Abstract
This paper reports a droplet-based microfluidics approach to fabricate a large number of monodisperse, portable microtissues, each in an individual drop. We use water-water-oil double emulsions as templates and spatially assemble hepatocytes in the core and fibroblasts in the shell, forming a 3D liver model in a drop.
Nearly all tissues are integrated three-dimensional (3D) structures of multiple types of cells and extracellular matrices (ECMs).1,2 The function of a tissue is typically governed by multiple cues, ranging from intercellular signalings to cell interactions with the surrounding ECMs.3-6 Liver, for example, consists of primary hepatocytes, hepatic stellate cells, Kupffer cells, endothelial cells, and fibroblasts, which are arranged in a 3D scaffold.7 Hepatocytes alone show very low levels of liver-specific functions, because conventional two-dimensional (2D) cultures of a monolayer of these cells on a plastic dish are poor mimics of tissues found in vivo.8 Instead, improved functionality of liver tissue mimics requires the development of 3D models that consist of simplified but similar scaffolds with multiple types of cells embedded in ECMs and that express improved functionality.9-12
Well-defined spatial distributions of different cells in 3D architectures made of biocompatible ECMs are required to construct artificial tissues that mimic the in vivo microenvironment. Miniaturized, multiwell culture systems,13-15 3D cell printing,16-18 and 3D cell molding,19 have each been used to develop 3D artificial liver models. In each case, hepatocytes are co-cultured with other cells in micro-patterned 3D scaffolds; they are separated in space while close enough to each other that they can interact biochemically. Therefore, different cells are able to coordinate both homotypic and heterotypic cell-cell interactions and show improved liver-specific functions.20-22 However, these technologies are limited in terms of uniformity, portability and quantity; versatile human microtissues composed of different cells in precisely controlled 3D structures are, as yet, unavailable.
Droplet-based microfluidics can produce monodisperse droplets whose size can be precisely controlled and whose internal structure can be tailored to incorporate different cells in designated locations.23-26 However, previous microfluidic-based research focused on biocompatible hydrogel microparticles templated from single water-in-oil (w/o) emulsions.27-31 These would lead to over-simplified 3D cellular microenvironment with limitations both in terms of impaired cell-cell contact and undefined spatial distribution of different cells in the drops. More sophisticated structures with well-defined 3D architectures are, therefore, required.
In this paper, we report the first study of reconstituting “organ in a droplet” by controlled assembly of heterotypic cells in biocompatible 3D core-shell hydrogel scaffolds. We use droplet-based microfluidics to produce highly monodisperse structures that enable multiple types of cells to be optimally arranged to create an artificial liver in a drop. We produce water-water-oil (w/w/o) double emulsions which we use as templates to successfully integrate the 3D core-shell scaffold with hepatocytes in the core surrounded by fibroblasts in the shell. We cross-link the shell of alginate hydrogel by triggered release of calcium cations; this method avoids exposure to harsh environments during fabrication and creates structures with good integrity and high permeability. Hepatocytes and fibroblasts co-cultured in the drop develop both homotypic and heterotypic cell-cell interactions, which cannot be achieved in a 2D culture system. Albumin secretion and urea metabolism, which are good assays of liver-specific functions, both exhibit higher activity in the microtissues as compared to that in hepatocytes alone.
To construct the core-shell scaffolds, we use w/w/o double emulsions as templates and generate core-shell droplets in a flow-focusing microfluidic device, as shown in Figure 1. The inner phase is cell culture medium and the middle phase is alginate aqueous solution, a well-studied biocompatible polymer that shows excellent cell function and survival in the network.32 The inner phase co-flows with the middle phase due to their low Reynolds numbers; a fluorinated carbon oil at the cross-junction forms monodisperse droplets consisting of an aqueous core and a hydrogel shell (Figures 1b and c). Instead of cross-linking by UV exposure, which is generally harmful to cells, we introduce an additional oil flow containing 0.15% acetic acid downstream and trigger release of Ca2+ from the Ca-EDTA complex in the alginate solution; the divalent Ca2+ subsequently binds to two different carboxylic groups of alginate chains, forming a cross-linked 3D network (Figure 1a). The in situ cross-linking locks alginate in the shell, which is directly visualized with fluorescent confocal microscopy by labeling alginate with fluorescein (see Supporting Information, Scheme S1), as shown in the inset of Figure 1d. Using these w/w/o double emulsions as templates, we are able to fabricate a large number of monodisperse core-shell droplets with well-controlled internal structure (Diameter=169 ± 6 μm, see Supporting Information, Figure S1).
Figure 1.
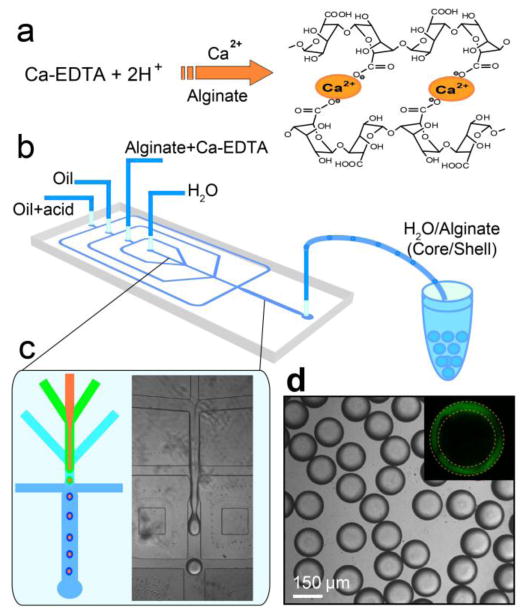
Construction of 3D scaffold in a drop consisting an aqueous core and a hydrogel shell. a) Cross-link of alginate network by triggered release of Ca2+ from Ca-EDTA complex. b) Schematic diagram of the PDMS device. c) Fabrication of core-shell droplets using w/w/o double emulsions as templates. Alginate in the shell is cross-linked by in situ triggered release of Ca2+. d) Monodisperse core-shell droplets generated using the droplet-based microfluidics. The shell of alginate hydrogel is clearly identified under confocal microscope when alginate is labeled with fluorescein, as shown in the inset.
To assemble hepatocytes in the core, we suspend hepatocytes in the cell culture medium and inject them with the inner phase (see Supporting Information, Figure S2). Hepatocytes are subsequently encapsulated in the core by the hydrogel shell and they start to form aggregates after several days' culture, as shown in Figure 2a. The aggregate is a common phenotype of healthy hepatocytes and is clear proof of direct cell-cell contacts due to homotypic cell-cell interactions. Similarly, we distribute fibroblasts in the shell by premixing them with the alginate solution and injecting them with the middle phase (see Supporting Information, Figure S3). The fibroblasts are then confined in the shell by the cross-linked alginate network and there are no cells observed in the core, as shown in Figure 2b. Random co-cultures of hepatocytes and fibroblasts in the same compartment will have lower liver-specific functions due to the lost balance of homotypic and heterotypic cell-cell interactions;13 therefore, to mimick the 3D structure of human liver in vivo and to develop an artificial liver in each drop, we simultaneously assemble hepatocytes (HepG2 cells) in the core surrounded by fibroblasts (NIH-3T3 cells) in the shell, forming a spheroid of different cells embedded in a 3D ECM (see Supporting Information, Figure S4 and Movie S1). In our study, the heterocellular spheroids are generated by one-step in situ cross-linking of alginate in the shell, after which the spheroids are quickly transferred to cell culture medium. The exposure of cells to mild acidic conditions during the whole process is limited and has little effect on the viability of cells, as evidenced by the predominance of live cells (green) and the complete lack of any dead cells (red) in the live-dead assay, as shown in Figure 2c.
Figure 2.
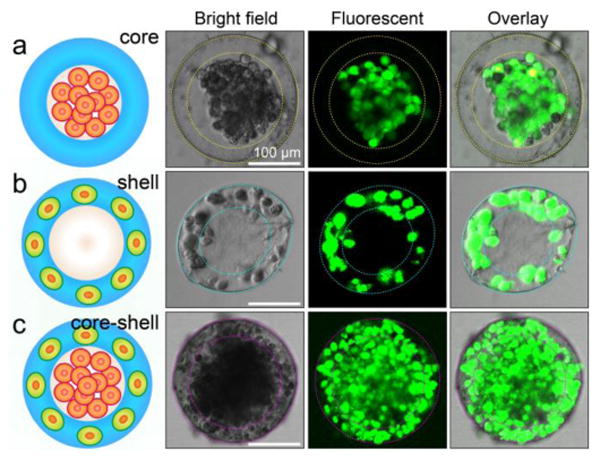
Spatial assembly of different cells in the 3D core-shell scaffold. a) HepG2 cells confined in the core by the hydrogel shell. b) NIH-3T3 fibroblasts immobilized by the cross-linked alginate network in the shell. c) Simultaneous assembly of hepatocytes in the core and fibroblasts in the shell, forming an artificial liver in a drop. Cell viability is characterized by Calcein AM/EthD-1 staining kit. The scale bars are 100 μm.
We culture the core-shell spheroids in the medium at 37°C. The spheroids maintain their spherical shape even for two weeks' culture, during which time the cells grow denser, as shown in Figures 3a-c. The distinct spherical shape is also observed in freeze-dried samples using scanning electron microscopy, as shown in Figure 3d and magnified in Figure 3e. Moreover, the cross-section of a freeze-dried spheroid reveals dense cell aggregates in the core and dispersed cells in the shell, as shown in Figure 3f. These observations suggest that the hydrogel shell of cross-linked alginate is mechanically strong. This strength, together with the portability of each microtissue in a drop enables us to store them at -80°C, thaw them at 37°C and culture them again without destroying the structure or affecting the viability of cells, as shown in Figure 3g. While the shell of alginate hydrogel is robust, it is also highly permeable, enabling nutrients and metabolites to pass through; therefore, cells in the spheroids can be cultured for long periods of time, as evidenced by the high viability of cells cultured in the spheroids. After 10 days' culture, cells are predominantly live (green) and there is complete lack of dead cells (red), as revealed by a live-dead staining kit (Figure 3h). During the period, cells have grown denser. However, because cells can't attach to unmodified alginate network, cells can't migrate from one layer to the other over time. Therefore, hepatocytes and fibroblasts remain spatially separated, which is beneficial for their liver-specific functions. Even after 14 days, a slight decrease in cell viability is observed; the percentage of dead cells is about 34%, as shown in Figure 3i. It is possible that a hypoxic core developed at day 14 due to cell proliferation and the resulting high cell density.
Figure 3.
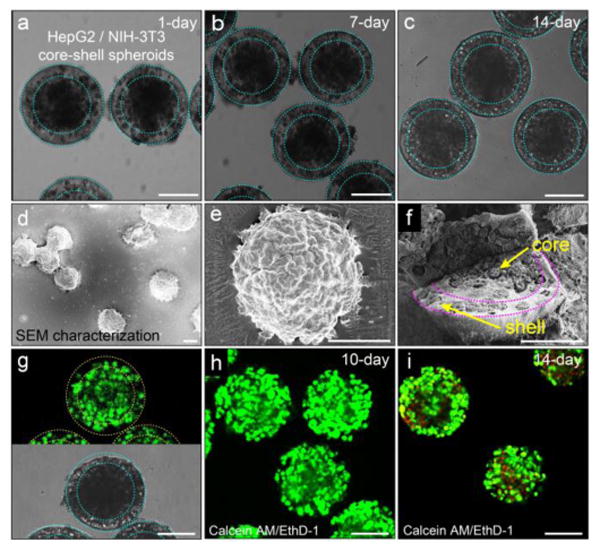
Co-culture of hepatocytes and fibroblasts in the core-shell spheroids. Morphology of the heterocellular spheroids and viability of the cells in the hydrogel scaffold. a-c) Co-culture of hepatocytes in the core and fibroblasts in the shell for 1 day, 7 days and 14 days, respectively. The heterocellular spheroids maintain their spherical morphology over time. d) SEM image and e) magnified image show individual spheroids that maintain their structural integrity after freeze-drying. f) Spatially confined cell ensembles in the core-shell structure. g) High viability of cells encapsulated in the spheroids after being frozen at -80°C for two weeks and then thawed at 37°C. Cells are predominantly alive (green) and there is complete lack of dead cells (red). h) Co-culture of cells encapsulated in the spheroids for 10 days shows high cell viability. i) The viability of cells cultured for 14 days decreases slightly, as evidenced by the appearance of dead cells (red). Cell viability is characterized by Calcein AM/EthD-1 staining kit. The scale bar is 100 μm in all images.
To measure the liver-specific functions of the microtissues in a drop, we monitor their albumin secretion and urea synthesis over time; the two metabolites are key biomarkers of the liver and this response has been widely used in drug screening.33 We culture two samples with same amount of hepatocytes, one in the core-shell structure with fibroblasts in the shell and the other without fibroblasts, and monitor the concentration of albumin and urea in the medium using albumin and urea assay kits, respectively. Compared to monotypic culture of hepatocytes alone, co-culture of hepatocytes and fibroblasts in the core-shell spheroids displays both increased albumin secretion and urea synthesis, as shown in Figures 4a and b. The enhanced liver-specific functions substantially differentiate our 3D microscale co-culture from conventional monotypic cell culture (statistically significant, *p<0.01). These results suggest that co-culture of hepatocytes and fibroblasts embedded in the 3D core-shell scaffold is a good model of the liver in vitro and has a balance of homotypic and heterotypic cell-cell interactions, which are beneficial for the expressions of their liver-specific functions. High-throughput assays of human tissue responses using these uniform, portable microtissues, each in a drop, will be valuable for rapid assessment of drugs, chemicals and cosmetics.
Figure 4.
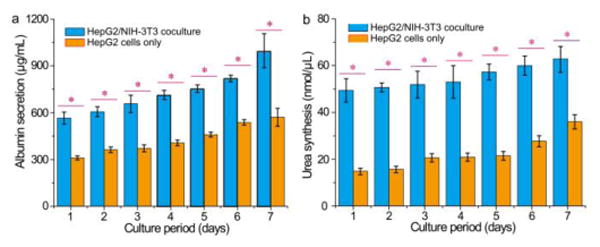
Comprehensive assays of liver-specific functions of hepatocyte/fibroblast co-culture and hepatocyte culture. a) Albumin secretion and b) urea synthesis of HepG2/NIH-3T3 co-culture and HepG2 culture measured over seven days. The liver-specific functions enhanced by the co-culture of hepatocytes and fibroblasts in the 3D core-shell spheroids are statistically significant (*p<0.01).
Conclusions
We use droplet-based microfluidics and hierarchically assemble hepatocytes and fibroblasts into a 3D core-shell scaffold, forming an artificial liver in each drop. The strong mechanical property and high permeability of the hydrogel shell enable cells in the spheroids to be cultured for long periods of time. Within the optimized microscale structure that promotes both homotypic and heterotypic cell-cell interactions, co-culture of hepatocytes in the core surrounded by fibroblasts in shell successfully expresses high-level liver-specific functions. Thousands of monodisperse microtissues, each in an individual drop, are achievable using the microfluidics technology, and each maintains enhanced liver-specific functions. In addition, hepatocytes and fibroblasts can easily be replaced with other cells to construct other tissue models and study cell-to-cell interaction in general. Thus, these structures are a promising in vitro liver model for high-throughput drug screening assays.34,35
Supplementary Material
Acknowledgments
Q. Chen acknowledges the China Scholarship Council (2012[3013]) and the staff of Experimental Soft Condensed Matter at Harvard University. This work was financially supported by the National Natural Science Foundation of China (Nos. 214350002, 91213305, and 81373373) and China Equipment and Education Resources System (No. CERS-1-75). This work was also supported by the National Science Foundation (DMR‐1310266), by the Harvard Materials Research Science and Engineering Center (DMR‐1420570) and the National Institutes of Health (R01EB014703). S. Utech was supported by Deutsche Forschungsgemeinschaft.
Footnotes
Electronic Supplementary Information (ESI) available: Preparation of the core-shell scaffold, controlled assembly of multiple cells into the 3D scaffold, cell viability analysis, morphology characterization and biomarker measurements. See DOI: 10.1039/x0xx00000x
References
- 1.Place ES, Evans ND, Stevens MM. Nat Mater. 2009;8:457–470. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- 2.Grolman JM, Zhang D, Smith AM, Moore JS, Kilian KA. Adv Mater. 2015;27:5512. doi: 10.1002/adma.201501729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel V, Sheetz M. Nature Reviews Molecular Cell Biology. 2006;7:265. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 4.Daley WP, Peters SB, Larsen M. Journal of Cell Science. 2008;121:255. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 5.Klein F, Richter B, Striebel T, Franz CM, von Freymann G, Wegener M, Bastmeyer M. Adv Mater. 2011;23:1341. doi: 10.1002/adma.201004060. [DOI] [PubMed] [Google Scholar]
- 6.Kraehenbuehl TP, Langer R, Ferreira LS. Nat Methods. 2011;8:731. doi: 10.1038/nmeth.1671. [DOI] [PubMed] [Google Scholar]
- 7.Kmieć Z. Adv Anat Embryol Cell Biol. 2001;161:1. doi: 10.1007/978-3-642-56553-3. [DOI] [PubMed] [Google Scholar]
- 8.Gidrol X, Fouque B, Ghenim L, Haguet V, Picollet-D'hahan N, Schaack B. Current Opinion in Pharmacology. 2009;9:664. doi: 10.1016/j.coph.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Moutos FT, Freed LE, Guilak F. Nat Mater. 2007;6:162. doi: 10.1038/nmat1822. [DOI] [PubMed] [Google Scholar]
- 10.Wong SF, No DY, Choi YY, Kim DS, Chung BG, Lee SH. Biomaterials. 2011;32:8087. doi: 10.1016/j.biomaterials.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 11.Chen Q, Wu J, Zhuang QC, Lin XX, Zhang J, Lin JM. Sci Rep. 2013;3:2433. doi: 10.1038/srep02433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 13.Khetani SR, Bhatia SN. Nat Biotechnol. 2008;26:120. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 14.Nakao Y, Kimura H, Sakai Y, Fujii T. Biomicrofluidics. 2011;5:022212. doi: 10.1063/1.3580753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane BJ, Zinner MJ, Yarmush ML, Toner M. Anal Chem. 2006;78:4291. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- 16.Matsusaki M, Sakaue K, Kadowaki K, Akashi M. Adv Healthcare Mater. 2013;2:534. doi: 10.1002/adhm.201200299. [DOI] [PubMed] [Google Scholar]
- 17.Murphy SV, Atala A. Nat Biotechnol. 2014;32:773. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 18.Hardin JO, Ober TJ, Valentine AD, Lewis JA. Adv Mater. 2015;27:3279. doi: 10.1002/adma.201500222. [DOI] [PubMed] [Google Scholar]
- 19.Matsunaga YT, Morimoto Y, Takeuchi S. Adv Mater. 2011;23:H90. doi: 10.1002/adma.201004375. [DOI] [PubMed] [Google Scholar]
- 20.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci USA. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillotin B, Guillemot F. Trends in Biotechnol. 2011;29:183. doi: 10.1016/j.tibtech.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Cho CH, Park J, Tilles AW, Berthiaume F, Toner M, Yarmush ML. Biotechniques. 2010;48:47. doi: 10.2144/000113317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C, Chung S, Kim YE, Lee KS, Lee SH, Oh KW, Kang JY. Lab Chip. 2011;11:246. doi: 10.1039/c0lc00036a. [DOI] [PubMed] [Google Scholar]
- 24.Utech S, Prodanovic R, Mao AS, Ostafe R, Mooney DJ, Weitz DA. Adv Healthcare Mater. 2015;4:1628. doi: 10.1002/adhm.201500021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joensson HN, Svahn HA. Angew Chem Int Ed. 2012;51:12176. doi: 10.1002/anie.201200460. [DOI] [PubMed] [Google Scholar]
- 26.Headen DM, Aubry G, Lu H, García AJ. Adv Mater. 2014;26:3003. doi: 10.1002/adma.201304880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo MT, Rotem A, Heyman JA, Weitz DA. Lab Chip. 2012;12:2146. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 28.Rossow T, Heyman JA, Ehrlicher AJ, Langhoff A, Weitz DA, Haag R, Seiffert S. J Am Chem Soc. 2012;134:4983. doi: 10.1021/ja300460p. [DOI] [PubMed] [Google Scholar]
- 29.Novak R, Zeng Y, Shuga J, Venugopalan G, Fletcher DA, Smith MT, Mathies RA. Angew Chem Int Ed. 2011;50:390. doi: 10.1002/anie.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinhilber D, Rossow T, Wedepohl S, Paulus F, Seiffert S, Haag R. Angew Chem Int Ed. 2013;52:13538. doi: 10.1002/anie.201308005. [DOI] [PubMed] [Google Scholar]
- 31.Yu L, Grist SM, Nasseri SS, Cheng E, Hwang YCE, Ni C, Cheung KC. Biomicrofluidics. 2015;9:024118. doi: 10.1063/1.4918754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Augst AD, Kong HJ, Mooney DJ. Macromol Biosci. 2006;6:623. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- 33.Uygun BE, Soto-Gutierrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, Milwid J, Kobayashi N, Tilles A, Berthiaume F, Hertl M, Nahmias Y, Yarmush ML, Uygun K. Nat Med. 2010;16:814. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esch EW, Bahinski A, Huh D. Nat Rev Drug Discov. 2015;14:248. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, He Z, Liu W, Lin X, Wu J, Li H, Lin JM. Adv Healthcare Mater. 2015;4:2291–2296. doi: 10.1002/adhm.201500383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


