Abstract
Protein carbonyls are protein oxidation products that are often used to measure the magnitude of protein oxidative damage induced by reactive oxygen or reactive nitrogen species. Protein carbonyls have been found to be elevated during aging and in age-related diseases such as stroke, diabetes, and neurodegenerative diseases. In the present article, we provide detailed protocols for detection of mitochondrial protein carbonyls labeled with biotin-hydrazide followed by 2-dimensional isoelectric focusing (IEF)/SDS-PAGE and Western blotting probed with horse-radish peroxidase-conjugated streptavidin. The presented procedures can also be modified for detection of carbonylation of non-mitochondrial proteins.
Keywords: biotin-hydrazide, protein carbonyls, carbonylation, IEF/SDS-PAGE, mitochondria, streptavidin
1. Introduction
Oxidative stress occurs due to redox imbalance and is caused by overwhelming production of reactive oxygen species and reactive nitrogen species [1, 2]. A direct consequence of this stress is oxidative modifications of proteins [3]. One type of protein modification is protein carbonyl formation (protein carbonylation) [4], which is an irreversible process and can occur on multiple amino acids residues such as histidine, lysine, cysteine, arginine, threonine, and proline [5]. Protein carbonyls have been widely used as a biomarker for oxidative stress during aging and under a variety of pathological conditions [4, 6-8]. Moreover, as protein carbonyls accumulate with progression of age- or disease-associated oxidative stress [9, 10], studying protein carbonylation may also help elucidating the mechanisms of oxidative stress and the nature of oxidative stress-induced impairment in protein function [11-14].
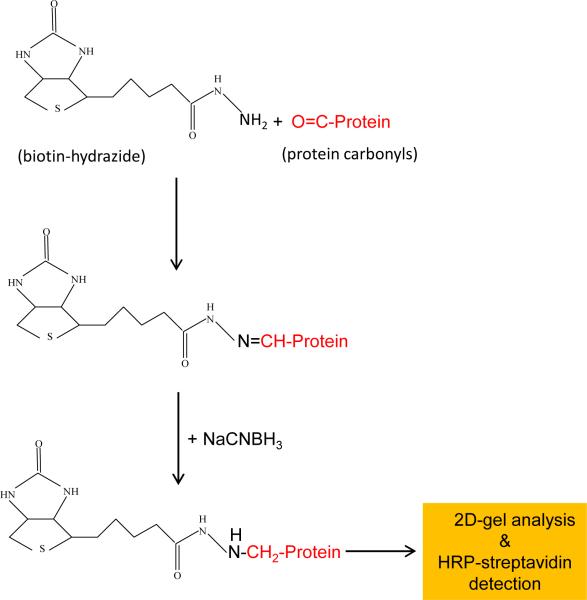
Protein carbonyls have been widely analyzed by the use of 2,4-dinitrophenylhydrazine (DNPH) [4], both spectrophotometrically and immunochemically due to the fact that DNPH itself has a maximum absorbance at 360 nm and that antibodies against DNPH are commercially available [3]. As the affinity of anti-DNPH antibodies varies widely from source to source and from batch to batch, reproducibility of the DNPH immunochemical assay could pose a potential problem [15]. Additionally, nonspecific anti-DNP detection could also occur [16]. Therefore, other affinity-based analysis of protein carbonyls has been developed. One such an assay is biotinylation of protein carbonyls using biotin-hydrazide probes in conjunction with detection by streptavidin [16-19] (Fig. 1). Biotinylated protein carbonyls can be analyzed by gel electrophoresis and western blot or purified by streptavidin beads or columns. In this article, we provide detailed procedures for 2-dimensional gel electrophoretic detection of mitochondrial protein carbonyls derivatized by biotin-hydrazide. The methods are also applicable for non-mitochondrial proteins.
Fig. 1.
Labeling reactions of protein carbonyls with biotin-hydrazide.
2. Materials and methods
2.1. Protocols for mitochondria isolation
In this article, we will use rat testis as the source of mitochondria, but the mitochondria isolation procedures [20] can also be used for other tissues such as liver, kidney, heart, and skeletal muscle. It should be noted that our focus in this article is placed more on sample preparation and labeling with the carbonyl probe biotin hydrazide than on gel electrophoretic procedures as both IEF and SDS-PAGE as well as Western blot can be performed by standard protocols. All chemicals are obtained from Sigma unless otherwise indicated.
2.1.1. Mitochondria isolation buffer (ice-cold)
70 mM sucrose (12 gram for 500 ml)
230 mM mannitol (20 gram for 500 ml)
15 mM MOPS, pH 7.2 (1.6 gram MOPS for 500 ml, adjust pH with KOH)
1 mM K2EDTA
2.1.2. Procedures
-
1)
Rinse tissues to remove any residue blood
-
3)
Homogenize (1 gram tissue/10 ml isolation buffer)
-
3)
Centrifuge the homogenate at 700 g for 10 min
-
4)
Centrifuge the resulting supernatant at 8,000 g for 10 min
-
5)
Resuspend the mitochondrial pellet with 10 ml of isolation buffer and centrifuge at 8,000 g for 10 min (this step serves the purpose of washing)
-
6)
Store the mitochondrial pellet at −80°C or use immediately
2.2
Protocols for labeling of protein carbonyls with biotin hydrazide [3]
2.2.1. Buffers and solutions
-
1)
100 mM sodium acetate, 20 mM NaCl, pH 6.0 containing 1% SDS
-
2)
60 mM biotin-hydrazide in DMSO. This stock solution is stable at −80°C for at least 6 months. (Biotin-hydrazide Sigma catalog number: B7639, MW: 258.34)
-
3)
30 mM cyanoborohydrazide in phosphate buffered saline (PBS)
-
4)
100% Trichloroacetic acid (TCA)
-
5)
Ethanol: ethyl acetate (1:1, v/v)
2.2.2. Procedures
-
1)
Dissolve mitochondrial pellet isolated as described above in 1 ml 100 mM sodium acetate, 20 mM NaCl, pH 6.0 containing 1% SDS. (1 ml/testis mitochondria pellet)
-
2)
Let the sample stand at room temperature for 10 min
-
3)
Clarify the sample by centrifugation at 13,000 g for 10 min. This step will remove any insoluble materials
-
4)
Keep the resulting supernatant and discard any pellet
-
5)
Add 60 mM biotin-hydrazide (in DMSO) to the supernatant to give a final concentration of 5 mM
-
6)
Incubate at room temperature on a shaker for 2 h
-
7)
Cool down on ice to 0°C
-
8)
Add equal volume of 30 mM sodium cyanoborohydrazide (prepared fresh in PBS)
-
9)
Keep on ice for 30 min with shaking
-
10)
Add 100% TCA to a final concentration of 10%, leave on ice for 10 min
-
11)
Centrifuge on bench top centrifuge (a floating pellet is always observed)
-
12)
Wash with ethanol: ethyl acetate (1:1, v/v) 3 times
2.3
First dimensional isoelectric focusing gel electrophoresis and second dimensional SDS-PAGE
2.3.1
Dissolve biotinylated mitochondrial pellet obtained above in the following solution:
40 mM Tris (48.4 mg/10 ml)
7 M urea (4.2 g/10 ml, FW=60)
2 M thiourea (1.52 g/10 ml, FW=76.12)
1% Triton X-100
4% CHAPS (0.4 g/10 ml)
1% DTT (100 mg/10 ml = 65 mM, FW=164.253)
2.3.2
Keep at room temperature for 60 minutes; clarify the solution by centrifugation at 20,000 g for 10 min.
2.3.3
Perform protein assay (Bio-Rad assay at 595 nm). (Make 5 fold dilutions just for protein assay purpose). None of the above chemicals interferes with the Bio-Rad protein assay (Bradford assay) kit that reads at 595 nm.
2.3.4
Mix protein sample with 2 X IEF sample buffer so that the final concentration would be 50 μg/125 μl.
IEF Sample buffer (10 ml, 2×):
7 M urea (4.2 g/10 ml)
2 M thiourea (1.52 g/10 ml)
4% CHAPS (0.4 g/10 ml)
50 mM DTT (77 mg/10 ml)
0.4% Biolyte (pH 3-10) (100 μl of 40%)
0.002% Bromophenol Blue
2.3.5
Perform rehydration and focusing (according to the instructions given by Bio-Rad)
-
1).
Rehydrate each IEF gel strip with 125 μl of protein mixture that contains a total of 2-4 mg proteins. Cover each strip with mineral oil and rehydrate at room temperature over night or at least 12 hours. The mineral oil used is also from Bio-Rad.
-
2).
Rinse each IEF strip with dd water and transfer the strip to focusing tray and focus for at least 12 hours. The wicks at each electrode should be wet with 10 μl of dd water. Cover each gel strip with mineral oil. Use preset settings (refer to the instruction manual) for focusing.
2.3.6
After IEF running is complete, rinse (with dd water) and equilibrate the IEF gel strip in 10 ml equilibration buffer I for 15 min by shaking
Equilibration buffer I (for 10 ml):
3.6 g urea
2.5 ml 1.5 M Tri-HCl, H 8.8
1 ml 20% SDS
1 ml 100% glycerol
2 ml 100% 2ME (or 200 mg DTT=130 mM)
2.3.7
Equilibrate in 10 ml equilibration buffer II for 15 min by shaking
Equilibration buffer II (for 10 ml):
3.6 g urea
2.5 ml 1.5 M Tri-HCl, H 8.8
1 ml 20% SDS
1 ml 100% glycerol
250 mg iodoacetamide (135 mM)
2.3.8
Perform second dimensional SDS-PAGE on a 10% resolving gel with 4% stacking gel according to standard procedures (a solution of 0.7% agarose containing 0.02% bromophenol blue is used to immobilize the gel strips in the stacking gel).
2.3.9
Electro-transfer of proteins from gels to Western blot polyvinyl difluoride (PVDF) or nitrocellulose membranes and perform Western blot detection using streptavidin-HRP according to standard procedures.
2.4. Results and notes
2.4.1. Results
A representative 2D detection of age-dependent increase in protein carbonylation of testis mitochondria is shown in Fig. 2. As can be seen in this figure, many proteins show age-related increase in their carbonyl content. Interestingly, some proteins also show an age-dependent decrease in their carbonyl content (such as spot number 7). If desired, proteins contained in these spots can be further analyzed and identified by mass spectrometry peptide sequencing, and individual proteins might be selected for functional analysis in the context of aging.
Fig. 2.
Representative 2D maps comparing intensities of protein carbonyls of rat testicular mitochondrial proteins between young and old rats. Numbered spots indicate those proteins showing apparent age-dependent changes in their carbonyl content.
2.4.2. Notes
-
1).
For mitochondria isolation, perform all procedures at low temperature such as tissue homogenization on ice and all centrifugations at 4°C.
-
2).
The IEF protocol described here is for the use of Bio-Rad Protean IEF cell. If a different IEF unit from another company is to be used, the instructions given by the manufacturer need to be followed.
-
3).
HRP-streptavidin can be diluted 1:50,000 in Tris-buffered saline containing 0.1% Tween-20 (TBST) and 0.2% BSA (pH 7.4). However, if the Western blot signal is too strong, BSA may be replaced with 5% non-fat dried milk in the same dilution buffer to hinder low affinity binding between Biotinylated (carbonylated) proteins and streptavidin. The presence of high concentrations of milk proteins is to compete with any nonspecific or low affinity bindings between biotinylated proteins and streptavidin.
-
4).
It has been reported [16] that sodium cyanoborohydride may not be needed to stabilize the resulting hydrazone bond (Fig.1). This is presumably because the next steps in sample preparations for both IEF gel and SDS-PAGE involve the use of reducing reagents such as DTT or β-mercaptoethanol that could still reduce and stabilize the hydrazone bond. In our lab, we have always used sodium cyanoborohydride to stabilize the hydrazone bonds before further process. There is a probe called N'-aminooxymethylcarbonylhydrazino-D-biotin [21] that can be used to label protein carbonyls without the use of sodium cyanoborohydride as the resulting labeling product is stable. A similar aldehyde-reactive biotin probe without the need of a reducing agent during labeling has also been introduced recently [22].
-
5).
If no hydrazone reduction by sodium cyanoborohydride is to be performed and if the sample is to be analyzed by 1D SDS-PAGE, the next step of TCA precipitation can be omitted. Instead, the Biotinylated protein samples can be directly loaded for SDS-PAGE analysis. This is because the small molecule biotin-hydrazide should not interfere with gel electrophoresis. However, for 2D PAGE, TCA precipitation and washing are still needed as the 1% SDS used in sample suspension and labeling will interfere with IEF gel electrophoresis.
-
6).
Iodoacetamide in section 2.3.7 is used to block disulfide-derived free sulfhydryl groups after DTT reduction.
-
7).
For further analysis such as protein identification by mass spectrometry peptide sequencing, an extra set of gels using the same samples can be run separately for protein staining by Coomassie blue or silver staining. The matched spots on the protein staining gel to those on the Western blot images can be excised for identification.
Highlights.
A general method is given for mitochondria isolation from non-muscle tissues
Mitochondrial proteins are labeled with biotin-hydrazide that specifically reacts with protein carbonyls that are products of protein oxidative damage
Protein carbonyls are detected by 2-dimensional IEF/SDS-PAGE and Western blotting probed with streptavidin-HRP
Representative results are given; and notes and precautions are discussed and provided
Acknowledgments
LJY was supported in part by the National Institutes of Health (Grant: R01NS079792)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None to be declared
References
- 1.Halliwell B, Gutteridge J. Free Radical in Biology and Medicine. 5th ed. Oxford University Press; 2015. [Google Scholar]
- 2.Luo X, Li R, Yan LJ. Roles of Pyruvate, NADH, and Mitochondrial Complex I in Redox Balance and Imbalance in β Cell Function and Dysfunction. Journal of diabetes research. 2015 doi: 10.1155/2015/512618. DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan LJ. Analysis of oxidative modification of proteins. Curr Protoc Protein Sci. 2009 doi: 10.1002/0471140864.ps1404s55. Chapter 14 Unit14 14. [DOI] [PubMed] [Google Scholar]
- 4.Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, Ahn BW, Shaltiel S, Stadtman ER. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 5.Cai Z, Yan LJ. Protein oxidative modifications: Beneficial roles in disease and health. Journal of Biochemical and Pharmacological Research. 2013;1:15–26. [PMC free article] [PubMed] [Google Scholar]
- 6.Fedorova M, Bollineni RC, Hoffmann R. Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom Rev. 2014;33:79–97. doi: 10.1002/mas.21381. [DOI] [PubMed] [Google Scholar]
- 7.Shacter E, Williams JA, Lim M, Levine RL. Differential susceptibility of plasma proteins to oxidative modification: examination by western blot immunoassay. Free Radic Biol Med. 1994;17:429–437. doi: 10.1016/0891-5849(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 8.Keller RJ, Halmes NC, Hinson JA, Pumford NR. Immunochemical detection of oxidized proteins. Chem Res Toxicol. 1993;6:430–433. doi: 10.1021/tx00034a007. [DOI] [PubMed] [Google Scholar]
- 9.Yan LJ, Sohal RS. Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc Natl Acad Sci USA. 1998;95:12896–12901. doi: 10.1073/pnas.95.22.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan LJ, Levine RL, Sohal RS. Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. Sci. USA. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 12.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 13.Shacter E. Quantification and significance of protein oxidation in biological samples. Drug Metab Rev. 2000;32:307–326. doi: 10.1081/dmr-100102336. [DOI] [PubMed] [Google Scholar]
- 14.Shacter E. Protein oxidative damage. Methods Enzymol. 2000;319:428–436. doi: 10.1016/s0076-6879(00)19040-8. [DOI] [PubMed] [Google Scholar]
- 15.Yan LJ, Forster MJ. Chemical probes for analysis of carbonylated proteins: A review. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:1308–1315. doi: 10.1016/j.jchromb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensley K. Detection of protein carbonyls by means of biotin hydrazide-streptavidin affinity methods. Methods Mol Biol. 2009;536:457–462. doi: 10.1007/978-1-59745-542-8_46. [DOI] [PubMed] [Google Scholar]
- 17.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 18.Yoo BS, Regnier FE. Proteomic analysis of carbonylated proteins in two-dimensional gel electrophoresis using avidin-fluorescein affinity staining. Electrophoresis. 2004;25:1334–1341. doi: 10.1002/elps.200405890. [DOI] [PubMed] [Google Scholar]
- 19.Hensley K. Detection of Protein Carbonyls by Means of Biotin Hydrazide-Streptavidin Affinity Methods. Methods Mol Biol. 2015;1314:95–100. doi: 10.1007/978-1-4939-2718-0_11. [DOI] [PubMed] [Google Scholar]
- 20.Navarro A, Boveris A. Rat brain and liver mitochondria develop oxidative stress and lose enzymatic activities on aging. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1244–1249. doi: 10.1152/ajpregu.00226.2004. [DOI] [PubMed] [Google Scholar]
- 21.Chung WG, Miranda CL, Maier CS. Detection of carbonyl-modified proteins in interfibrillar rat mitochondria using N'-aminooxymethylcarbonylhydrazino-D-biotin as an aldehyde/keto-reactive probe in combination with Western blot analysis and tandem mass spectrometry. Electrophoresis. 2008;29:1317–1324. doi: 10.1002/elps.200700606. [DOI] [PubMed] [Google Scholar]
- 22.Bollineni RC, Fedorova M, Bluher M, Hoffmann R. Carbonylated plasma proteins as potential biomarkers of obesity induced type 2 diabetes mellitus. J Proteome Res. 2014;13:5081–5093. doi: 10.1021/pr500324y. [DOI] [PubMed] [Google Scholar]