Pdcd6ip |
| 16 | regulation of cell death | 4.1E-07 | 16 | | Hells| Sh3rf1| Birc6| Mmp9| Brca1| Skp2| Tgm2| Tsc22d3| Bak1| Wwox| Gnaq| Traf4| Pdcd6ip |
| 17 | intracellular signaling pathway | 3.6E-06 | 17 | Brca2| Racgap1| Plek| Spsb2| Asb2| Ar| Ect2| Chn2| Ss18| Brca1| Tgm2| Taf7| Gnaq| Stmn1| Cdk1| Pdcd6ip |
| 18 | chromosome organization | 6.8E-06 | 11 | Brca2| Parp1| Nusap1| Whsc1l1| Hells| Kat7| Cenpv| H1f0| Cdk1| |
| 19 | embryonic development | 2.1E-05 | 14 | Arnt| Racgap1| Ar| Birc6Bub1| Ncapg2| Top1| Brca1| Phgdh| Prdm1| Ell| Gnaq |
| 20 | cell proliferation | 2.4E-05 | 9 | Brca2| Racgap1| Hells| Dock2| Bak1Mki67| Pafah1b1 |
| 21 | positive regulation of apoptosis | 5.3E-05 | 9 | Brca1| Mmp9| Skp2| Tgm2| Bak1| Wwox| Sh3rf1| Pdcd6ip |
| 22 | positive regulation of programmed cell death | 5.7E-05 | 9 | Brca1| Mmp9| Skp2| Tgm2| Bak1| Wwox| Sh3rf1| Pdcd6ip |
| 23 | positive regulation of cell death | 6.4E-05 | 9 | Brca1| Mmp9| Skp2| Tgm2| Bak1| Wwox| Sh3rf1| Pdcd6ip |
| 24 | regulation of cell proliferation | 6.4E-05 | 13 | Ar| Birc6Nos3| Skp2| Tgm2| Cdc20| Bak1| Ccna2| Gnaq| Cdk1| Rrm2 |
| 25 | cytoskeleton organization | 6.6E-05 | 9 | Brca1| Nusap1| Dock2| Ckap5| Ss18| Kif11| Pafah1b1| Stmn1 |
| 26 | positive regulation of cell proliferation | 7.7E-05 | 10 | Skp2| Cdc20| Tgm2| Ccna2| Birc6| Gnaq| Cdk1| Rrm2 |
| 27 | macromolecular complex subunit organization | 1.3E-04 | 10 | Ap1b1| Tgm2| Cenpv| Taf7| H1f0| Stmn1| Cdk1| Rrm2 |
| 28 | regulation of catalytic activity | 1.8E-04 | 13 | Dock2| Ar| Chn2| Pafah1b1| Nos3| Dock10| Tgm2| Bak1| Gnaq| Dock11 |
| 29 | chordate embryonic development | 2.2E-04 | 10 | Brca2| Brca1| Phgdh| Arnt| Prdm1| Ar| Birc6| Ell |
| 30 | embryonic development ending in birth or egg hatching | 2.4E-04 | 10 | Brca2| Brca1| Phgdh| Arnt| Prdm1| Ar| Birc6| Ell |
| 31 | macromolecular complex assembly | 3.8E-04 | 9 | Ap1b1Tgm2| Cenpv| Taf7| H1f0| Cdk1| Rrm2 |
| 32 | regulation of molecular function | 6.8E-04 | 13 | Dock2| Ar| Chn2| Pafah1b1| Nos3| Dock10| Tgm2| Bak1| Gnaq| Dock11 |
| 33 | cellular component biogenesis | 8.8E-04 | 11 | Hells| Ap1b1| Tgm2| Cenpv| Taf7| H1f0| Pafah1b1| Cdk1| Rrm2| |
| 34 | cellular component assembly | 1.1E-03 | 10 | Ap1b1| Tgm2| Cenpv| Taf7| H1f0| Pafah1b1| Cdk1| Rrm2 |
| 35 | cellular response to stimulus | 1.5E-03 | 11 | Brca2| Brca1| Mmp9| Parp1| Ar| Fen1| Cdk1| Pole| Pdcd6ip| |
| 36 | programmed cell death | 1.7E-03 | 9 | Bak1| Tctn3| Wwox| Bub1b| Serpina3g| Traf4| Pdcd6ip| Bub1 |
| 37 | positive regulation of gene expression | 2.5E-03 | 10 | Arnt| Prdm1| Ar| Ccna2| Maf| Rfc1| Klf6| Cdk1 |
| 38 | cell death | 3.0E-03 | 9 | Bak1| Tctn3| Wwox| Bub1b| Serpina3g| Traf4| Pdcd6ip| Bub1 |
| 39 | death | 3.3E-03 | 9 | Bak1| Tctn3| Wwox| Bub1b| Serpina3g| Traf4| Pdcd6ip| Bub1 |
| 40 | regulation of developmental process | 3.3E-03 | 11 | Mmp9| Cdc20| H2-Aa| Ar| Maf| Gnaq| Pafah1b1| Cdk1 |
| 41 | positive regulation of macromolecule metabolic process | 4.5E-03 | 11 | Arnt| Prdm1| Ar| Ccna2| Maf| Rfc1| Klf6| Cdk1 |
| 42 | positive regulation of metabolic process | 9.4E-03 | 11 | Arnt| Prdm1| Ar| Ccna2| Maf| Rfc1| Klf6| Cdk1 |
Table 3.
GO pathways identified in uniquely down-regulated genes using BiNGO.
| RANK | GO_Term | P-value | Nr. Genes | Associated Gene Symbol |
|---|---|---|---|---|
| 1 | negative regulation of metabolic process | 3.0E-05 | 10 | Prkcd| Foxp1| Skil| Satb1| Tgfb1| Jun| Ago1| Nr1h2| Msh2 |
| 2 | negative regulation of cellular metabolic process | 9.5E-05 | 9 | Prkcd| Foxp1| Skil| Satb1| Tgfb1| Jun| Nr1h2| Msh2| |
| 3 | negative regulation of macromolecule metabolic process | 1.2E-04 | 9 | Prkcd| Foxp1| Skil| Satb1| Tgfb1| Jun| Ago1| Nr1h2| Msh2 |
| 4 | regulation of cellular protein metabolic process | 1.9E-04 | 7 | Casc3| Prkcd| Csnk1e| Tgfb1| Jun| Ago1| Nr1h2| |
| 5 | intracellular signaling pathway | 2.4E-04 | 10 | Il6st| Prkcd| Rcan1| Xpc| Sos1| Prkci| Nr1h2| Msh2| Cdc42ep4 |
| 6 | immune system process | 2.4E-04 | 9 | Prkcd| Foxp1| Skil| C1qa| Bcl2l11| Satb1| Tgfb1| Msh2| Polr3e |
| 7 | regulation of protein metabolic process | 5.0E-04 | 7 | Casc3| Prkcd| Csnk1e| Tgfb1| Jun| Ago1| Nr1h2| |
| 8 | regulation of developmental process | 5.1E-04 | 9 | Il6st| Iqcb1| Rcan1| Skil| Bcl2l11| Tgfb1| Jun| Ntn4| Cdc42ep4 |
| 9 | regulation of transcription from RNA polymerase II promoter | 1.0E-03 | 8 | Ahr| Foxp1| Skil| Satb1| Tgfb1| Jun| Nr1h2| Pbx2| |
| 10 | cellular response to stimulus | 1.2E-03 | 8 | Ahr| Ash2l| Xpc| Tgfb1| Jun| Prkci| Msh2| |
| 11 | cell death | 1.4E-03 | 7 | Ahr| Mef2a| Bcl2l11| Tgfb1| Jun| Pacs2| Msh2| |
| 12 | death | 1.5E-03 | 7 | Ahr| Mef2a| Bcl2l11| Tgfb1| Jun| Pacs2| Msh2| |
| 13 | positive regulation of transcription | 2.2E-03 | 7 | Ahr| Mef2a| Zscan21| Tgfb1| Jun| Nr1h2| Pbx2| |
| 14 | positive regulation of macromolecule metabolic process | 2.5E-03 | 8 | Ahr| Mef2a| Zscan21| Csnk1e| Tgfb1| Jun| Nr1h2| Pbx2| |
| 15 | positive regulation of gene expression | 2.7E-03 | 7 | Ahr| Mef2a| Zscan21| Tgfb1| Jun| Nr1h2| Pbx2| |
| 16 | positive regulation of cellular metabolic process | 2.8E-03 | 8 | Ahr| Mef2a| Zscan21| Csnk1e| Tgfb1| Jun| Nr1h2| Pbx2| |
| 17 | positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 3.0E-03 | 7 | Ahr| Mef2a| Zscan21| Tgfb1| Jun| Nr1h2| Pbx2| |
| 18 | cellular localization | 3.3E-03 | 7 | Casc3| Khsrp| Vps26b| Tgfb1| Prkci| Copa| |
| 19 | positive regulation of nitrogen compound metabolic process | 3.4E-03 | 7 | Ahr| Mef2a| Zscan21| Tgfb1| Jun| Nr1h2| Pbx2| |
| 20 | positive regulation of macromolecule biosynthetic process | 3.4E-03 | 7 | Ahr| Mef2a| Zscan21| Tgfb1| Jun| Nr1h2| Pbx2| |
| 21 | macromolecule localization | 3.5E-03 | 8 | Rab11fip5| Casc3| Khsrp| Vps26b| Tgfb1| Rab3ip| Copa| |
| 22 | positive regulation of metabolic process | 3.8E-03 | 8 | Ahr| Mef2a| Zscan21| Csnk1e| Tgfb1| Jun| Nr1h2| Pbx2| |
| 23 | positive regulation of cellular biosynthetic process | 4.2E-03 | 7 | Ahr| Mef2a| Zscan21| Tgfb1| Jun| Nr1h2| Pbx2| |
| 24 | positive regulation of biosynthetic process | 4.5E-03 | 7 | Ahr| Mef2a| Zscan21| Tgfb1| Jun| Nr1h2| Pbx2| |
| 25 | phosphate metabolic process | 4.7E-03 | 8 | Prkcd| Tnks| Csnk1e| Dusp2| Tgfb1| Prkci| Dusp4| Msh2| |
| 26 | phosphorus metabolic process | 4.7E-03 | 8 | Prkcd| Tnks| Csnk1e| Dusp2| Tgfb1| Prkci| Dusp4| Msh2| |
Discussion
In this study, we focused on specific transcriptional profile analysis of differentially expressed genes of dnTGFβRII Tregs compared to WT Tregs controls. Our purpose was not only to identify TGFβ related Treg genetic abnormalities, but also to identify aberrant Treg gene expression that could be operative in human PBC (affected independently by other, non-TGFβ pathways). Our results, at least in terms of phenotypic markers, are partially consistent with a recently identified cell subset coined “inflammation induced activation T regulatory cells”. However, this subset reportedly contains highly upregulated Foxp3, CTLA-4, ICOS and CD25, and potent suppressive function, while our previous data implies that dnTGFβRII Tregs have impaired suppressive activity towards pathogenic CD8+ T cells [14, 15, 31]. To understand these differences, we systematically analyzed the differentially expressed genes. Tregs from dnTGFβRII mice have upregulated inflammatory factors such as Granzyme B, IFN-γ and CCL5. Importantly, however, dnTGFβRII Tregs retain capacity to secrete IL-10. Therefore, dnTGFβRII Tregs should be considered Tregs with compromised suppressive function. Our data is also consistent with the previous observation that reduction of IL-10 [32] or ablation of CD4+ T cells in dnTGFβRII mice leads to more severe hepatic inflammation (our unpublished data).
We found transcription factors alterations in dnTGFβRII Tregs, including Eos (Ikzf4), KLF2 (Klf2), Ahr, Foxp1, and Helios (Ikzf2). Helios was initially described as a critical mediator of Foxp3-dependent gene silencing in Tregs; it directly interacts with Foxp3. Knockdown of Eos leads to up-regulation of pro-inflammatory genes, including IL-2 and IFN-γ [33]. Recent papers also suggest that CD69+CD38+CD103− Tregs, termed “Eos labile” Tregs, easily lose Eos expression and can be reprogrammed to express CD40L and IL-2 in tumor bearing draining lymph nodes, in a IL-6 dependent manner. These “reprogrammed Tregs” may help antigen presenting cells promote selective antigen specific proliferation of CD8+ T cells [34]. Hence, down regulation of these transcription factors, especially Eos, in dnTGFβRII Tregs may contribute to their “reprogramming” and acquisition of pro-inflammatory function. Hence, dnTGFβRII Tregs can down regulate CD103 but up regulate CD69 and IFN-γ, consistent with the common phenotype of reprogrammed Tregs.
Transcription factor Krüppel-like factor 2 (KLF2) has been demonstrated to regulate the quiescence, survival, activation, and migration of T cells. KLF2 is constitutively expressed in naïve T cells, then rapidly lost upon the T cell stimulation, and re-expressed in recall immune responses [35, 36]. CD4 specific KLF2 knockout mice display profound peripheral lymphopenia and spontaneous T cell activation. Furthermore, KLF2 is required for the production of peripheral but not thymus derived Tregs [37]. Thus, future studies should focus on the regulatory capacity of KLF2 in PBC.
FoxP1 has four isoforms: FoxP1A, FoxP1B, FoxP1C, FoxP1D. In T cell lineages, FoxP1 plays an important roles in the generation of naive T cells in thymus and maintenance of naive T cell quiescence in the periphery [38]. FoxP1 can also serve as a critical negative regulator for follicular helper T cell differentiation [39]. CD4-Cre;Foxp1f/f mice demonstrate slight activation of periphery CD4+ T cells and CD8+ T cells by a cell-autonomous mechanism, but Foxp1-deficient Tregs are generated normally [40]. Hence, the role of down-regulated Foxp1 in dnTGFβRII mice is not clear.
We also note distinctive differences in the transcriptional profiles of cell adhesion and chemotaxis genes in dnTGFβRII mice, including, for example, down regulation of the adhesion molecules CD103 and Vcam1. A deficiency of CD103 has been reported in the absence of TGFβ signaling in T cells [14, 41, 42]. Down regulation of CD103 and Vcam1 may lead to reduction of normal homing of T regulatory cells to mucosal tissues, promoting cholangitis in dnTGFβRII mice.
Conventional CD4+Foxp3+ Treg populations are composed of thymus-derived Tregs (t-Tregs) and peripheral Tregs (p-Tregs) that are generated in lymphoid organs upon the stimulation of specific antigen, and cytokines such as TGFβ. The t-Treg specific transcription factor Helios was up-regulated in dnTGFβRII Tregs, and a newly found p-Treg related surface marker IGFBP4 [43] was down-regulated in dnTGFβRII Tregs. These data are noteworthy and require further study. Since Foxp3 is a lineage specific transcription factor for regulatory T cells, more detailed analysis of its interactions with other transcription factors should be done. Our data suggests two possibilities. One possibility is that T regulatory populations alter their functional status and may even promote autoimmunity based upon their changes from suppressive to effector or inflammatory states following up-regulation of activated phenotypes, including, for example, CD25, GITR, ICOS, and CTLA-4. The unresolved question is why upregulated Foxp3 here does not compensate for down regulation of Eos, IGFBP4 or other transcription factors.
To comprehensively analyze the differentially expressed genes between dnTGFβRII Tregs and WT Tregs, we constructed a gene-gene interaction network. At least two topologic hubs were identified in the simplified network, Foxp3 in the up-regulated gene group and Iqcb1 in the down-regulated gene group. Furthermore, these up-regulated or down-regulated genes were clustered into biological processes according to GO and the enrichment of different pathways, in which the up-regulated and down-regulated genes involved, were analyzed separately to produce a visual network. Interestingly, immune cell differentiation, homeostatic processes, and immune response processes were identified in the down-regulated gene pathway network. Virtually all of the down-regulated processes include TGFβ1, whereas none of the upregulated processes do (Tables 2, 3). This may be a major clue to how the dominant negative TGFβR transgene disrupts tolerance: the primary defect is in regulation of immunity, which is down-regulated, leading to an upregulation of multiple other pathways including inflammation. The down regulation of these important immune “regulatory” features is correlated with defective Treg function and might well explain the key processes that are aberrant in dnTGFβRII Tregs. These pathway analyses are consistent with human studies although no human pathway analysis of Tregs has yet been performed [8, 44].
In summary, we have identified a variety of T regulatory genes and gene pathways that could have relevance, and functionally contribute to human PBC. These genes/ pathways could be affected both dependently and independently from abnormalities in TGFβ signaling, thus suggesting that dnTGFβRII modeling can identify immunogenic pathways of broad interest in PBC. Targeted analyses of human PBC Tregs to test for overlapping pathway defects that can explain the role of Tregs in PBC pathogenesis should be performed to extend these analyses. Pathway analysis will be critical in human autoimmunity, particularly when done in combination with genome-wide association studies [44, 45].
Acknowledgments
Financial support provided by the National Basic Research Program of China (973 Program-2013CB944900, 2010CB945300), the National Natural Science Foundation of China (81130058, 81430034), Research Fund for the Doctoral Program of Higher Education of China (RFDP 20133402110015) and the National Institutes of Health grant DK090019.
Abbreviations
- dnTGFβRII
Dominant negative transforming growth factor β receptor II
- PBC
primary biliary cirrhosis
- Tregs
regulatory T cells
- mLN
mesenteric lymph node
- WT
wild type
- MNC
mononuclear cells
- IFN-γ
interferon-γ
Contributor Information
Yin-Hu Wang, Email: wangyh8@mail.ustc.edu.cn.
Wei Yang, Email: ywei0702@mail.ustc.edu.cn.
Jing-Bo Yang, Email: yjingb@mail.ustc.edu.cn.
Yan-Jie Jia, Email: jyanjie@mail.ustc.edu.cn.
Wei Tang, Email: wayne012@mail.ustc.edu.cn.
M. Eric Gershwin, Email: megershwin@ucdavis.edu.
William M. Ridgway, Email: wridg1@gmail.com.
Zhe-Xiong Lian, Email: zxlian1@ustc.edu.cn.
References
- 1.Gershwin ME, Mackay IR, Sturgess A, Coppel RL. Identification and specificity of a cDNA encoding the 70 kd mitochondrial antigen recognized in primary biliary cirrhosis. Journal of immunology. 1987;138:3525–31. [PubMed] [Google Scholar]
- 2.Yang CY, Ma X, Tsuneyama K, Huang S, Takahashi T, Chalasani NP, et al. IL-12/Th1 and IL-23/Th17 biliary microenvironment in primary biliary cirrhosis: implications for therapy. Hepatology. 2014;59:1944–53. doi: 10.1002/hep.26979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawata K, Yang GX, Ando Y, Tanaka H, Zhang W, Kobayashi Y, et al. Clonality, activated antigen-specific CD8(+) T cells, and development of autoimmune cholangitis in dnTGFbetaRII mice. Hepatology. 2013;58:1094–104. doi: 10.1002/hep.26418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rong GH, Yang GX, Ando Y, Zhang W, He XS, Leung PS, et al. Human intrahepatic biliary epithelial cells engulf blebs from their apoptotic peers. Clinical and experimental immunology. 2013;172:95–103. doi: 10.1111/cei.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen RC, Naiyanetr P, Shu SA, Wang J, Yang GX, Kenny TP, et al. Antimitochondrial antibody heterogeneity and the xenobiotic etiology of primary biliary cirrhosis. Hepatology. 2013;57:1498–508. doi: 10.1002/hep.26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhirapong A, Yang GX, Nadler S, Zhang W, Tsuneyama K, Leung P, et al. Therapeutic effect of cytotoxic T lymphocyte antigen 4/immunoglobulin on a murine model of primary biliary cirrhosis. Hepatology. 2013;57:708–15. doi: 10.1002/hep.26067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan MM, Gershwin ME. Primary biliary cirrhosis. The New England journal of medicine. 2005;353:1261–73. doi: 10.1056/NEJMra043898. [DOI] [PubMed] [Google Scholar]
- 8.Hirschfield GM, Gershwin ME. The immunobiology and pathophysiology of primary biliary cirrhosis. Annual review of pathology. 2013;8:303–30. doi: 10.1146/annurev-pathol-020712-164014. [DOI] [PubMed] [Google Scholar]
- 9.Oertelt S, Lian ZX, Cheng CM, Chuang YH, Padgett KA, He XS, et al. Anti-mitochondrial antibodies and primary biliary cirrhosis in TGF-beta receptor II dominant-negative mice. Journal of immunology. 2006;177:1655–60. doi: 10.4049/jimmunol.177.3.1655. [DOI] [PubMed] [Google Scholar]
- 10.Selmi C, Meda F, Kasangian A, Invernizzi P, Tian Z, Lian Z, et al. Experimental evidence on the immunopathogenesis of primary biliary cirrhosis. Cellular & molecular immunology. 2010;7:1–10. doi: 10.1038/cmi.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–87. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Lan RY, Cheng C, Lian ZX, Tsuneyama K, Yang GX, Moritoki Y, et al. Liver-targeted and peripheral blood alterations of regulatory T cells in primary biliary cirrhosis. Hepatology. 2006;43:729–37. doi: 10.1002/hep.21123. [DOI] [PubMed] [Google Scholar]
- 13.Aoki CA, Roifman CM, Lian ZX, Bowlus CL, Norman GL, Shoenfeld Y, et al. IL-2 receptor alpha deficiency and features of primary biliary cirrhosis. Journal of Autoimmunity. 2006;27:50–3. doi: 10.1016/j.jaut.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka H, Zhang W, Yang GX, Ando Y, Tomiyama T, Tsuneyama K, et al. Successful Immunotherapy of Autoimmune Cholangitis by Adoptive Transfer of Foxp3 Regulatory T cells. Clinical and experimental immunology. 2014 doi: 10.1111/cei.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang W, Kachapati K, Adams D, Wu Y, Leung PS, Yang GX, et al. Murine autoimmune cholangitis requires two hits: cytotoxic KLRG1(+) CD8 effector cells and defective T regulatory cells. J Autoimmun. 2014;50:123–34. doi: 10.1016/j.jaut.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–41. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Yang W, Yao Y, Yang YQ, Lu FT, Li L, Wang YH, et al. Differential Modulation by IL-17A of Cholangitis versus Colitis in IL-2Ralpha Deleted Mice. PloS one. 2014;9:e105351. doi: 10.1371/journal.pone.0105351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–20. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Yao Y, Yang W, Yang YQ, Ma HD, Lu FT, Li L, et al. Distinct from its canonical effects, deletion of IL-12p40 induces cholangitis and fibrosis in interleukin-2Ralpha(−/−) mice. J Autoimmun. 2014;51:99–108. doi: 10.1016/j.jaut.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Gene Ontology C. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–6. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with cytoscape 3. Curr Protoc Bioinformatics. 2014;47:8 13 1–8 24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–23. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–9. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 26.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, et al. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–5. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 28.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nature protocols. 2006;1:1559–82. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 30.Hilscher C, Vahrson W, Dittmer DP. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic acids research. 2005;33:e182. doi: 10.1093/nar/gni181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arvey A, van der Veeken J, Samstein RM, Feng Y, Stamatoyannopoulos JA, Rudensky AY. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nature immunology. 2014;15:580–7. doi: 10.1038/ni.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang SS, Bloom SM, Norian LA, Geske MJ, Flavell RA, Stappenbeck TS, et al. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS medicine. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–6. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma MD, Huang L, Choi JH, Lee EJ, Wilson JM, Lemos H, et al. An inherently bifunctional subset of Foxp3+ T helper cells is controlled by the transcription factor eos. Immunity. 2013;38:998–1012. doi: 10.1016/j.immuni.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Z, Sun X, Icli B, Wara AK, Feinberg MW. Role of Kruppel-like factors in leukocyte development, function, and disease. Blood. 2010;116:4404–14. doi: 10.1182/blood-2010-05-285353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hart GT, Hogquist KA, Jameson SC. Kruppel-like factors in lymphocyte biology. Journal of immunology. 2012;188:521–6. doi: 10.4049/jimmunol.1101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pabbisetty SK, Rabacal W, Maseda D, Cendron D, Collins PL, Hoek KL, et al. KLF2 is a rate-limiting transcription factor that can be targeted to enhance regulatory T-cell production. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9579–84. doi: 10.1073/pnas.1323493111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng X, Wang H, Takata H, Day TJ, Willen J, Hu H. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nature immunology. 2011;12:544–50. doi: 10.1038/ni.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Geng J, Wen X, Bi E, Kossenkov AV, Wolf AI, et al. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat Immunol. 2014;15:667–75. doi: 10.1038/ni.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng X, Ippolito GC, Tian L, Wiehagen K, Oh S, Sambandam A, et al. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood. 2010;115:510–8. doi: 10.1182/blood-2009-07-232694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang LY, Lin YC, Kang CW, Hsu CY, Chu YY, Huang CT, et al. The indispensable role of CCR5 for in vivo suppressor function of tumor-derived CD103+ effector/memory regulatory T cells. Journal of immunology. 2012;189:567–74. doi: 10.4049/jimmunol.1200266. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds LA, Maizels RM. Cutting edge: in the absence of TGF-beta signaling in T cells, fewer CD103+ regulatory T cells develop, but exuberant IFN-gamma production renders mice more susceptible to helminth infection. Journal of immunology. 2012;189:1113–7. doi: 10.4049/jimmunol.1200991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. The Journal of experimental medicine. 2012;209:1723–42. S1. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kar SP, Seldin MF, Chen W, Lu E, Hirschfield GM, Invernizzi P, et al. Pathway-based analysis of primary biliary cirrhosis genome-wide association studies. Genes and immunity. 2013;14:179–86. doi: 10.1038/gene.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lleo A, Zhang W, McDonald WH, Seeley EH, Leung PS, Coppel RL, et al. Shotgun proteomics: identification of unique protein profiles of apoptotic bodies from biliary epithelial cells. Hepatology. 2014;60:1314–23. doi: 10.1002/hep.27230. [DOI] [PMC free article] [PubMed] [Google Scholar]


