Abstract
In this study, we develop and test a method to determine the rate of particle transfer and kinetic isotope effects in enzymatic reactions, specifically yeast alcohol dehydrogenase (YADH), from first-principles. Transition path sampling (TPS) and normal mode centroid dynamics (CMD) are used to simulate these enzymatic reactions without knowledge of their reaction coordinates and with the inclusion of quantum effects, such as zero-point energy and tunneling, on the transferring particle. Though previous studies have used TPS to calculate reaction rate constants in various model and real systems, it has not been applied to a system as large as YADH. The calculated primary H/D kinetic isotope effect agrees with previously reported experimental results, within experimental error. The kinetic isotope effects calculated with this method correspond to the kinetic isotope effect of the transfer event itself. The results reported here show that the kinetic isotope effects calculated from first-principles, purely for barrier passage, can be used to predict experimental kinetic isotope effects in enzymatic systems.
Graphical abstract
INTRODUCTION
Particle transfer plays a crucial role in a breadth of enzymatic reactions, including hydride, hydrogen atom, electron, and proton-coupled electron transfer reactions.1 The transfer of the relevant particle or particles from donor to acceptor can involve tunneling through the barrier to reaction,2 coupled motion,3 or both.4 Kinetic isotope effects, the ratio of two rates of reactions that are identical except for an isotopic substitution, are powerful tools in the determination of the contributions of these phenomena to enzymatic mechanism, as well as the geometry of the transition state.5 The difference in rates arises from differences in zero point energy and vibrational frequency between different isotopes, and the effects of the isotopic substitution provide information on the reaction’s progress to the transition state. Of particular interest to this study, kinetic isotope effects are important tools in the study of hydride-transfer enzymes, as kinetic isotope effects can indicate whether or not tunneling is a component of the reaction.6 Computational determination of these kinetic isotope effects would allow for this mechanistic detail to be revealed without the use of experiments.
A review of the inclusion of nuclear quantum effects (NQEs) into computational methods to simulate enzyme kinetics can be found in Gao and Truhlar.7 A short review of some of these methods follows. One of the first methods to incorporate NQEs into the computational study of enzymes is the quantized classical path (QCP) method,8,9 which utilizes a dispersed polaron model within a path integral method to approximate the quantum rate constant from the quantum component of the free energy change. A related method is the staging method from the Chandler group.10 Another method that uses an energy gap formalism like QCP is a hybrid quantum-classical approach of Hammes-Schiffer and co-workers, which models the transferring particle nucleus as a multidimensional vibrational wave function.11,12 This method, however, is computationally expensive when including multiple quantum particles.13 Additionally, variations on transition state theory (TST), such as ensemble-averaged variational transition state theory with multidimensional tunneling (EA-VTST/MT),14 have also been used to calculate kinetic isotope effects in enzymatic reactions. However, this method relies on an assumption of the dividing surface and, in many practical implementations, relies on a correctly chosen reaction coordinate for use in umbrella sampling. In an effort to reduce the error of the calculation of kinetic isotope effects, Major and Gao developed a path integral and free energy perturbation/umbrella sampling method (PI-FEP/UM),15,16 which allows for the inclusion of quantum effects to the calculation of potential of mean force (PMF) in condensed phase hydrogen transfer reactions. By including a mass perturbation method into the calculation of the kinetic isotope effect, Major and Gao were able to calculate the kinetic isotope effect in one simulation, reducing the resulting error.
In contrast to many of the previously mentioned methods, this study seeks to develop a method to calculate the relative rates of enzymes from first-principles, with a fully dynamical description of the reaction, and without the need to rely on an a priori assumption of a reaction coordinate or dividing surface. To do so, we utilize transition path sampling (TPS),17,18 a Monte Carlo method which samples reactive trajectory space, to study enzymatic reactions without prior knowledge of a reaction coordinate. Additionally, quantum propagation of the transferring particle with normal mode centroid molecular dynamics (CMD)19–23 allows for the approximate inclusion of zero-point energy effects, as well as possible tunneling of the transferring particle through the reaction barrier, and has been shown to allow for the efficient propagation of more than one quantum particle in the same simulation.24
Yeast alcohol dehydrogenase-1 (YADH) presents a good model system on which to test new methods, as it is well studied experimentally25–30 and computationally,24,31 and has a high-resolution crystal structure.32 YADH catalyzes the interconversion of alcohols and aldehydes via the transfer of a proton to a histidine through a hydrogen bonding chain and the transfer of a hydride from the substrate alcohol to the nicotinamide ring of the nicotinamide adenine dinucleotide (NAD) cofactor.33 It should be noted that most computational work, including this current study, starts with the proton already transferred from the substrate,24 as it is a pre-equilibrium phenomenon and is very rapid relative to the hydride transfer.26 Prior work indicates that YADH may utilize hydride tunneling25,26,28 and coupled motion29 as main facets of its chemical reaction, though recent computational work disputes the contribution of hydride tunneling.24 It is well-known that aromatic substrates isolate the chemical step as rate-determining.25,26 Furthermore, motions coupled to the reaction coordinate that promote the chemical reaction, called rate-promoting vibrations, have been identified in horse liver alcohol dehydrogenase (HLDH).34 With this context, we seek to develop a method using transition path sampling and normal mode centroid dynamics to calculate the rate of particle transfer in enzymatic reactions. To test the method, the primary H/D kinetic isotope effect in the conversion of benzyl alcohol to benzaldehyde in yeast alcohol dehydrogenase is calculated to determine if the kinetic isotope effects of hydride transfer agree with measurements on the full enzymatic reaction.
THEORY AND METHODS
System Setup
The starting structure for simulations was yeast alcohol dehydrogenase from Saccharomyces cerevisiae, crystallized with nicotinamide-8-iodo-adenine-dinucleotide and trifluoroethanol (pdb: 2HCY).32 Preparation of this structure for use in dynamics was done previously by Dzierlenga et al.24 For completeness, the preparation is described here. The substrate analogue, trifluoroethanol, was changed to benzyl alcohol, and the iodine of the coenzyme analogue, nicotinamide-8-iodo-adenine-dinucleotide (NAD), was removed to create NAD+. Force field parameters for benzyl alcohol were generated using CGenFF.35 The system was then solvated explicitly with a 130 Å diameter sphere of TIP3P water molecules and neutralized with 20 K+ ions. All simulations were performed using the CHARMM36,37 molecular dynamics package and the CHARMM27 force field.38 In order to perform QM/MM simulations, the system was partitioned into quantum mechanical and molecular mechanical regions and stitched together using the generalized hybrid orbital (GHO) method of Gao and co-workers.39 The quantum region consists of the active site of one monomer, consisting of the benzyl alcohol substrate, the nicotinamide ring of the NAD+, the catalytic zinc, and the side chains of the three zinc-coordinating amino acids (two thiolate form cysteines and a histidine). The GHO linker atoms were the α-carbon atoms of the zinc-coordinating amino acids and the NC1 carbon of the nicotinamide ring. In the simulations, this quantum region was simulated using a PM3 method40,41 modified to better model metalloenzymatic zinc ions.42 Minimization proceeded with 100 steps of steepest descent minimization, followed by 1000 steps of adopted-basis Newton–Raphson minimization. During minimization, constraints were added to the protein (20 (kcal/mol)/Å2), quantum region (50 (kcal/mol)/Å2), and coenzyme, substrate, and catalytic zinc (20 (kcal/mol)/Å2), which were progressively relaxed over the course of minimization. The system was then heated to 300 K from 0 K over 300 ps, followed by 300 ps of equilibration.
Transition Path Sampling
TPS was used to study the particle transfer event. TPS is a powerful method to study rare events; it allows for the study of these systems without prior knowledge of a reaction coordinate, a significant advantage in complex systems such as enzymes. First, an order parameter, a metric describing the progress of the reaction, is determined. This order parameter need not be the reaction coordinate itself for TPS to work correctly. However, it must accurately describe the course of the particle transfer in order for the trajectories to capture the nature of the reaction. An initial reactive trajectory is created by constraining the donor and acceptor to force the system to react. A time slice from this initial trajectory is chosen at random, and the momenta are perturbed randomly in accordance with a Boltzmann distribution. From this set of coordinates and randomly perturbed momenta, a new, unconstrained trajectory is propagated. This new trajectory is accepted if it starts in the reactant well and ends in the product well, i.e., if the trajectory is reactive, in either direction. By doing this in succession, using the previous trajectory as a seed for each new trajectory, an ensemble that obeys Boltzmann statistics and contains all paths from reactants to products is created. This method has been used previously to study enzyme dynamics and kinetics, such as in YADH,24 HLDH,43,44 and dihydrofolate reductase.45
Rate Calculation Algorithm
In order to examine the kinetics of the particle transfer event, we utilized an algorithm developed to calculate reaction rates within the TPS framework.46,47 A short description follows. Due to the fact that, in systems where reactants and product wells are stable states, transitions between the two are rare, the rate constant can be equated to a correlation function, C(t) ≈ kR→Pt. Consequently, the time derivative of the correlation function is the reaction rate constant in the region in which it plateaus. The correlation function, C(t), of product formation from reactants is calculated (eq 1), where hR and hP are Heaviside functions for the reactant and product wells, respectively, t is the length of the trajectory, and 〈…〉 denotes an ensemble average. It should be noted that the Heaviside functions will be 1 if the trajectory is a member of the respective well and 0 otherwise.
| (1) |
The time derivative of this correlation function is the rate, k(t). The correlation function can be factorized47 into two terms to calculate the reaction rate (eq 2), where 〈hP(t)〉RP and 〈hP(t′)〉RP are the ensemble average values of hP(t) and hP(t′) in the ensemble FRP(x0,T) ≡ ρ(x0) hR(x0) HP(x0,T), with HP(x0,T) denoting trajectories which visit, but do not necessarily end in, the product well in the interval [0:T]. C(t′) is a correlation function at a specific time t′ in the interval [0:t]. This t′ can be chosen to be much smaller than t,47 in order to make the large quantity of trajectories acquired more manageable.
| (2) |
Thus, the computationally prohibitive calculation of a correlation function at every time slice along a trajectory has been reduced to two simpler calculations: the calculation of the first term, 〈ḣP(t)〉AB, via one TPS simulation to a plateau, and the calculation of the correlation function at a specific time, t′, which is done with a method described here. For trajectories starting in the reactant region, R, at time t = 0, the distribution of the order parameter, λ, at time t is
| (3) |
In a rare event system, this distribution is very small in the product region, so direct calculation is not feasible. This can be circumvented by defining a series of overlapping windows W[i], where x ∈ W[i] → λmin[i] ≤ λ(x) ≤ λmax[i] and the union of all windows W[i] yields the entirety of phase space. The distribution of the order parameter in the windows W[i] is then calculated separately,
| (4) |
The full distribution function P(λ,t) can then be obtained by matching the individual histograms P(λ,t;i), using the overlapping regions of each window W[i] where available, to refine the matching. The correlation function can then be calculated by integration of the product region of this full histogram:
| (5) |
Variations of this method have previously been used to calculate rates in simpler systems; combined with umbrella sampling, it has been used to calculate the rate of sodium chloride dissociation at air–water48 and organic–aqueous interfaces,49 the rate of diffusion of hydrocarbons in a silica zeolite,50 and the component of mechanical stress in the base-catalyzed hydrolysis of tetraglycine.51
Centroid Molecular Dynamics
In order to account for zero point energy differences between hydride and deuteride and the possible contribution of tunneling, normal mode centroid molecular dynamics (CMD)19–23 was applied to the propagation of the transferring particle in the simulations. Full details of the integration of CMD into TPS can be found in previous works.24,52 In CMD formalism, the motion of the quantum particle is described by the motion of its centroid (Ri) and its mass (mi) in a path integral average, 〈…〉c.
| (6) |
The position of the centroid is the center of mass of B beads,
| (7) |
where particles are denoted by Latin subscripts and beads are denoted by Greek subscripts. The centroid force, Fi (eq 8) is the sum of forces on all beads with mass mi/B.
| (8) |
The path integral average of eq 6 is weighted by a factor of exp(−βVeff) where Veff is the effective bead potential
| (9) |
with neighboring beads in the polymer chain connected by harmonic potentials with a spring constant of ki = miB(kBT)2. To simplify the calculation of the centroid force, a normal mode transformation is applied to eq 9 to separate the faster motion of the beads from the motion of the centroid
| (10) |
This normal mode transformation allows for the path integral average value of the force to be replaced by the instantaneous force of the propagation of the centroid, a technique analogous to Carr–Parrinello molecular dynamics.53 CMD was implemented into CHARMM directly into the source code; during the energy calculation of each time step, the CMD code calculates the force on the quantum particle over several shorter bead propagation time steps, i.e., a multistep propagation. The CHARMM calculated force on the quantum particle is then replaced by the centroid calculated force. For the purposes of this study, eight beads were chosen as a compromise between accuracy and speed; an increase in the number of beads was found to substantially increase the computation time required for little change in the accuracy of the calculated centroid force. One method to determine the validity of the CMD modeled particles is to examine the radius of the necklace of beads;54 the radius of the deuteride particle should be approximately a factor of smaller than the radius of the hydride particle, which we observed in our CMD implementation. While CMD allows for the inclusion of zero point energy differences between isotopes, it comes at the cost of increased CPU time for calculations; each trajectory with quantum propagation requires 3–4 times more CPU hours than trajectories with classical propagation. The issue of convergence in such calculations is always of concern. We have run test dynamics calculations at multiple numbers of bead discretizations. In all cases we find particle “necklace” geometries that converge to the appropriate width (hydride √2 greater width than deuteride) and average forces that converge to the same distribution. Examples of such calculations over 1000 time steps are shown in the Supporting Information.
Implementation of the Rate Algorithm
To calculate the first term of the rate equation (eq 2), we performed microcanonical TPS at 300 K. To start the simulation, an initial reactive trajectory was obtained from the equilibrated enzyme structure by placing harmonic constraints on the donor and acceptor distance and the transferring particle, hydride or deuteride, and the acceptor distance in the quantum region. These constraints were kept relatively small, 7 and 12 (kcal/mol)/A2, in order to allow for quick decorrelation of the subsequent trajectories. The order parameter was chosen to be the fractional distance of the hydride to donor and acceptor, rD−hydr/(rD−hydr − rA−hydr), which varies between ~0.17 and ~0.80, depending on the progress of the chemical reaction. This order parameter eliminates oscillation due to compression of the donor and acceptor during the particle transfer, which would be present in an order parameter based on absolute distance. An initial TPS trajectory, without constraints, was then obtained from the initial constrained trajectory. Each subsequent TPS trajectory was obtained by shooting from a random time slice on the previous trajectory. Each trajectory obtained was 500 fs in length, with a time step of 0.5 fs and a CMD time step of 0.05 fs. This smaller time step was required in order for the centroid force to converge properly. The perturbation of momenta was kept relatively high to ensure that the system decorrelated from the initial, constrained trajectory after 10–15 trajectories, with an acceptance ratio (reactive trajectories/total trajectories) of approximately 0.25. Dellago et al.46 argue that 0.40 is the optimal acceptance ratio for smaller systems, but a smaller acceptance ratio allows for quicker decorrelation and the circumvention of convergence issues. These trajectories populated a cumulative histogram of 〈hP(t)〉RP. A trajectory was accepted, and hP was unity, if the order parameter exceeded 0.60. The ratio of this and the ensemble average value of hP at time t′, 125 fs, was fit to a linear regression in the linear region of the histogram, from 800 time steps to the end of the trajectory. The slope of the linear regression is the first term value required in eq 2.
As mentioned above, calculation of the second term of the rate equation, C(t′), is a more involved process than that of the first term. Practically, the creation of a properly sampled histogram for a complicated system such as yeast alcohol dehydrogenase is a laborious process. A trajectory from the calculation of 〈ḣP(t)〉AB was chosen, which was then partitioned into multiple windows based on the previously mentioned order parameter. We then ran a separate TPS simulation for each of the windows, with the trajectories starting in the reactant well and accepted if they end within the determined window. These trajectories are t′, 125 fs, in length. Each window was then divided into multiple bins in which the end points of the trajectories it contained are deposited. In an ideal situation, bin width and number of bins are chosen to represent the underlying data structure.55 To do so, these parameters are chosen in order to minimize local maxima within the window. A more complete description of the windows and bin width associated with them can be found below. Windows are matched by multiplying the values of the following histogram by the ratio of the values of the two histograms’ overlap bin. In situations in which an overlap bin does not exist, due to different bin sizes in neighboring windows, the last bin of the window is matched with the first bin of the following window. The full histogram was then fit using a Hermite interpolation and normalized such that the integral was unity. The data is smoothed with a Gaussian weighting function, and the product region of the normalized, smoothed histogram is then integrated to obtain the value of C(t′).
RESULTS AND DISCUSSION
Calculation of the Transfer Rates
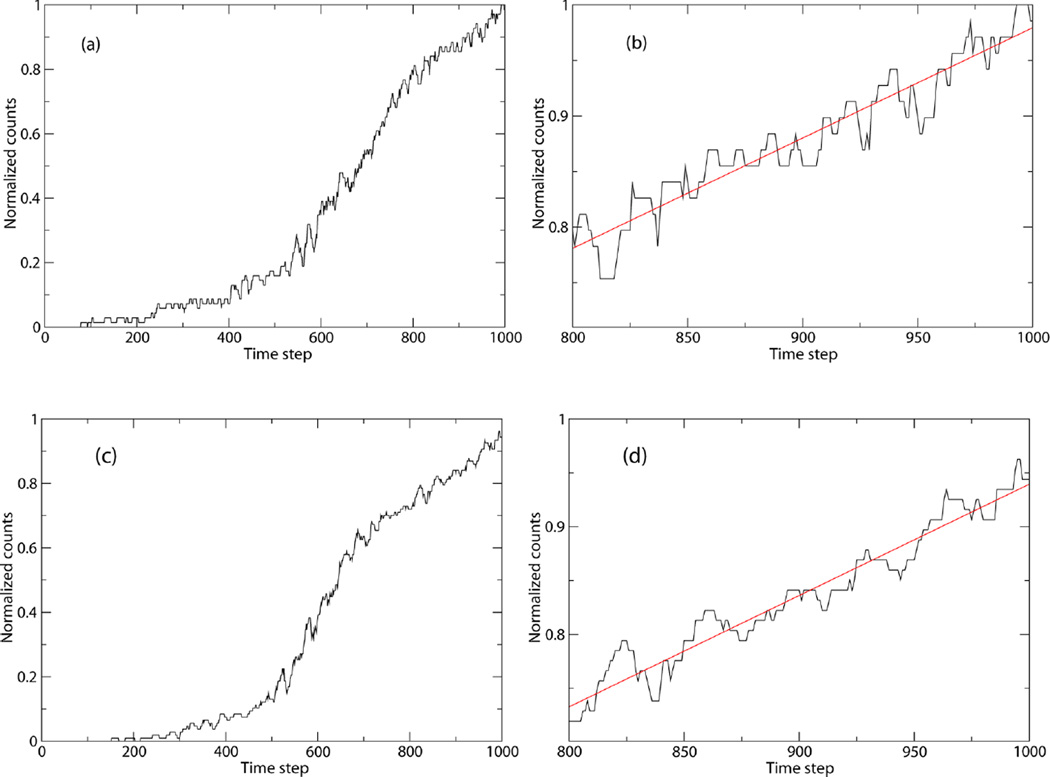
The TPS simulations for the calculation of 〈ḣP(t)〉AB yielded 79 and 117 reactive trajectories, with acceptance ratios of 24.7% and 30.5% for the hydride and deuteride systems, respectively. Of these trajectories, the first ten from each ensemble were discarded to account for decorrelation from the initial, constrained trajectory. These trajectories then were used to populate a cumulative histogram, 〈hP(t)〉AB, as shown in Figures 1a and 1c. As can be seen in both cumulative histograms, a linear region appears after ~800 time steps. This region was fit to a linear regression, and the time derivative was taken to yield 〈ḣP(t)〉AB/〈hP(t′)〉AB (Figures 1b and 1d). From these regression slopes, first term values of 9.901 × 10−4 fs−1 and 1.033 × 10−3 fs−1 were obtained for hydride and deuteride systems, respectively.
Figure 1.
Cumulative histograms for the calculation of the first term for hydride (a) and deuteride (c) systems, normalized to unity integral. Fits of the linear region, from 800 to 1000 time steps (400 to 500 fs), are shown for hydride (b) and deuteride (d) systems. The slopes of these linear fits are the values for the first term of the rate calculation. Note that a time step corresponds to 1/2 fs.
As discussed above, calculation of C(t′) requires careful consideration of the bin width within each window. For the hydride system, seven windows with bins of 0.010λ, except window 2, which had a bin of 0.020λ and four bins of 0.010λ, each produced histograms most consistent with their placement along the reaction coordinate. The simulations produced >8000 trajectories, of which ~900 were accepted. As they were obtained using an already decorrelated trajectory as a seed, no trajectories were discarded. Windows 4 and 5 were difficult to populate with trajectories, with acceptance ratios of 3.10% and 2.7%, respectively (Table 1). This is likely due to the fact that transitions between reactants and products in enzymatic reactions, while being rare, are very quick and on the order of 10–15 fs.44 This can also be seen in the trajectories obtained for yeast alcohol dehydrogenase; the trajectories spend most of the time in each of the stable wells, with quick transitions between the two. Being that windows 4 and 5 are near the top of the free energy barrier, it is unlikely that a trajectory will be within one of these windows at t′. It should also be noted that the high acceptance ratio for windows 1 and 2 is likely due to these two windows being the reactant well and just after the reactant well, meaning that most trajectories obtained will still be in these states at time t′.
Table 1.
Windows and Their Member Trajectories for the Calculation of C(t′) for the Hydride System
| window | range of order parameter (λ) |
accepted trajectories |
total trajectories |
acceptance ratio, % |
|---|---|---|---|---|
| 1 | 0.200–0.360 | 216 | 493 | 43.8 |
| 2 | 0.350–0.410 | 179 | 487 | 36.8 |
| 3 | 0.400–0.460 | 167 | 1087 | 15.4 |
| 4 | 0.450–0.510 | 68 | 2066 | 3.10 |
| 5 | 0.500–0.560 | 56 | 2072 | 2.70 |
| 6 | 0.550–0.610 | 111 | 1303 | 8.51 |
| 7 | 0.600–0.900 | 125 | 811 | 15.4 |
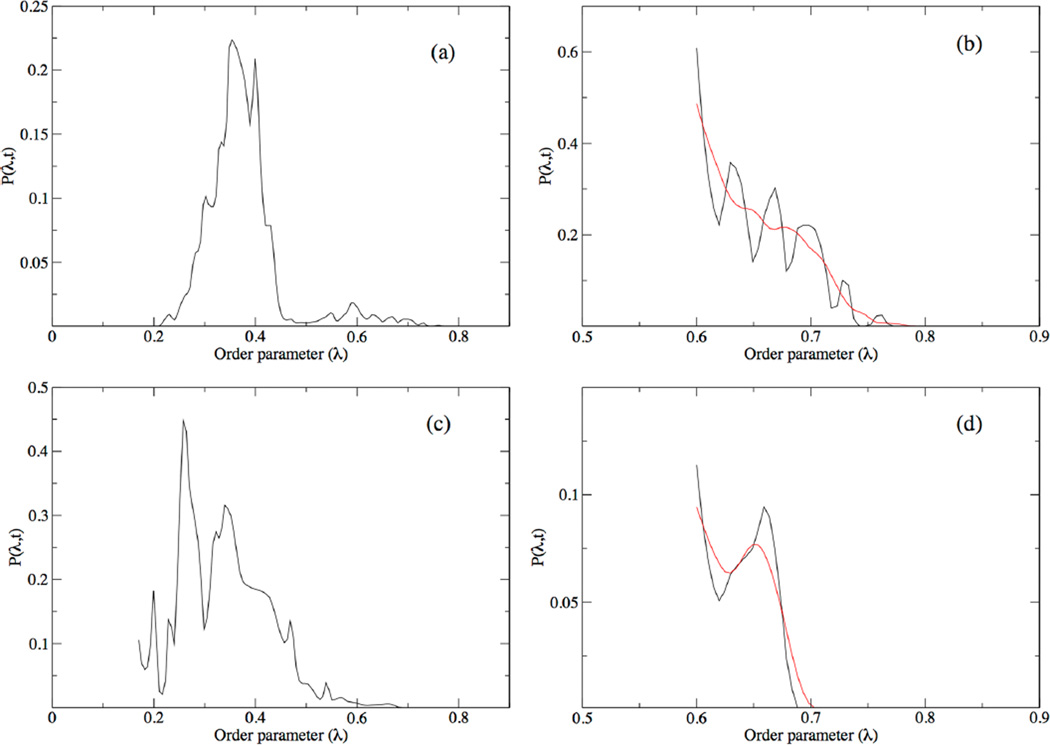
The normalized histogram, with the product region moving average fit, can be seen in Figures 2a and 2b. The histogram shows the expected data structure, with a large peak in the reactant well and a dip at the top of the barrier, with a smaller peak in the product well. Integration of the Gaussian-smoothed window 7 histogram, the product region, resulted in 3.16 × 10−5 as the value of C(t′) for the hydride system. Combined with the hydride 〈ḣP(t)〉AB/〈hP(t′)〉AB, this produced 3.13 × 10−5 fs−1 for the rate constant for the transfer of hydride.
Figure 2.
Normalized histograms depicting the Hermite interpolation of full distribution function, P(λ,t), for the calculation of C(t′) for the hydride (a) and deuteride (c) systems. The last window, P(λ,t;i), of each distribution ((b) for the hydride system and (d) for the deuteride system) was denoted as the product region, and they were smoothed with a Gaussian function (shown in red) which was then integrated to determine C(t′).
Proper sampling of all windows in the deuteride system required a different histogram creation strategy. The chosen trajectory from the calculation of 〈ḣP(t)〉AB was split into 9 windows of equal width, except the first and last windows, which were wider and represent the reactant and product wells. The nine TPS simulations yielded >12000 trajectories, of which >900 were accepted (Table 2). All windows had bins of 0.010λ in length, except windows 4 and 5, which each had two bins of width 0.025λ, window 7, which had a bin of width 0.020λ and 3 bins of width 0.010λ, and window 8, which had a bin of width 0.030λ and 2 bins of 0.010λ. Windows 3, 4, and 5 do not have the usual 0.010λ overlap window due to their different width bins at the edges. This binning strategy provided the best trade-off between resolution and accurate representation of data trends.
Table 2.
Windows and Their Member Trajectories for the Calculation of C(t′) for the Deuteride System
| window | range of order parameter (λ) |
accepted trajectories |
total trajectories |
acceptance ratio, % |
|---|---|---|---|---|
| 1 | 0.170–0.260 | 201 | 517 | 38.9 |
| 2 | 0.250–0.310 | 150 | 506 | 29.6 |
| 3 | 0.300–0.350 | 113 | 481 | 23.5 |
| 4 | 0.350–0.400 | 138 | 744 | 18.5 |
| 5 | 0.400–0.450 | 40 | 885 | 4.52 |
| 6 | 0.450–0.510 | 46 | 2513 | 1.83 |
| 7 | 0.500–0.560 | 59 | 2451 | 2.35 |
| 8 | 0.550–0.610 | 94 | 2630 | 3.57 |
| 9 | 0.600–0.900 | 101 | 1458 | 6.9 |
Figures 2c and 2d shows the completed and normalized histogram for the deuteride system, as well as the moving average fit to the product region. This histogram shows relatively the same structure as the histogram for the hydride system, with a large peak in the reactant well, a dip at the barrier, and a smaller product well compared to the hydride histogram. Integration of the product well, window 9, yielded 5.80 × 10−3 for C(t). Combined with the deuteride 〈ḣP(t)〉AB/〈hP(t′)〉AB, this produced 5.99 × 10−6 fs−1 for the rate constant for the transfer of deuteride. Thus, the calculated primary H/D kinetic isotope effect for YADH is 5.22. We posit that the rates and kinetic isotope effects calculated using this method correspond only to the rate of particle transfer across the barrier, as discussed below.
Discussion of Calculated Kinetic Isotope Effect
Experimentally, the primary H/D kinetic isotope effect of benzyl alcohol conversion via YADH has been determined to be 3.4.26 Although the kinetic isotope effect as reported in this study, 5.22, is within the error of the experimentally determined value, the larger kinetic isotope effect relative to that previously reported is expected, when differences in what exactly is being calculated are taken into account. Following the standard view of the course of enzymatic reactions, once the enzyme–substrate complex, also known as the Michaelis complex, has been formed, the system undergoes a conformational search to reach a reactive conformation, which then proceeds through the reaction. Noncompetitive kinetic isotope effect studies, such those performed by Klinman,25,26 obtain the kinetic isotope effect on the step from the Michaelis complex to the first irreversible step,56 or the transition state in the case of YADH. However, transition path sampling requires the system to begin at, or near, a reactive conformation,57 excluding the conformational search for the reactive conformation that occurs after the formation of the Michaelis complex. We contend that instead of kcat or kchem, as is often experimentally determined, what is calculated using the method described in this study is ktrans, the rate of transfer over the barrier to reaction. The inclusion of the conformational search in the calculation of the kinetic isotope effect could mask a large kinetic isotope effect when that conformational search does not change with isotopic substitution.58 Thus, the kinetic isotope effects calculated using transition path sampling are likely to be larger than an experimentally determined value. However, our results indicate that the magnitude of this masking effect on the intrinsic kinetic isotope effect is small. By eliminating the conformational search step from simulations, we can determine the kinetic isotope effect of the particle transfer step alone.
Recently published work24 calculated the work applied to the transferring particle to obtain free energy barriers to particle transfer, which were then used to obtain an approximate primary H/D kinetic isotope effect. This approximate kinetic isotope effect agreed with the experimentally calculated kinetic isotope effect,26 and thus was slightly lower than our calculated result. However, this is in fact due to differences in the focus of the two methods; the intention of the calculation in the aforementioned paper was solely to calculate the barrier felt by hydride or deuteride as the particle transferred. Along the course of an enzymatic reaction, the donor–acceptor distance compresses and the wells and the barrier broaden and flatten,59 decreasing the difference in zero-point energy between hydride and deuteride. Due to the focus of the study on the mechanics of the transfer event itself, as well as on the plausibility of tunneling contributing to the reaction, Dzierlenga et al.24 did not calculate the work applied to the entire system along the entire trajectory, only 5–10 fs of the trajectory encompassing the transfer event. Because of this, the calculated barriers likely are already broadened and flattened. Thus, the kinetic isotope effect approximated from barrier height difference would be smaller relative to a method that takes into account the energy surface of the entire chemical step.
CONCLUSIONS
We have detailed a method using transition path sampling with normal mode centroid molecular dynamics to determine the kinetic isotope effects in enzymatic reactions. Using the conversion of benzyl alcohol to benzaldehyde in yeast alcohol dehydrogenase as a model system, the primary H/D kinetic isotope effect of particle transfer was determined to be 5.22. While elevated compared to the mean value of those reported in experimental studies, it falls within the bounds of experimental error. This shows that the kinetic isotope effect for the entire enzymatic chemical step can be accurately predicted from first-principles calculations. In addition to calculating kinetic isotope effects, this method could be used to determine how changes within the enzyme, structural or otherwise, change the mechanics of particle transfer.
Supplementary Material
Acknowledgments
The authors would like to thank Dimitri Antoniou and Michael Dzierlenga for their CMD code and many useful conversations.
Funding
NIH Grants GM068036 and GM102226.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Information on the 8 bead CMD calculated forces relative to a 16 bead system (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Hammes-Schiffer S. Comparison of hydride, hydrogen atom, and proton-coupled electron transfer reactions. Chem Phys Chem. 2002;3(1):33–42. doi: 10.1002/1439-7641(20020118)3:1<33::AID-CPHC33>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Nagel ZD, Klinman JP. Tunneling and dynamics in enzymatic hydride transfer. Chem. Rev. 2006;106(8):3095–3118. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz SD. Protein dynamics and the enzymatic reaction. Top. Curr. Chem. 2013;337:189–208. doi: 10.1007/128_2012_412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammes-Schiffer S. Hydrogen tunneling and protein motion in enzyme reactions. Acc. Chem. Res. 2006;39:93–100. doi: 10.1021/ar040199a. [DOI] [PubMed] [Google Scholar]
- 5.Cleland WW. Isotope effects: Determination of enzyme transition state structure. Methods Enzymol. 1995;249(1982):341–373. doi: 10.1016/0076-6879(95)49041-8. [DOI] [PubMed] [Google Scholar]
- 6.Roston D, Islam Z, Kohen A. Kinetic isotope effects as a probe of hydrogen transfers to and from common enzymatic cofactors. Arch. Biochem. Biophys. 2014;544:96–104. doi: 10.1016/j.abb.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J, Truhlar DG. Quantum mechanical methods for enzyme kinetics. Annu. Rev. Phys. Chem. 2002;53(1):467–505. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]
- 8.Hwang JK, Chu ZT, Yadav A, Warshel A. Simulations of quantum mechanical corrections for rate constants of hydride-transfer reactions in enzymes and solutions. J. Phys. Chem. 1991;95(22):8445–8448. [Google Scholar]
- 9.Hwang JK, Warshel A. A quantized classical path approach for calculations of quantum mechanical rate constants. J. Phys. Chem. 1993;97(39):10053–10058. [Google Scholar]
- 10.Sprik M, Klein ML, Chandler D. Staging: A sampling technique for the Monte Carlo evaluation of path integrals. Phys. Rev. B: Condens. Matter Mater. Phys. 1985;31(7):4234–4244. doi: 10.1103/physrevb.31.4234. [DOI] [PubMed] [Google Scholar]
- 11.Billeter SR, Webb SP, Iordanov T, Agarwal PK, Hammes-Schiffer S. Hybrid approach for including electronic and nuclear quantum effects in molecular dynamics simulations of hydrogen transfer reactions in enzymes. J. Chem. Phys. 2001;114(15):6925. [Google Scholar]
- 12.Billeter SR, Webb SP, Agarwal PK, Iordanov T, Hammes-Schiffer S. Hydride transfer in liver alcohol dehydrogenase: Quantum dynamics, kinetic isotope effects, and role of enzyme motion. J. Am. Chem. Soc. 2001;123(45):11262–11272. doi: 10.1021/ja011384b. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Hammes-Schiffer S. Hybrid quantum/classical path integral approach for simulation of hydrogen transfer reactions in enzymes. J. Chem. Phys. 2006;125(18):184102. doi: 10.1063/1.2362823. [DOI] [PubMed] [Google Scholar]
- 14.Truhlar DG, Gao J, Garcia-Viloca M, Alhambra C, Corchado J, Sanchez ML, Poulsen TD. Ensemble-averaged variational transition state theory with optimized multidimensional tunneling for enzyme kinetics and other condensed-phase reactions. Int. J. Quantum Chem. 2004;100(6):1136–1152. [Google Scholar]
- 15.Major DT, Gao J. Implementation of the bisection sampling method in path integral simulations. J. Mol. Graphics Modell. 2005;24(2):121–127. doi: 10.1016/j.jmgm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Major DT, Gao J. An integrated path integral and free-energy perturbation - umbrella sampling method for computing kinetic isotope effects of chemical reactions in solution and in enzymes. J. Chem. Theory Comput. 2007;3:949–960. doi: 10.1021/ct600371k. [DOI] [PubMed] [Google Scholar]
- 17.Dellago C, Bolhuis PG, Chandler D. Efficient transition path sampling: Application to Lennard-Jones cluster rearrangements. J. Chem. Phys. 1998;108(22):9236–9245. [Google Scholar]
- 18.Bolhuis PG, Dellago C, Chandler D. Sampling ensembles of deterministic transition pathways. Faraday Discuss. 1998;110:421–436. [Google Scholar]
- 19.Cao J, Voth GA. The formulation of quantum statistical mechanics based on the Feynman path centroid density. I. Equilibrium properties. J. Chem. Phys. 1994;100(7):5093–5104. [Google Scholar]
- 20.Cao J, Voth GA. The formulation of quantum statistical mechanics based on the Feynman path centroid density. II. Dynamical properties. J. Chem. Phys. 1994;100(7):5106–5117. [Google Scholar]
- 21.Cao J, Voth GA. The formulation of quantum statistical mechanics based on the Feynman path centroid density. III. Phase space formalism and analysis of centroid molecular dynamics. J. Chem. Phys. 1994;101(7):6157–6167. [Google Scholar]
- 22.Cao J, Voth GA. The formulation of quantum statistical mechanics based on the Feynman path centroid density. IV. Algorithms for centroid molecular dynamics. J. Chem. Phys. 1994;101(7):6168–6183. [Google Scholar]
- 23.Cao J, Voth GA. The formulation of quantum statistical mechanics based on the Feynman path centroid density. V. Quantum instantaneous normal mode theory of liquids. J. Chem. Phys. 1994;101(7):6184–6192. [Google Scholar]
- 24.Dzierlenga MW, Antoniou D, Schwartz SD. Another look at the mechanisms of hydride transfer eenzymes with quantum and classical transition path sampling. J. Phys. Chem. Lett. 2015;6(7):1177–1181. doi: 10.1021/acs.jpclett.5b00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinman JP. The mechanism of enzyme-catalyzed reduced nicotinamide adenine dinucleotide-dependent reductions. Substituent and isotope effects in the yeast alcohol dehydrogenase reaction. J. Biol. Chem. 1972;247(24):7977–7987. [PubMed] [Google Scholar]
- 26.Klinman JP. Isotope effects and structure-reactivity correlations in the yeast alcohol dehydrogenase reaction. A study of the enzyme-catalyzed oxidation of aromatic alcohols. Biochemistry. 1976;15(9):2018–2026. doi: 10.1021/bi00654a032. [DOI] [PubMed] [Google Scholar]
- 27.Welsh KM, Creighton DJ, Klinman JP. Transition-state structure in the yeast alcohol dehydrogenase reaction: The magnitude of solvent and alpha-secondary hydrogen isotope effects. Biochemistry. 1980;19(10):2005–2016. doi: 10.1021/bi00551a001. [DOI] [PubMed] [Google Scholar]
- 28.Cha Y, Murray CJ, Klinman JP. Hydrogen tunneling in enzyme reactions. Science. 1989;243(4896):1325–1330. doi: 10.1126/science.2646716. [DOI] [PubMed] [Google Scholar]
- 29.Roston D, Kohen A. Elusive transition state of alcohol dehydrogenase unveiled. Proc. Natl. Acad. Sci. U. S. A. 2010;107(21):9572–9577. doi: 10.1073/pnas.1000931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wratten CC, Cleland WW. Product inhibition studies on yeast and liver alcohol dehydrogenases. Biochemistry. 1963;2(5):935–941. doi: 10.1021/bi00905a007. [DOI] [PubMed] [Google Scholar]
- 31.Rucker J, Klinman JP. Computational study of tunneling and coupled motion in alcohol dehydrogenase-catalyzed reactions: Implication for measured hydrogen and carbon isotope effects. J. Am. Chem. Soc. 1999;121(10):1997–2006. [Google Scholar]
- 32.Raj SB, Ramaswamy S, Plapp BV. Yeast alcohol dehydrogenase structure and catalysis. Biochemistry. 2014;53(36):5791–5803. doi: 10.1021/bi5006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Smidt O, du Preez JC, Albertyn J. The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS Yeast Res. 2008;8(7):967–978. doi: 10.1111/j.1567-1364.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- 34.Caratzoulas S, Mincer JS, Schwartz SD. Identification of a protein-promoting vibration in the reaction catalyzed by horse liver alcohol dehydrogenase. J. Am. Chem. Soc. 2002;124(13):3270–3276. doi: 10.1021/ja017146y. [DOI] [PubMed] [Google Scholar]
- 35.Vanommeslaeghe K, MacKerell AD. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012;52(12):3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4(2):187–217. [Google Scholar]
- 37.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Field MJ, Bash PA, Karplus M. A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulations. J. Comput. Chem. 1990;11(6):700–733. [Google Scholar]
- 39.Gao J, Amara P, Alhambra C, Field MJ. A Generalized Hybrid Orbital (GHO) method for the treatment of boundary atoms in combined QM/MM calculations. J. Phys. Chem. A. 1998;102(24):4714–4721. [Google Scholar]
- 40.Stewart JJP. Optimization of parameters for semiempirical methods I. Method. J. Comput. Chem. 1989;10(2):209–220. [Google Scholar]
- 41.Stewart JJP. Optimization of parameters for semiempirical methods II. Applications. J. Comput. Chem. 1989;10(2):221–264. [Google Scholar]
- 42.Brothers EN, Suarez D, Deerfield DW, Merz KM. PM3-compatible zinc parameters optimized for metalloenzyme active sites. J. Comput. Chem. 2004;25(14):1677–1692. doi: 10.1002/jcc.20086. [DOI] [PubMed] [Google Scholar]
- 43.Masterson JE, Schwartz SD. Changes in protein architecture and subpicosecond protein dynamics impact the reaction catalyzed by lactate dehydrogenase. J. Phys. Chem. A. 2013;117(32):7107–7113. doi: 10.1021/jp400376h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masterson JE, Schwartz SD. The enzymatic reaction catalyzed by lactate dehydrogenase exhibits one dominant reaction path. Chem. Phys. 2014;442(17):132–136. doi: 10.1016/j.chemphys.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dametto M, Antoniou D, Schwartz SD. Barrier crossing in dihydrofolate reductase does not involve a rate-promoting vibration. Mol. Phys. 2012;110(9–10):531–536. doi: 10.1080/00268976.2012.655337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dellago C, Bolhuis PG, Csajka FS, Chandler D. Transition path sampling and the calculation of rate constants. J. Chem. Phys. 1998;108(5):1964–1977. [Google Scholar]
- 47.Dellago C, Bolhuis PG, Chandler D. On the calculation of reaction rate constants in the transition path ensemble. J. Chem. Phys. 1999;110(14):6617–6625. [Google Scholar]
- 48.Wick CD. NaCl dissociation dynamics at the air–water interface. J. Phys. Chem. C. 2009;113(6):2497–2502. [Google Scholar]
- 49.Wick CD, Dang LX. Computational investigation of the influence of organic-aqueous interfaces on NaCl dissociation dynamics. J. Chem. Phys. 2010;132(4):044702. doi: 10.1063/1.3299279. [DOI] [PubMed] [Google Scholar]
- 50.Boulfelfel SE, Ravikovitch PI, Sholl DS. Modeling Diffusion of Linear Hydrocarbons in Silica Zeolite LTA Using Transition Path Sampling. J. Phys. Chem. C. 2015;119(27):15643–15653. [Google Scholar]
- 51.Xia F, Bronowska AK, Cheng S, Gräter F. Base-catalyzed peptide hydrolysis is insensitive to mechanical stress. J. Phys. Chem. B. 2011;115(33):10126–10132. doi: 10.1021/jp202162r. [DOI] [PubMed] [Google Scholar]
- 52.Antoniou D, Schwartz SD. Approximate inclusion of quantum effects in transition path sampling. J. Chem. Phys. 2009;131(22):224111. doi: 10.1063/1.3272793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Car R, Parrinello M. Unified approach for molecular dynamics and Density-Functional Theory. Phys. Rev. Lett. 1985;55(22):2471–2474. doi: 10.1103/PhysRevLett.55.2471. [DOI] [PubMed] [Google Scholar]
- 54.Lobaugh J, Voth GA. The quantum dynamics of an excess proton in water. J. Chem. Phys. 1996;104(5):2056. [Google Scholar]
- 55.Wand PM. Data-based choice of histogram bin width. Am. Stat. 1997;51(1):59–64. [Google Scholar]
- 56.Simon H, Palm D. Isotope effects in organic chemistry and biochemistry. Angew. Chem., Int. Ed. Engl. 1966;5(11):920–933. [Google Scholar]
- 57.Bolhuis PG, Chandler D, Dellago C, Geissler PL. Transition path sampling: Throwing ropes Over rough mountain passes, in the dark. Annu. Rev. Phys. Chem. 2002;53(1):291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 58.Schramm VL. Enzymatic transition States, transition-state analogs, dynamics, thermodynamics, and lifetimes. Annu. Rev. Biochem. 2011;80:703–732. doi: 10.1146/annurev-biochem-061809-100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamerlin SCL, Mavri J, Warshel A. Examining the case for the effect of barrier compression on tunneling, vibrationally enhanced catalysis, catalytic entropy and related issues. FEBS Lett. 2010;584(13):2759–2766. doi: 10.1016/j.febslet.2010.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.