Abstract
Oncogenic transformation in Ewing sarcoma tumors is driven by the fusion oncogene EWS-FLI1. However, despite the well-established role of EWS-FLI1 in tumor initiation, the development of models of Ewing sarcoma in human cells with defined genetic elements has been challenging. Here, we report a novel approach to model the initiation of Ewing sarcoma tumorigenesis that exploits the developmental and pluripotent potential of human embryonic stem cells. The inducible expression of EWS-FLI1 in embryoid bodies, or collections of differentiating stem cells, generates cells with properties of Ewing sarcoma tumors, including characteristics of transformation. These cell lines exhibit anchorage-independent growth, a lack of contact inhibition and a strong Ewing sarcoma gene expression signature. Furthermore, these cells also demonstrate a requirement for the persistent expression of EWS-FLI1 for cell survival and growth, which is a hallmark Ewing sarcoma tumors.
Keywords: Ewing sarcoma, embryonic stem cell, embryoid body, transformation, tumor initiation
INTRODUCTION
Ewing sarcoma is an aggressive bone and soft tissue tumor that affects children and young adults1. Ewing sarcoma is defined by a recurrent chromosomal translocation between the EWSR1 gene and various ETS genes2. The most common fusion, EWS-FLI1, is present in 85% of cases. In each case, the transcriptional activation domain from EWSR1 is fused to the DNA-binding domain of an ETS transcription factor, consistent with experimental evidence suggesting that EWS-FLI1 functions as an aberrant transcription factor3–6. Importantly, Ewing sarcoma tumors are dependent on EWS-FLI1 and require the persistent expression of this oncogene to maintain the transformed phenotype7–10.
Additional genomic alterations in Ewing sarcoma tumors, other than the EWS-FLI1 translocation, are often minimal11–14. However, some tumors do exhibit mutations in TP53, deletions of the CDKN2A locus or mutations in STAG211–13. Specifically, mutations in TP53 and STAG2 occur in ~5–10% and ~15–20% of tumors, respectively11–13,15. Interestingly, almost all Ewing sarcoma cell lines exhibit mutations in p53, or members of the p53 pathway, which has led to the hypothesis that loss of p53 is required for the in vitro culture of Ewing sarcoma cells16.
Although the initiating oncogene in Ewing sarcoma, EWS-FLI1, was first identified over twenty years ago, the cell-of-origin17 in Ewing sarcoma is still unknown and a source of considerable debate. There is experimental support for both neural crest and mesenchymal origins in Ewing sarcoma18–21. Multiple experiments have demonstrated that the effects of EWS-FLI1 expression are strongly dependent on the cellular background. For example, EWS-FLI1 causes a p53-dependent growth arrest and toxicity in human and mouse fibroblasts, but is tolerated in some human mesenchymal and neural crest cells18–23. However, mesenchymal and neural crest cells, unlike Ewing sarcoma tumors, do not require EWS-FLI1 for growth and, thus, fail to recapitulate the critical hallmark of the dependency on persistent EWS-FLI1 expression for cell survival.
One significant difficulty in developing a model system of Ewing sarcoma has been the uncertainty regarding the cell-of-origin and the resulting lack of an appropriate cell type in which to study the EWS-FLI1 oncogene. To circumvent this problem, we have developed a novel approach to model Ewing sarcoma that exploits the differentiation potential of human stem cells and the cellular diversity of embryoid bodies. Embryoid bodies, which are three-dimensional aggregates of differentiating stem cells, contain cells from all three germ cell layers and are the in vitro equivalent of a teratoma. Our hypothesis was that embryoid bodies, due to their cellular diversity, could contain an appropriate cell-of-origin for Ewing sarcoma. In this work, we demonstrate that the doxycycline-inducible expression of EWS-FLI1 in embryoid bodies derived from human embryonic stem cells (hESC) with knockdown of p53 generates cells with an Ewing sarcoma-like phenotype, including properties of transformation and dependency on persistent EWS-FLI1 expression for survival.
RESULTS
Human embryoid bodies are permissive for EWS-FLI1 expression
The molecular pathogenesis of Ewing sarcoma remains poorly understood, despite the underlying association with the EWS-FLI1 oncogene16,24. In order to develop a model of Ewing sarcoma with defined genetic elements in human cells, we used a lentiviral vector to generate H1 human embryonic stem cells that express EWS-FLI1 (EF1) and green fluorescent protein (GFP) under the control of a doxycycline-inducible element (pLVX-EF1-IRES-GFP). This lentiviral vector was also modified, as described in the Materials and Methods section, to constitutively express an shRNA targeting p53 because loss of this tumor suppressor is relevant to a subset of Ewing sarcoma tumors. Data are shown for the modified H1 stem cell line (referred to as EF cells), but similar results were obtained with an independent stem cell line (WA25, WiCell Research Institute) (Supplemental Figure S1).
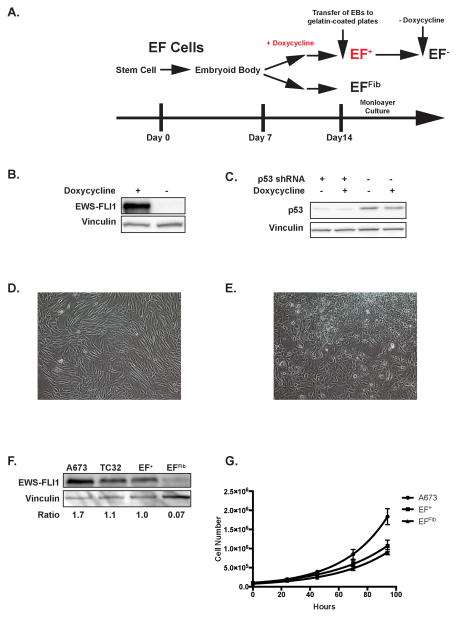
A schematic of the differentiation protocol is shown in Figure 1A. The EF cells, when cultured as embryoid bodies (Supplemental Figure S2A) under non-adherent conditions, spontaneously differentiate to cells from all three germ layers, as demonstrated by RT-qPCR for lineage specific genes (Supplemental Figure S2B). Addition of doxycycline to the embryoid body cultures after 7 days of culture results in the expression of EWS-FLI1, as demonstrated by western blot analysis (Figure 1B). Similarly, western blot analysis also demonstrates constitutive knockdown of p53 (Figure 1C). Dissociation of the embryoid bodies with trypsin followed by flow cytometry for GFP shows an approximately 100-fold range of expression levels in the embryoid body cells (Supplemental Figure S2C).
Figure 1.
EWS-FLI1 expression in embryoid bodies. (A) Schematic diagram illustrating the differentiation protocol of the human embryonic stem cells. The culture protocol consists of the three steps: 1) embryoid body formation under non-adherent conditions; 2) transfer of embryoid bodies to gelatin-coated plates; 3) monolayer culture of the cells that outgrow from the embryoid bodies. Doxycycline is added to the embryoid bodies after seven days of culture. (B) Western blot analysis showing the inducible expression of EWS-FLI1 in the embryoid bodies in the presence, but not absence, of doxycycline. (C) Western blot analysis showing the constitutive knockdown of p53 by an shRNA in embryoid bodies. Knockdown efficiency is not affected by doxycycline. (D) EFFib cells exhibit a morphology similar to fibroblast cells. (E) The EF+ cells exhibit a distinct morphology and are more rounded and less elongated than the EFFib cells. (F) Western blot analysis of EWS-FLI1 expression in Ewing sarcoma cell lines (A673 and TC32), EF+ cells and EFFib cells. The relative expression level of EWS-FLI1 for each cell line compared to EF+ is shown below the blot. (G) The growth rate of the EF+, EFFib and A673 cells was measured using trypan blue exclusion and cell counting. The data were fit to an exponential curve using GraphPad Prism. Error bars indicate the standard deviation of three replicates.
Cell Phenotype
After 10–14 days of suspension culture the embryoid bodies were plated on gelatin-coated plates for monolayer culture to assay for transformation. In the absence of doxycycline, adherent cells with a fibroblast morphology (Figure 1D), referred to as EFFib cells, grew out of the embryoid bodies and could be expanded and propagated in culture. These cells were non-immortalized and could be grown in vitro for approximately 6–8 weeks before undergoing senescence. In the presence of doxycycline, rare colonies of cells with a unique morphology (Figure 1E), referred to as EF+ cells, emerged after several weeks of culture, usually in a background of more fibroblast-like cells. These EF+ cells exhibited a round shape, rather than the more elongated, spindle shape of the EFFib cells. Repeated passaging of these cells in the presence of doxycycline (see Material and Methods section) resulted in progressive enrichment for the round cells and, eventually, a homogenous population of cells. The expression level of EWS-FLI1 in these cells was comparable to the levels in the ES cell lines A673 and TC32 (Figure 1F). The EF+ cells grow with a doubling time of approximately 28 hours (Figure 1G) and have now been in continuous cell culture for >10 months without undergoing senescence. In contrast to the EF+ cells, addition of doxycycline to the EFFib cells results in a significant decrease in proliferation, consistent with reports that EWS-FLI1 induces a growth arrest in fibroblast cells (Supplemental Figure S3)22.
The karyotype of the parental H1 embryonic stem cells is 46, XY, which is maintained by the EFFib cells (Supplemental Figure S4A). Karyotype results from the EF+ cells, however, were more variable. In one experiment, the EF+ cells showed a stable karyotype identical to the parental cells (Supplemental Figure S4B and S4E). However, a second experiment showed a tetraploid karyotype, as assessed by G-band karyotype (Supplemental Figure S4C and S4F) and interphase fluorescent in situ hybridization (FISH) (Supplemental Figure S4D). This tetraploid karyotype was observed at passages 15 and 37, although within this population of tetraploid cells there were some cells with gains and losses of individual chromosomes (data not shown). This is consistent with data demonstrating that tetraploidy can lead to chromosomal instability25.
Immunophenotype
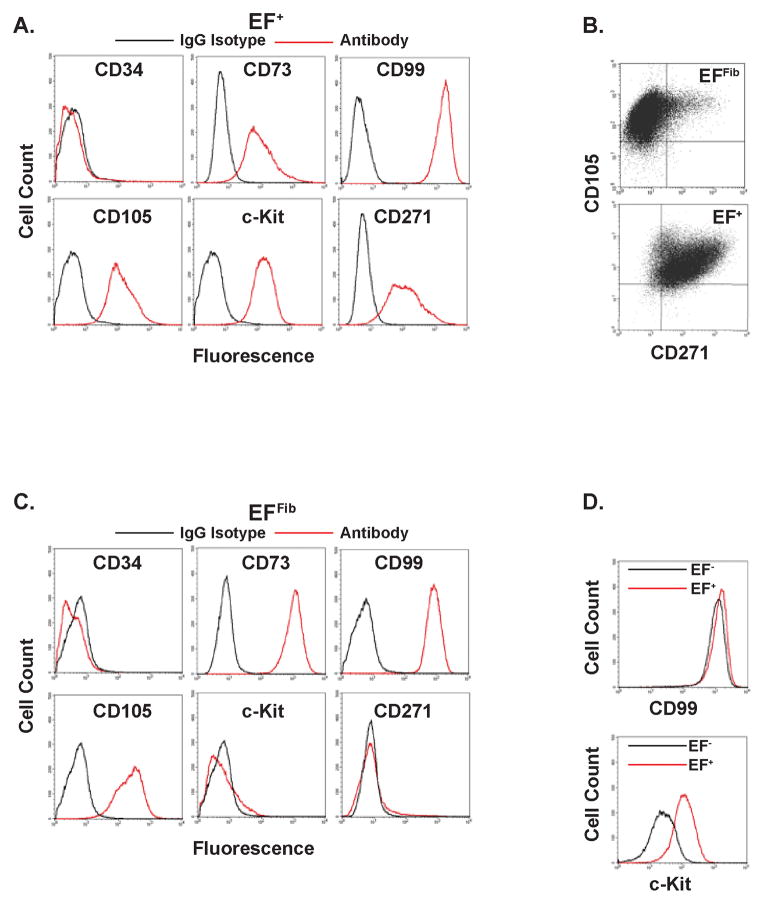
Ewing sarcoma tumors can express both mesenchymal markers, including CD73 and CD105, and neural crest markers, including CD27126,27. We found that the EF+ cells, similar to Ewing sarcoma tumors, express the markers CD73, CD105 and CD271 (Figure 2A). Multiparameter flow cytometry was used to confirm that individual EF+ cells were positive for both CD105 and CD271 (Figure 2B). In contrast, the EFFib cells express the mesenchymal cell markers CD73 and CD105, consistent with the differentiation protocol used in this study and their fibroblast-like morphology (Figure 1D), but do not express the neural crest cell marker CD271. None of the cells express CD34, a hematopoietic cell marker.
Figure 2.
The immunophenotype of EF+ (A) and EFFib (C) cells as determined by flow cytometry. Cells were labeled with APC- and PE-labeled antibodies against CD34, CD73, CD99, CD105, c-Kit, CD271 and isotype-matched control antibodies. The isoptype control is shown in black and the specific antibody is shown in red. (B) Multiparameter flow cytometry with EFFib and EF+ cells for CD105 and CD271. (D) FACS analysis comparing expression of CD99 and c-Kit in EF+ cells (red line) and EF− cells (black line), four days after removal of doxycycline.
Common diagnostic markers in Ewing sarcoma that may reflect lineage and/or transformation include CD99 and c-Kit. CD99 is a highly sensitive, but not specific, marker for Ewing sarcoma that is also expressed on mesenchymal cells28. The c-Kit receptor is expressed in 30–50% of patient-derived Ewing sarcoma tumors, but is not expressed in mesenchymal cells29,30. The EFFib cells express CD99, but do not express c-Kit (Figure 2C). The EF+ cells, on the other hand, express both CD99 and c-Kit (Figure 2A), recapitulating the characteristics of primary Ewing sarcoma tumors. Although CD99 is a sensitive marker for Ewing sarcoma, data suggest that its expression is not strongly regulated by EWS-FLI17,8,28. In agreement with this data, the removal of doxycycline from the EF+ cells had only a minimal effect on CD99 expression, but did lead to a 10-fold loss of c-Kit expression (Figure 2D). In summary, the cell surface marker analysis of the EF+ cells was highly consistent with an Ewing sarcoma tumor expression pattern.
EF+ cells exhibit properties of transformation
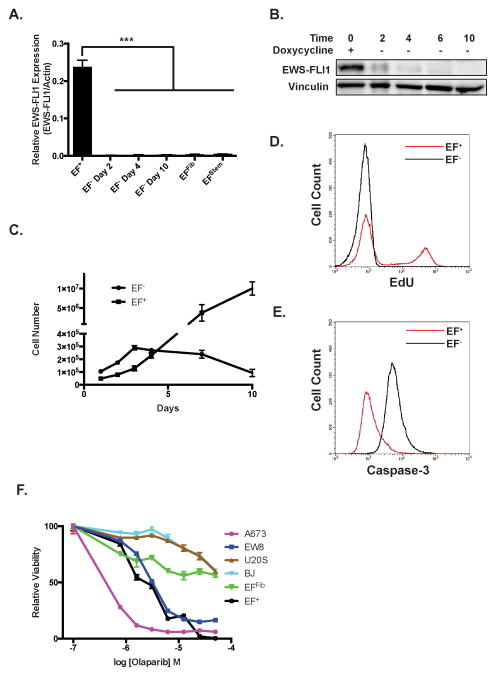
We removed doxycycline from the EF+ cells to evaluate for dependency on continued EWS-FLI1 expression for growth, a feature of most Ewing sarcoma cells lines. RT-qPCR (Figure 3A) and western blotting (Figure 3B) were used to verify the rapid and complete depletion of EWS-FLI1 after drug removal. A morphological change was noted in the cells approximately three-to-five days after doxycycline removal, with the EF− cells exhibiting a larger, flatter and more spindle-shaped morphology (data not shown). After doxycycline removal, the EF− cells exhibit decreased proliferation (Figure 3C). Notably, EdU labeling demonstrated a complete lack of S-phase cells after four days of doxycycline removal (Figure 3D). Culture for additional days in the absence of doxycycline resulted in the loss of cell adhesion and cell death. This coincided with the expression of caspase-3, a marker of apoptosis, in the EF− cells (Figure 3E). Recent work has demonstrated that Ewing sarcoma cell lines exhibit sensitivity to PARP1 inhibitors, including olaparib31,32. The EF+ cells show enhanced sensitivity to olaparib treatment (IC50 2.37 μM; 95% confidence interval 2.1–2.7 μM) relative to the EFFib cells (IC50 54.2 μM; 95% confidence interval 23.6–124.4 μM) (Figure 3F). Similarly, the EF+ cells are also more sensitive than the EFFib cell to treatment with additional drugs with reported lethality toward Ewing sarcoma cell lines, including YK-4-279 (inhibitor of the interaction between RNA helicase A and EWS-FLI1), mithramycin (DNA-binding drug) and LY294002 (PI3K pathway inhibitor) (Supplemental Figure S5)33–35.
Figure 3.
The EF+ cells exhibit oncogene addiction. (A) RT-qPCR for the EWS-FLI1 fusion in the EF+ cells and the EF− cells after 2, 4, and 10 days of doxycycline removal. For comparison, expression of EWS-FLI1 in the EFFib cells and EF stem cells is also shown. Expression of EWS-FLI1 is shown relative to actin expression. Error bars indicate the standard error of the mean of three experiments (***, p<0.001). (B) Western blot of EWS-FLI1 in the EF+ cells and the EF− cells after 2, 4, 6, and 10 days of doxycycline removal. (C) Growth of EF+ cells and EF− cells. Cell growth was measured using trypan blue exclusion and a Vi-CELL Cell Viability Analyzer. Error bars indicate the standard error of the mean of three experiments. (D) Flow cytometry analysis of EdU incorporation by EF+ cells (red line) and EF− cells (black line) after four days without doxycycline. (E) Flow cytometry analysis of caspase-3 activation in EF+ cells (red line) and EF− cells (black line) after ten days without doxycycline. (F) Dose-response curves of EFFib, EF+, A673 (Ewing sarcoma), EW8 (Ewing sarcoma), U2OS (osteosarcoma) and BJ-fibroblast (non-transformed fibroblast) cells treated with olaparib for three days. Cell viability was measured using CellTiter-Glo luminescence. Data were log-transformed and normalized. Error bars indicate the standard error of the mean of three experiments.
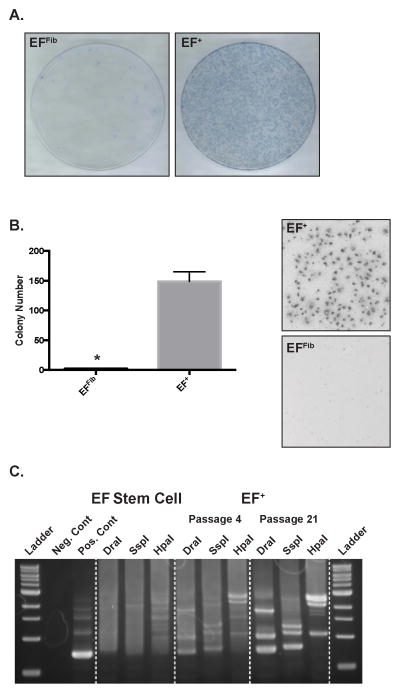
The EF+ cells, unlike the EFFib cells, exhibit a lack of contact inhibition. After culture for fourteen days, staining with methylene blue revealed many dense foci of EF+ cells, whereas the EFFib cells maintained a monolayer growth pattern with contact inhibition, expected for non-transformed fibroblast cells (Figure 4A). The EF+ cells also exhibited anchorage-independent growth and formed colonies in a soft agar assay (Figure 4B). In contrast, the EFFib control cells did not form colonies in soft agar. Despite exhibiting hallmarks of transformation, the EF+ cells did not form tumors when injected subcutaneously into immunocompromised mice in a xenograft experiment (see Material and Methods).
Figure 4.
The EF+ cells exhibit properties of transformation. (A) The EFFib and EF+ cells were cultured for fourteen days without passaging and then colonies were stained with methylene blue. (B) Soft-agar assay for anchorage-independent growth of EFFib and EF+ cells. Error bars indicate the standard error of the mean of three experiments (*, p<0.05). (C) Lentiviral integration analysis. Genomic DNA was isolated from the parental stem cells and EF+ cells at passage 4 and 21. Integration sites were then analyzed in three digestion libraries using a genome walking approach.
Since the EF+ cells were created using lentiviral vectors we were able to assess the EF+ cells lines for a clonal growth pattern, a common characteristic of transformed cells, using lentiviral integration site analysis and a genome walking approach. The EF+ cells were generated using two lentiviral vectors, one expressing rtTA and the other expressing EWS-FLI1 under the control of the tetracycline responsive element (TRE) promoter. Integration analysis of the parental, EF stem cells shows an indistinct pattern consistent with multiple lentiviral integration sites, or clones (Figure 4C). The EF+ cells at passage 4 also demonstrate a pattern consistent with multiple clones, but there is the early appearance of a limited number of dominant bands, or integration sites, in each digestion library. By passage 21, however, the integration analysis shows a very discrete pattern consistent with a limited number of clones.
EF+ cells exhibit a highly specific Ewing sarcoma gene expression profile
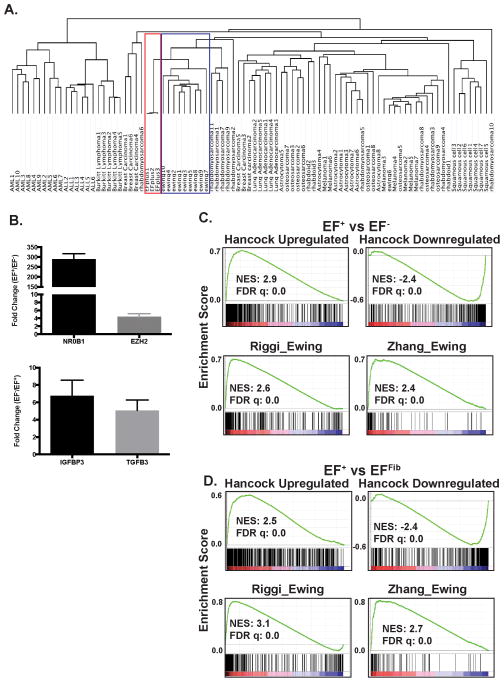
Next we addressed whether the EF+ cells exhibited an Ewing sarcoma gene expression signature and, if so, whether our doxycycline-inducible, isogenic system would be advantageous in identifying gene expression changes related to EWS-FLI1. Unsupervised hierarchical clustering was performed with the EF+ cells and 88 cancer cell lines across 12 different tumor types (Figure 5A). Notably, the EF+ cells clustered most closely with the Ewing sarcoma cell lines relative to the other sarcomas and tumor types.
Figure 5.
EF+ cells exhibit an Ewing sarcoma gene expression signature. (A) Unsupervised hierarchical clustering of 88 cancer cell lines, across 12 different tumor types, and the EF+ cells. The Ewing sarcoma cell line cluster is outlined in blue and the EF+ cell line cluster is outlined in red. (B) RT-qPCR of select upregulated and downregulated genes. The error bars indicate the standard error of the mean from three experiments. The upper panel shows genes that are upregulated in the EF+ cells and the lower panel shows genes upregulated in the EF− cells. (C and D) Gene set enrichment analysis (GSEA) shows enrichment of the Hancock et al. upregulated and downregulated gene sets in the gene expression data from the EF+ versus EF− (C) and EF+ versus EFFib (D) comparisons. GSEA plots are also shown for the curated gene sets RIGGI_EWING and ZHANG_TARGETS_OF_EWSR1_FLI1 from the MSigDB collection. The normalized enrichment scores (NES) and FDR q-values are shown.
The gene expression of the EF+ cells was then compared to both the EF− cells and the EFFib cells. The EF+ versus EF− analysis is the most direct comparison, but the advantage of the EF+ versus EFFib comparison is that the EFFib cells proliferate normally, unlike the EF− cells that undergo cell cycle arrest and apoptosis after the removal of doxycycline. Furthermore, a number of other studies have compared the gene expression of Ewing sarcoma tumors to mesenchymal cells, the lineage of the EFFib cells based on immunophenotype (Figure 2C), in order to identify genes regulated by EWS-FLI16,20,36,37. A potential limitation of this comparison, though, is that the EF+ and EFFib cells may derive from different precursor cells. However, we anticipated that the intersection of these different gene lists could be useful for identifying a core set of genes regulated by EWS-FLI1.
A comparison of gene expression between EF+ and EF− cells, four days after doxycycline removal, revealed 659 genes with differential expression (Fold > 3 and FDR < 0.01) (Supplemental Table S3). A total of 1175 genes (Fold > 3 and FDR < 0.01) were differentially expressed between the EF+ and EFFib cells (Supplemental Table S4). Importantly, both analyses identified the upregulation and downregulation of well-validated targets of EWS-FLI1, including NR0B13, NKX2-25,7, CCND138, BCL11B39, EZH240, and GRP41. Furthermore, in both comparisons, multiple genes that are highly expressed in Ewing sarcoma tumors, including NR0B13, GRP41 and NPY42, demonstrated greater than a 50-fold change in expression level, validating the utility of this inducible, isogenic and dual comparison approach. Select genes were validated with RT-qPCR (Figure 5B) and the twenty-five most highly upregulated genes from each comparison are shown in Table 1. Notably, these gene expression signatures are also significantly enriched in the Ewing sarcoma cell lines in the Cancer Cell Line Encyclopedia (Broad Institute) compared to the ~1000 other tumor cell lines, as assessed using the analysis tool Enrichr43 (Table 2 and Supplemental Table S5).
Table 1.
The most highly upregulated genes from the EF+ versus EF− and EF+ versus EFFib comparisons.
| EF+ versus EF− | EF+ versus EFFib | ||
|---|---|---|---|
| Gene | Fold Change | Gene | Fold Change |
| GRP | 407 | RBM11 | 722 |
| SYNPR | 332 | LIPI | 455 |
| LOC400796 | 324 | NELL2 | 383 |
| NR0B1 | 220 | SLAIN1 | 371 |
| KIAA0101 | 175 | LOC400796 | 314 |
| TRDN | 166 | SYNPR | 313 |
| F2RL1 | 144 | NPY1R | 310 |
| RRM2 | 142 | SYCP1 | 289 |
| CDH12 | 101 | OTX2 | 283 |
| RBM11 | 99 | NPY | 263 |
| KCNE3 | 95 | GRP | 239 |
| RP11 | 83 | NR0B1 | 223 |
| PBK | 81 | AKR1C3 | 176 |
| KIF20A | 74 | TRDN | 173 |
| NPY | 73 | EPHA3 | 157 |
| DLG7 | 68 | FLJ25076 | 156 |
| TOP2A | 63 | APCDD1 | 154 |
| CXCR7 | 60 | NKX2-2 | 137 |
| PCDH20 | 60 | CLDN1 | 133 |
| CDC2 | 58 | HSD17B2 | 121 |
| NUSAP1 | 55 | OLFM3 | 119 |
| LOC144997 | 51 | ITM2A | 114 |
| LOC647248 | 51 | PTGER3 | 107 |
| BHLHB5 | 48 | HOXD10 | 103 |
| GAS1 | 46 | BCL11B | 95 |
Table 2.
Enrichment of the EF+ versus EF− upregulated gene signature in different cell lines in the Cancer Cell Line Encyclopedia. Enrichment was assessed using the Enrichr tool43. Ewing sarcoma cell lines are shown in bold.
| Rank | Cell Line | P-Value | Z-Score | Combined Score |
|---|---|---|---|---|
| 1 | A673_BONE | 1.67e-48 | −1.79 | 185.41 |
| 2 | SKNMC_BONE | 3.97e-44 | −1.81 | 169.91 |
| 3 | MHHES1_BONE | 3.01e-41 | −1.86 | 162.84 |
| 4 | SKES1_BONE | 4.22e-38 | −1.86 | 150.52 |
| 5 | TC71_BONE | 2.56e-37 | −1.81 | 143.24 |
| 6 | RDES_BONE | 3.30e-33 | −1.85 | 129.07 |
| 7 | CADOES1_BONE | 1.61e-18 | −1.75 | 63.42 |
| 8 | NCIH446_LUNG | 2.71e-8 | −1.75 | 22.40 |
| 9 | NCIH889_LUNG | 6.55e-7 | −1.81 | 17.66 |
| 10 | NCIH1092_LUNG | 2.17e-6 | −1.87 | 16.23 |
| 11 | CORL47_LUNG | 3.03e-6 | −1.83 | 15.55 |
| 12 | NCIH1385_LUNG | 4.22e-6 | −1.83 | 15.10 |
| 13 | NCIH1184_LUNG | 3.04e-6 | −1.74 | 14.79 |
| 14 | NCIH2081_LUNG | 7.15e-6 | −1.81 | 14.37 |
| 15 | NCIH1930_LUNG | 4.61e-6 | −1.69 | 13.97 |
To more systematically evaluate our gene expression results, we used gene set enrichment analysis (GSEA) to determine whether a “core EWS-FLI1 gene expression signature,” generated by Hancock et al. using a meta-analysis approach that combined data from thirteen independent studies, was enriched in our data sets44. The Hancock et al. upregulated and downregulated EWS-FLI1 gene sets (Figure 5C and 5D) were significantly (FDR q-value = 0.0) enriched in the EF+ cells in both of our gene expression comparisons. A second set of EWS-FLI1 core genes, generated by Kauer at al. from different experimental data than the Hancock et al. data, showed similar enrichment in the EF+ cells (Supplemental Figure S6)36.
GSEA was also performed using gene sets from the MSigDB collection, which is a collection of annotated gene sets available for use with GSEA45. Multiple, published EWS-FLI1 gene sets in this collection, including RIGGI_EWING, STAEGE_EWING, ZHANG_TARGETS_OF_EWSR1_FLI1, and MIYAGAWA_TARGETS_OF_EWSR1_ETS, showed significant (FDR q-value = 0.0) enrichment in our expression data (Figure 5C and 5D). In addition to these EWS-FLI1 data sets, the EF+ cells also showed significant enrichment for other gene sets in the MSigDB (Supplemental Table S6 and S7). In the EF+ versus EF− comparison, the most enriched gene sets from the gene ontology collection were related to the cell cycle and mitosis (Supplemental Table S6), which is likely reflective of the arrest in cell proliferation that is observed after the removal of doxycycline (Figures 3C and 3D). In the EF+ versus EFFib comparison, there was significant enrichment (FDR q-value = 0.0) for gene ontology sets related to the mitochondria, oxidative phosphorylation, mitochondria and electron transport (Supplemental Table S7).
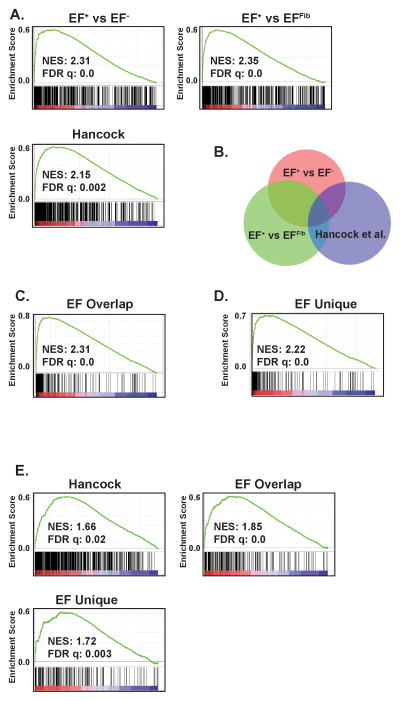
Next, we evaluated whether the differentially expressed genes identified in our inducible and isogenic system were enriched in Ewing sarcoma cell lines and primary tumors. In the same analysis, we also compared how our gene sets performed relative to the Hancock et al. gene set. Gene expression data from the Cancer Cell Line Encyclopedia (CCLE, http://www.broadinstitute.org/ccle/home) was used to compare ten Ewing sarcoma cells lines to 88 other cell lines46. These other cell lines included all sarcoma samples in the CCLE, as well as a spectrum of other tumor types (Supplemental Table S8). GSEA was performed using the upregulated genes from EF+ versus EF−, EF+ versus EFFib and Hancock et al. All gene sets showed significant enrichment in the Ewing sarcoma cell lines relative to the other tumor types (Figure 6A). The normalized enrichment scores (NES), which allow for the comparison of analysis results across gene sets, for the EF+ versus EF−, EF+ versus EFFib and Hancock et al. gene sets were 2.31, 2.35 and 2.15, respectively.
Figure 6.
The EF+ gene sets are enriched in Ewing cell lines. (A) GSEA shows enrichment of the EF+ versus EF− gene set, EF+ versus EFFib gene set, and the Hancock et al. gene set in the gene expression data from the Ewing sarcoma cell lines versus other tumor types comparison. (B) Venn diagram showing the overlap between the EF+ versus EF− gene set, EF+ versus EFFib gene set, and the Hancock et al. gene set. (C) GSEA shows enrichment of the EF Overlap upregulated gene set in the gene expression data from the Ewing sarcoma cell lines versus other tumor types comparison. (D) GSEA shows enrichment of the EF Unique upregulated gene set in the gene expression data from the Ewing sarcoma cell lines versus other tumor types comparison. (E) GSEA shows enrichment of the Hancock et al., EF Overlap and EF Unique gene sets in primary Ewing sarcoma tumors, compared to primary osteosarcoma tumors. The normalized enrichment scores (NES) and FDR q-values are shown.
The gene list from EF+ versus EF− was then compared to the gene list from EF+ versus EFFib to identify an overlapping set of genes with differential expression, referred to as the EF Overlap gene set (Figure 6B and Supplemental Table S9). This set contains 273 genes and includes many genes with established roles and/or expression profiles in Ewing sarcoma, including BCL11B39, CCND138, NKX2-25,7, NR0B13, NPY42, NPY1R20, STEAP147. The EF Overlap upregulated gene list exhibits significant enrichment (FDR q-value = 0.0; NES 2.31) in the Ewing sarcoma cell lines compared to the other tumor types (Figure 6C). There were forty-three upregulated genes in common between the EF Overlap and Hancock et al. gene sets (Figure 6B, Supplemental Table S10). This included the well-validated genes NR0B13, NKX2-25,7 and EZH240. The next question we addressed was whether these common genes were primarily responsible for the enrichment of our gene set in the Ewing sarcoma cell lines so we repeated the GSEA analysis using the set of upregulated genes that was unique to the EF Overlap list and not included on the Hancock et al. list. These unique genes (EF Unique, Supplemental Table S11) were significantly (FDR q-value = 0.0; NES 2.22) enriched in the Ewing sarcoma cell lines compared to the other tumor types (Figure 6D). Similar results were obtained when the EF gene lists were compared to the Kauer et al. gene list (data not shown)36.
Finally, using published microarray data, we evaluated whether the EWS-FLI1 gene sets identified in our system were significantly enriched in primary Ewing sarcoma tumors48 relative to primary osteosarcoma tumors49 (Figure 6E). Again, similar to the cell lines results, we observed significant (FDR q-value = 0.0–0.003) enrichment of our gene sets in the primary Ewing sarcoma tumor expression data. Overall, these gene expression data provide further evidence for an Ewing sarcoma-like phenotype for the EF+ cells and also identify new genes that are potentially regulated by EWS-FLI1.
DISCUSSION
The use of model organisms, including mice and zebrafish, has been critical in exploring the role of the cell-of-origin in tumorigenesis (reviewed in Visvader et al. 17). In human cells, however, it is often difficult to express an oncogene in a specific cell type at the appropriate developmental stage. In this work, we demonstrate that the expression of EWS-FLI1, in combination with the knockdown of p53, in embryoid bodies generates cells that recapitulate a number of important aspects of Ewing sarcoma biology. We chose to knockdown p53 in our model system because mutations in this tumor suppressor occur in vivo in a subset of patients and, importantly, almost all Ewing sarcoma cell lines exhibit mutations in p53.
The EF+ cells demonstrate a dependency on EWS-FLI1 expression for cell survival, or oncogene addiction. To our knowledge, this is the first model of Ewing sarcoma in human cells that exhibits a requirement for EWS-FLI1 expression. The removal of doxycycline from the EF+ cells leads to cell cycle changes and apoptosis, which parallels what occurs after knockdown of EWS-FLI1 in Ewing sarcoma cell lines8,24. The EF+ cells also exhibit other properties of transformation. This includes both anchorage-independent growth and a lack of contact inhibition. Furthermore, the EF+ cells, relative to the EFFib cells, exhibit significantly enhanced sensitivity to the PARP inhibitor olaparib, which is also reported for Ewing sarcoma cells lines relative to other cancer cell lines and non-transformed cells31,32. This drug toxicity data suggest that the EF+ and EFFib cells may be useful in small molecule and/or RNAi screens to identify novel compounds/genes with selective toxicity to Ewing sarcoma cells.
In one experiment, the EF+ cells became tetraploid with a complete duplication of their chromosome content. Tetraploidy is a common feature of tumors and transformed cells. A recent study that included a spectrum of eleven different tumor types identified whole genome doublings in 37% of tumors50. In Ewing sarcoma, tetraploidy has been reported in both primary tumors and cell lines51–53. Although the impact of genome doubling in Ewing sarcoma tumors is not known, tetraploidy is also consistent with a transformed phenotype in the EF+ cells. Notably, in our system, the knockdown of p53 may have also contributed to the propagation of the EF+ tetraploid cells since p53 is a well-described barrier to tetraploidy54.
Although the EF+ cells exhibit multiple characteristics of transformation, the cells did not form tumors in immunocompromised mice. Importantly, the engraftment efficiencies in mice of primary, patient-derived tumors is highly variable, with some human tumors failing to form any tumors in mice55. Consequently, the inability of the EF+ cells to form xenograft tumors does not exclude the possibility that the cells are transformed. However, additional reasons could also explain the inability of the EF+ cells to form tumors in vivo. For example, the lack of xenograft tumors could reflect the need for additional genetic events, such as the loss of STAG2 or trisomy 8, for growth of the cells in the mouse.
The gene expression experiments identified many known targets of EWS-FLI1, including NR0B1, NKX2-2, and BCL11B. A number of these differentially regulated genes exhibit >50-fold changes in expression level, likely reflecting the advantages of an isogenic system and the ability to tightly regulate the expression of EWS-FLI1 using doxycycline. Although, there is not a gold standard for an Ewing sarcoma gene expression signature, the differentially expressed genes identified in this study are significantly enriched for EWS-FLI1 “core genes,” as identified in the analysis of multiple studies36,44.
In addition to Ewing sarcoma gene sets, GSEA also identified enrichment for cell cycle gene sets in the EF+ versus EF− comparison. Cell cycle deregulation is well described in Ewing sarcoma (reviewed in Kowalewski et al.56) and enrichment of cell cycle genes has been identified in other gene expression experiments36. Cyclin D1, for example, is an established target of EWS-FLI1 and this gene was upregulated 8-fold in the EF+ cells relative to the EF− cells 38. However, the alteration in the expression of some of the other cell cycle genes is likely an indirect effect due to the arrest in proliferation seen after the removal of doxycycline (Figure 3D), rather than direct targeting by EWS-FLI1. This conclusion is supported by the observation that the strong enrichment for the cell cycle and mitotic gene sets, described for the EF+ versus EF− comparison, was not observed in the EF+ versus EFFib comparison, where both the EF+ and EFFib cells proliferate normally. By contrast, Cyclin D1 remains overexpressed (8-fold) in the EF+ cells compared to the EFFib cells, as might be predicted for a direct target of EWS-FLI1 38.
GSEA analysis also identified enrichment for oxidative phosphorylation and mitochondrial gene sets in the EF+ versus EFFib comparison. Alterations in metabolism are well described in cancer cells57. Furthermore, Lin28b, an important regulator of cellular metabolism58, is 3.5-fold and 40-fold overexpressed in the EF+ cells relative to the EF− and EFFib cells, respectively. Importantly, the overexpression of Lin28b in Ewing sarcoma cell lines, relative to mesenchymal cells, and alterations in the level of Let-7a, a known target of Lin28, have also been reported in Ewing sarcoma cell lines59.
Many known targets of EWS-FI1, as well as novel genes, were identified in our analysis. Notably, when tested using a set of Ewing sarcoma cell lines and primary tumors our gene expression signatures perform better, based on NES scores, than a core Ewing sarcoma gene signature generated from a meta-analysis of thirteen studies. Our isogenic and inducible system avoids the experimental variation that is likely one cause of the only modest overlap of Ewing sarcoma gene signatures that are generated in different studies44. Furthermore, the genes that are unique to our gene expression data are still significantly enriched in Ewing sarcoma tumor cell lines even if the common genes are removed from the analysis. This suggests that these “unique” genes may have a role in Ewing sarcoma pathophysiology.
In summary, we have developed a novel approach to model tumorigenesis in human cells that utilizes the developmental and pluripotent potential of stem cells. We anticipate these model cell lines and the gene expression data will be useful for investigating the pathophysiology of Ewing sarcoma. Furthermore, the cell-of-origin question in Ewing sarcoma could be investigated in a modification our approach by using lineage specific promoters, rather than the doxycycline-inducible promoter. We also expect that it may be possible to extend our experimental approach to additional types of cancer and to other biological systems where the cell-of-origin is critical.
MATERTIALS AND METHODS
Ethics statement
The Embryonic Stem Cell Research Oversight Committee at the Dana-Farber Cancer Institute approved the stem cell experiments. The Institutional Animal Care and Usage Committee at the Dana-Farber Cancer Institute approved the animal studies.
Stem cell culture
Cell lines were maintained at 37°_C in a 5% CO2 atmosphere. H1 stem cells (WA01, WiCell Research Institute) were cultured in mTESR1 media (Stemcell Technologies) on plates coated with matrigel (BD Biosciences). The stem cells were passaged every 4–6 days using versene (Gibco).
Embryoid body formation
Embryoid bodies were formed using AggreWell plates (Stemcell Technologies) according to the manufacturer’s instructions. Briefly, stem cells were collected using Accutase (Stemcell Technologies), centrifuged at 300 r.c.f. and re-suspended in AggreWell media (Stem Cell Technologies) with 10 μm ROCK inhibitor (R & D Systems). Cells (1.0 x 107) were added to one well of an AggreWell 400ex plate and spun at 100 r.c.f. After centrifugation, the plates were transferred to an incubator at 37°_C. After 24 hours, the embryoid bodies were collected with gentle pipetting, allowed to sediment by gravity, re-suspended in DMEM/F12 media (Gibco) with 20% Knock-out Serum Replacement (Life Sciences) and cultured in very low adhesion plates (Corning). After 7 days of suspension culture, doxycycline (1 μg ml−1) was added to the media and 3–7 days later the embryoid bodies were transferred to gelatin-coated plates. The media was then switched to DMEM supplemented with 10% FBS, 100_IU_ml 1 penicillin, 100 μg_ml 1 streptomycin and 1 μgml−1 doxycycline.
Gene Expression
For the microarray experiments, RNA was collected from three biological replicates of EF+, EF− and EFFib cells using trizol (Life Technologies) and an RNeasy kit (Qiagen). The samples were then prepared for analysis and hybridized to Human Genome U133+ v2.0 chips (Affymetrix) by the Microarray Core at Dana-Farber Cancer Institute. GenePattern release 3.8.0 was used to normalize the raw microarray data using the robust multichip average (RMA) algorithm, preprocess the normalized data using default parameters and find differentially expressed probe sets. Gene set enrichment analysis (GSEA) was performed using the GSEA platform (www.broadinstitute.org/gsea) and Enrichr (amp.pharm.mssm.edu/Enrichr). The gene expression CEL files were deposited in the Gene Expression Omnibus (GEO) Repository under the accession number GSE64686. Venn diagrams were prepared using BioVenn (http://www.cmbi.ru.nl/cdd/biovenn/)60. Published data sets used in the gene expression analysis included GSE3462048 (Ewing sarcoma), GSE1482749 (osteosarcoma), and the Cancer Cell Line Encyclopedia46 (http://www.broadinstitute.org/ccle/home).
Statistical Analysis
Data are presented as mean ± SEM. Student’s t test was used to calculate p values. Statistical analyses were conducted using GraphPad Prism 5.0.
Supplementary Material
Acknowledgments
We would like to thank Hubo Li for helpful discussions and Kelli Goss for assistance with the drug dose-response experiments. D.J.G. is supported by a NCI K08 award (5K08 CA160346-03), a CureSearch Award and a Sarcoma Foundation of America Award. D.P. is a HHMI investigator and supported by NIH grant GM083299.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
Supplementary information accompanies the paper on the Oncogene website (http:///www.nature.com/onc).
References
- 1.Balamuth NJ, Womer RB. Ewing's sarcoma. Lancet Oncology. Elsevier Ltd. 2010 Feb 1;11(2):184–92. doi: 10.1016/S1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- 2.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992 Sep 10;359(6391):162–5. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 3.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol Cancer Res. 2006 Nov;4(11):851–9. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 4.May WA, Gishizky ML, Lessnick SL, Lunsford LB, Lewis BC, Delattre O, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci USA. 1993 Jun 15;90(12):5752–6. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owen LA, Kowalewski AA, Lessnick SL. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing's sarcoma. PLoS ONE. 2008;3(4):e1965. doi: 10.1371/journal.pone.0001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suvà ML, et al. EWS-FLI1 Utilizes Divergent Chromatin Remodeling Mechanisms to Directly Activateor Repress Enhancer Elements in Ewing Sarcoma. Cancer Cell. Elsevier Inc. 2014 Oct 27;:1–14. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006 May;9(5):405–16. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 Silencing and Gene Profiling of Ewing Cells Reveal Downstream Oncogenic Pathways and a Crucial Role for Repression of Insulin-Like Growth Factor Binding Protein 3. Molecular and Cellular Biology. 2004 Jul 28;24(16):7275–83. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chansky HA, Barahmand-Pour F, Mei Q, Kahn-Farooqi W, Zielinska-Kwiatkowska A, Blackburn M, et al. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing's sarcoma cells in vitro. J Orthop Res. 2004 Jul;22(4):910–7. doi: 10.1016/j.orthres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Kovar H, Aryee DN, Jug G, Henöckl C, Schemper M, Delattre O, et al. EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells in vitro. Cell Growth Differ. 1996 Apr;7(4):429–37. [PubMed] [Google Scholar]
- 11.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent STAG2 Mutation. In: Horwitz MS, editor. PLoS Genet. 7. Vol. 10. 2014. Jul 10, p. e1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic Landscape of Ewing Sarcoma Defines an Aggressive Subtype with Co-Association of STAG2 and TP53 Mutations. Cancer Discov. 2014 Nov 2;4(11):1342–53. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The Genomic Landscape of Pediatric Ewing Sarcoma. Cancer Discov. 2014 Nov 2;4(11):1326–41. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 14.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. Nature Publishing Group. 2013 Jun 16;:1–5. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011 Aug 19;333(6045):1039–43. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladanyi M. EWS-FLI1 and Ewing's sarcoma: recent molecular data and new insights. Cancer Biol Ther. 2002 Jul;1(4):330–6. [PubMed] [Google Scholar]
- 17.Visvader JE. Cells of origin in cancer. Nature. 2011 Jan 20;469(7330):314–22. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 18.Levetzow von C, Jiang X, Gwye Y, Levetzow von G, Hung L, Cooper A, et al. Modeling initiation of Ewing sarcoma in human neural crest cells. PLoS ONE. 2011;6(4):e19305. doi: 10.1371/journal.pone.0019305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potikyan G, France KA, Carlson MRJ, Dong J, Nelson SF, Denny CT. Genetically defined EWS/FLI1 model system suggests mesenchymal origin of Ewing's family tumors. Lab Invest. 2008 Oct 6;88(12):1291–302. doi: 10.1038/labinvest.2008.99. [DOI] [PubMed] [Google Scholar]
- 20.Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S, et al. EWS-FLI-1 Expression Triggers a Ewing's Sarcoma Initiation Program in Primary Human Mesenchymal Stem Cells. Cancer Res. 2008 Apr 1;68(7):2176–85. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 21.Leacock SW, Basse AN, Chandler GL, Kirk AM, Rakheja D, Amatruda JF. A zebrafish transgenic model of Ewing's sarcoma reveals conserved mediators of EWS-FLI1 tumorigenesis. Disease Models & Mechanisms. 2011 Dec 28;5(1):95–106. doi: 10.1242/dmm.007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lessnick SL, Dacwag CS, Golub TR. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002 May;1(4):393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 23.Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010 May 3;24(9):916–32. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lessnick SL, Ladanyi M. Molecular Pathogenesis of Ewing Sarcoma: New Therapeutic and Transcriptional Targets. Annu Rev Pathol Mech Dis. 2012 Feb 28;7(1):145–59. doi: 10.1146/annurev-pathol-011110-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007 Apr;17(2):157–62. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Miyagawa Y, Okita H, Nakaijima H, Horiuchi Y, Sato B, Taguchi T, et al. Inducible Expression of Chimeric EWS/ETS Proteins Confers Ewing's Family Tumor-Like Phenotypes to Human Mesenchymal Progenitor Cells. Molecular and Cellular Biology. 2008 Mar 7;28(7):2125–37. doi: 10.1128/MCB.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amaral AT, Manara MC, Berghuis D, Ordóñez JL, Biscuola M, Lopez-García MA, et al. Characterization of human mesenchymal stem cells from ewing sarcoma patients. Pathogenetic implications. PLoS ONE. 2014;9(2):e85814. doi: 10.1371/journal.pone.0085814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocchi A, Manara MC, Sciandra M, Zambelli D, Nardi F, Nicoletti G, et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J Clin Invest. 2010 Mar;120(3):668–80. doi: 10.1172/JCI36667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro S, Giraudo P, Karseladze AI, Smirnov A, Petrovichev N, Savelov N, et al. Immunophenotypic profile of biomarkers related to anti-apoptotic and neural development pathways in the Ewing's family of tumors (EFT) and their therapeutic implications. Anticancer Res. 2007 Jul;27(4B):2457–63. [PubMed] [Google Scholar]
- 30.Scotlandi K. c-kit Receptor Expression in Ewing's Sarcoma: Lack of Prognostic Value but Therapeutic Targeting Opportunities in Appropriate Conditions. Journal of Clinical Oncology. 2003 May 15;21(10):1952–60. doi: 10.1200/JCO.2003.11.111. [DOI] [PubMed] [Google Scholar]
- 31.Brenner JC, Feng FY, Han S, Patel S, Goyal SV, Bou-Maroun LM, et al. PARP-1 inhibition as a targeted strategy to treat Ewing's sarcoma. Cancer Res. 2012 Apr 1;72(7):1608–13. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012 Mar 29;483(7391):570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-Rotenberg JS, Yuan L, et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma. Nature Medicine. 2009 Jul 5;15(7):750–6. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen QR, Yeung C, et al. Identification of an Inhibitor of the EWS-FLI1 Oncogenic Transcription Factor by High-Throughput Screening. JNCI Journal of the National Cancer Institute. 2011 Jun 21;103(12):962–78. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benini S, Manara MC, Cerisano V, Perdichizzi S, Strammiello R, Serra M, et al. Contribution of MEK/MAPK and PI3-K signaling pathway to the malignant behavior of Ewing's sarcoma cells: Therapeutic prospects. Int J Cancer. 2003;108(3):358–66. doi: 10.1002/ijc.11576. [DOI] [PubMed] [Google Scholar]
- 36.Kauer M, Ban J, Kofler R, Walker B, Davis S, Meltzer P, et al. A molecular function map of Ewing's sarcoma. PLoS ONE. 2009;4(4):e5415. doi: 10.1371/journal.pone.0005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007 May;11(5):421–9. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez G, Bittencourt D, Laud K, Barbier J, Delattre O, Auboeuf D, et al. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proceedings of the National Academy of Sciences. 2008 Apr 22;105(16):6004–9. doi: 10.1073/pnas.0710748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiles ET, Lui-Sargent B, Bell R, Lessnick SL. BCL11B is up-regulated by EWS/FLI and contributes to the transformed phenotype in Ewing sarcoma. PLoS ONE. 2013;8(3):e59369. doi: 10.1371/journal.pone.0059369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richter GHS, Plehm S, Fasan A, Rössler S, Unland R, Bennani-Baiti IM, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proceedings of the National Academy of Sciences. 2009 Mar 31;106(13):5324–9. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawlor ER, Lim JF, Tao W, Poremba C, Chow CJ, Kalousek IV, et al. The Ewing tumor family of peripheral primitive neuroectodermal tumors expresses human gastrin-releasing peptide. Cancer Res. 1998 Jun 1;58(11):2469–76. [PubMed] [Google Scholar]
- 42.Tilan JU, Lu C, Galli S, Izycka-Swieszewska E, Earnest JP, Shabbir A, et al. Hypoxia shifts activity of neuropeptide Y in Ewing sarcoma from growth-inhibitory to growth-promoting effects. Oncotarget. 2013 Dec;4(12):2487–501. doi: 10.18632/oncotarget.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle. 2008 Jan 15;7(2):250–6. doi: 10.4161/cc.7.2.5229. [DOI] [PubMed] [Google Scholar]
- 45.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005 Oct 25;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012 Mar 29;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grunewald TGP, Diebold I, Esposito I, Plehm S, Hauer K, Thiel U, et al. STEAP1 is associated with the invasive and oxidative stress phenotype of Ewing tumors. Molecular Cancer Research. 2012 Jan;10(1):52–65. doi: 10.1158/1541-7786.MCR-11-0524. [DOI] [PubMed] [Google Scholar]
- 48.Postel-Vinay S, Véron AS, Tirode F, Pierron G, Reynaud S, Kovar H, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012 Mar;44(3):323–7. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, et al. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Molecular Cancer Therapeutics. 2010 Mar;9(3):535–44. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- 50.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013 Sep 26;45(10):1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowal-Vern A, Walloch J, Chou P, Gonzalez-Crussi F, Price J, Potocki D, et al. Flow and image cytometric DNA analysis in Ewing's sarcoma. Mod Pathol. 1992 Jan;5(1):56–60. [PubMed] [Google Scholar]
- 52.Roberts P, Burchill SA, Brownhill S, Cullinane CJ, Johnston C, Griffiths MJ, et al. Ploidy and karyotype complexity are powerful prognostic indicators in the Ewing“s sarcoma family of tumors: A study by the United Kingdom Cancer Cytogenetics and the Children”s Cancer and Leukaemia Group. Genes Chromosomes Cancer. 2008;47(3):207–20. doi: 10.1002/gcc.20523. [DOI] [PubMed] [Google Scholar]
- 53.Maire G, Brown CW, Bayani J, Pereira C, Gravel DH, Bell JC, et al. Complex rearrangement of chromosomes 19, 21, and 22 in Ewing sarcoma involving a novel reciprocal inversion-insertion mechanism of EWS-ERG fusion gene formation: a case analysis and literature review. Cancer Genet Cytogenet. 2008 Mar;181(2):81–92. doi: 10.1016/j.cancergencyto.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Ganem NJ, Cornils H, Chiu S-Y, O'Rourke KP, Arnaud J, Yimlamai D, et al. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014 Aug 14;158(4):833–48. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams SA, Anderson WC, Santaguida MT, Dylla SJ. Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Lab Invest. 2013 Sep;93(9):970–82. doi: 10.1038/labinvest.2013.92. [DOI] [PubMed] [Google Scholar]
- 56.Kowalewski AA, Randall RL, Lessnick SL. Cell Cycle Deregulation in Ewing's Sarcoma Pathogenesis. Sarcoma. 2011;2011:598704. doi: 10.1155/2011/598704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009 May 22;324(5930):1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, et al. The Lin28/let-7 AxisRegulates Glucose Metabolism. Cell. Elsevier Inc. 2011 Sep 30;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Vito C, Riggi N, Suvà M-L, Janiszewska M, Horlbeck J, Baumer K, et al. Let-7a is a direct EWS-FLI-1 target implicated in Ewing's sarcoma development. PLoS ONE. 2011;6(8):e23592. doi: 10.1371/journal.pone.0023592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.