Abstract
Herpes simplex virus 1 (HSV-1) expresses infected cell protein 0 (ICP0), a multi-functional protein with E3 ubiquitin ligase activity and a critical regulator of the viral life cycle. To obtain novel insights into the molecular mechanism by which ICP0 regulates HSV-1 replication, we analyzed HEp-2 cells infected with HSV-1 by tandem affinity purification and mass spectrometry-based proteomics. This screen identified 50 host-cell proteins that potentially interact with ICP0, including ubiquitin-specific protease 9X (USP9X). The interaction between ICP0 and USP9X was confirmed by co-immunoprecipitation. Notably, USP9X depletion increased the ICP0 abundance and promoted viral replication. These results suggest that USP9X-dependent regulation of ICP0 expression is part of a complex feedback mechanism that facilitates optimal HSV-1 replication.
Keywords: deubiquitylating enzyme, E3 ubiquitin ligase, herpes simplex virus, ICP0, USP9X
Herpesviruses are widely distributed in nature, and most animal species are susceptible to at least one herpesvirus. The most important veterinary herpesviruses belong to the Alphaherpesvirinae subfamily. The pseudorabies virus, the causative agent of Aujeszky’s disease in pigs, is in this subfamily, as is the bovine herpesvirus 1, which causes infectious rhinotrachetis and pustular vulvovaginitis in cattle. Chickens infected with gallid herpesvirus 2 or Marek’s disease virus develop Marek’s disease, while horses infected with equine herpes virus type 1 and 4 miscarry and suffer from rhinopneumonitis, respectively [19]. On the other hand, herpes simplex virus 1 (HSV-1) is an important human pathogen that causes a variety of diseases, such as mucocutaneous diseases, keratitis, skin diseases and encephalitis [19]. HSV-1 is also one of the best-studied members of the Alphaherpesvirinae, and results from HSV-1 studies typically provide a springboard for parallel studies on veterinary alphaherpesviruses.
HSV-1 virions consist of a linear double-stranded DNA packed into an icosahedral capsid, which is encased in a tegument layer and an envelope on which surface glycoproteins are displayed [19]. The HSV-1 genome encodes at least 84 proteins that are tightly regulated and coordinated in a cascade to ensure efficient viral replication [19]. One of these proteins, infected cell protein 0 (ICP0), is a multifunctional protein with a zinc-stabilized RING finger domain. This domain is a functional E3 ubiquitin ligase [3, 10, 16]. ICP0 regulates various cellular pathways by targeting a number of host proteins for degradation, some of which are involved in cellular defenses that restrict viral infection [3]. While ICP0 targets in the host cell have become increasingly clear, the functional significance of ICP0 complexes remains to be fully characterized. To investigate novel role (s) of ICP0 in cells infected with HSV-1, we sought to identify new ICP0 binding partners by tandem affinity purification coupled to mass spectrometry. Of the putative ICP0-interacting proteins we identified, we focused on ubiquitin-specific protease 9X (USP9X), since ICP0 has been reported to interact with another ubiquitin-specific proteinase USP7 [5, 8]. In this study, we characterized the biological relevance of the ICP0-USP9X complex in HSV-1 infection.
MATERIALS AND METHODS
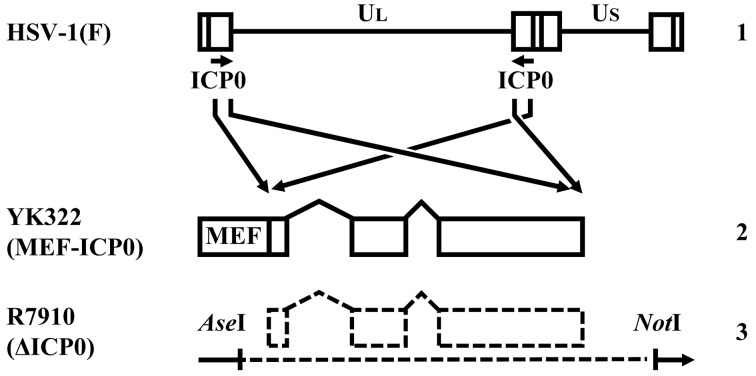
Cells and viruses: Vero, rabbit skin, Plat-GP, HEp-2 and HeLa cells were cultured as described previously [12, 20, 21]. U-2 OS cells (ATCC HTB-96) (ATCC, Manassas, VA, U.S.A.) were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum. The wild-type strain HSV-1 (F) and the recombinant strain R7910 (ΔICP0) lacking both copies of the α0 gene (Fig. 1) have been described [14, 21]. All viruses were grown in Vero cells, except in cases where R7910 (ΔICP0) was used. This recombinant virus as well as the control viruses wild-type HSV-1 (F) was propagated and titrated in U-2 OS cells [24].
Fig. 1.
The genomic structure of wild-type HSV-1 (F) and recombinant strains. Line 1, wild-type HSV-1 (F) genome; line 2, the recombinant strain YK322, in which both copies of ICP0 are fused to a fragment consisting of a Myc epitope, a tobacco etch virus protease cleavage site and a Flag epitope (MEF); line 3, the recombinant strain R7910, from which both copies of ICP0 gene have been deleted.
Plasmids: A vector encoding short hairpin RNA (shRNA) against the 3′ untranslated region of USP9X was constructed. Briefly, oligonucleotides with sequence 5′- TTTGAGAGTTTATTCACTGTCTTAGCTTCCTGTCACTAAGACAGTGAATAAACTCTCTTTTTTG-3′ and 5′- AATTCAAAAAAGAGAGTTTATTCACTGTCTTAGTGACAGGAAGCTAAGACAGTGAATAAACTCT-3′ were annealed and cloned into the BbsI and EcoRI sites in pmU6 [1]. To generate pSSCN, Hygromycin resistant gene of pSSCH [11] was substituted with Neomycin resistant gene of pMXs-IN [15]. The BamHI-EcoRI fragment of the resulting plasmid, which contains the U6 promoter and the shRNA against USP9X, was cloned into the corresponding restriction sites in pSSCN. A vector encoding an shRNA against β-galactosidase (LacZ), was constructed in a similar manner, using oligonucleotides with sequence 5′- TTTGTCGAAAACCCGAAACTGTGGGCTTCCTGTCACCCACAGTTTCGGGTTTTCGACTTTTTTG-3′ and 5′- AATTCAAAAAAGTCGAAAACCCGAAACTGTGGGTGACAGGAAGCCCACAGTTTCGGGTTTTCGA-3′.
Generation of recombinant, mutated HSV-1: The recombinant strain YK322 was engineered to fuse both copies of ICP0 to a fragment consisting of a Myc epitope, a tobacco etch virus (TEV) protease cleavage site and a Flag epitope (Fig. 1). This virus was generated by two-step Red-mediated mutagenesis in E. coli GS1783 carrying pYEbac102 [21], a full-length infectious HSV-1 (F) clone as described previously [21], except using primers 5′-CGACCCCCAGGGACCCTCCGTCCGCGACCCTCCAACCGCATACGACCCCCATGGAGCAAAAGCTCATTTC-3′ and 5′-TGGGGGCGGCCCTCAGGCCGGCGGGTACTCGCTCCGGGGCGGGGCTCCATATCTTTGTCATCGTCGTCCT-3′ (Fig. 1). Progeny viruses from the transfection were plaque-isolated and analyzed by PCR using primers 5′-TCTCCGCATCACCACAGAAG-3′ and 5′-GACCACCATGACGACGACTC-3′ to verify insertion of tags to both copies of α0. These isolates were plaque-purified on Vero cells an additional two times.
Identification of proteins that interact with ICP0: HEp-2 cells were infected with YK322 at multiplicity of infection (MOI) 5, harvested 8 hr postinfection and lysed in 0.1% NP-40 buffer (50 mM Tris-HCl pH 8.0, 120 mM NaCl, 50 mM NaF and 0.1% NP-40) containing protease and phosphatase inhibitor cocktails (Nacalai Tesque, Kyoto, Japan). After clearing by centrifugation, lysates were immunoprecipitated with a monoclonal antibody against Myc, and immunoprecipitates were digested with AcTEV protease (Invitrogen, Whaltham, MA, U.S.A.) for 1 hr at room temperature. Digests were then centrifuged, and supernatants were immunoprecipitated a second time with anti-Flag M2 Affinity Gel. The final immunoprecipitates were washed three times with 0.1% NP-40 buffer and twice with 50 mM Tris-HCl pH 8.0, 120 mM NaCl and 50 mM NaF. Bound proteins were eluted for 2 hr at 4°C with rotary shaking in 50 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.5 mg/ml Flag peptide. A tenth of the eluate was analyzed by denaturing electrophoresis and silver staining (Fig. 2), while the remaining 90% was digested with trypsin and analyzed by nano-liquid chromatography tandem mass spectrometry (nanoLC-MS/MS) as described previously [17].
Fig. 2.
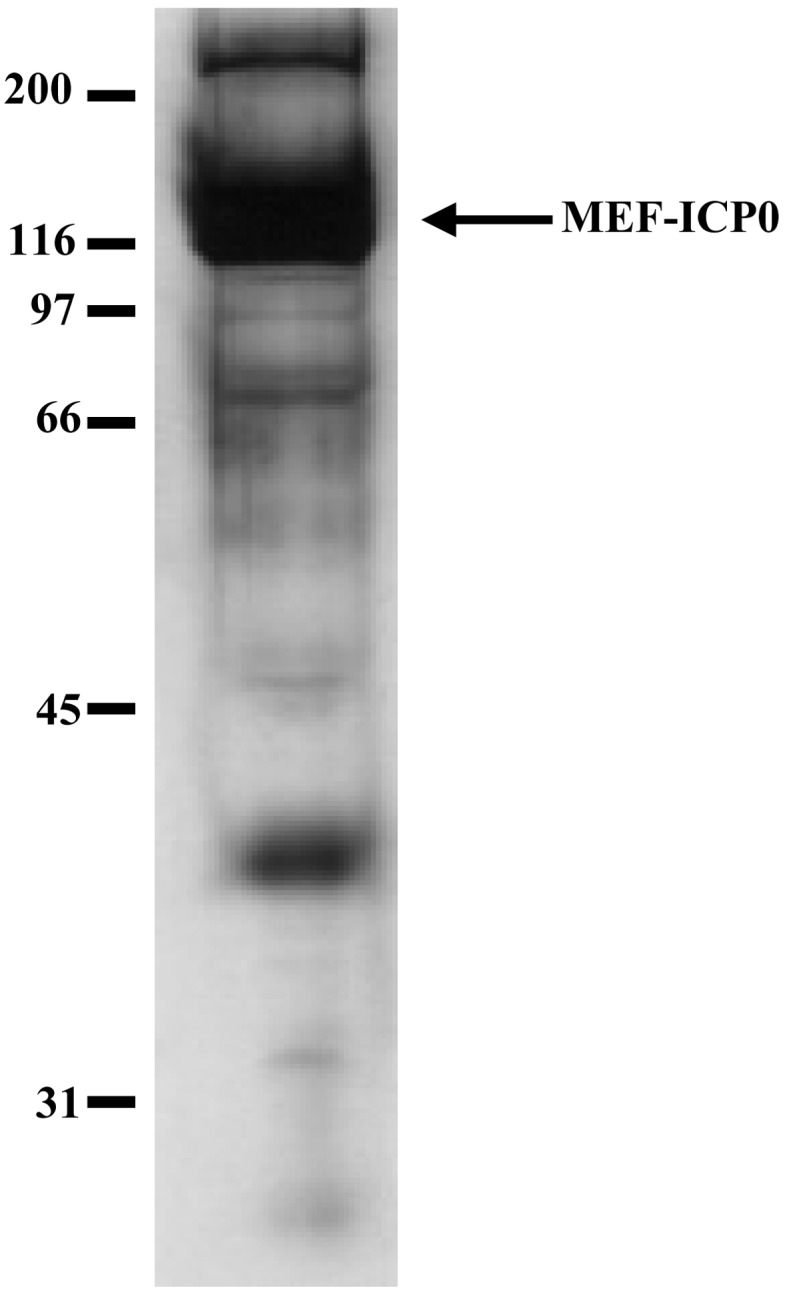
Identification of proteins that interact with ICP0. HEp-2 cells were infected with the recombinant HSV-1 strain YK322 at MOI 5, harvested 8 hr postinfection and immunoprecipitated with antibodies against Myc and with anti-Flag M2 Affinity Gel. Immunoprecipitates were separated on a denaturing gel and silver stained. The arrow marks Myc- and Flag-tagged ICP0, and molecular mass markers are indicated.
A Q-STAR Elite system (AB SCIEX, Tokyo, Japan) coupled to a Dina system (KYA Technologies, Tokyo, Japan) was used for nanoLC-MS/MS. MS/MS signals were then matched against the 32,968 protein sequences in the RefSeq human protein database (as of 12 Sep 2011; National Center for Biotechnology Information [NCBI]) and the 996,046 protein sequences in the non-redundant viral protein database (as of 23 Dec 2012; NCBI) using Mascot algorithm version 2.4.1 (Matrix Science, London, UK). Protein identification was based on at least one MS/MS signal with a Mascot score that exceeded the threshold (P<0.05).
Antibodies: Mouse monoclonal antibodies against Flag (M2), USP9X (1C4) and β-actin (AC15) were obtained from Sigma (St. Louis, MO, U.S.A.). Mouse monoclonal antibodies against Myc (PL14), ICP0 (1112) and ICP4 (58S) were procured from MBL (Nagoya, Japan), Goodwin Institute (Plantation, FL, U.S.A.) and ATCC, respectively. Rabbit polyclonal antibodies against UL12 were described previously [9].
Immunoprecipitation: Infected cells were lysed in 0.1% NP-40 buffer supplemented with protease and phosphatase inhibitor cocktails (Nacalai Tesque). Cell debris was separated by centrifugation, and lysates were precleared by incubation with protein A-Sepharose beads at 4°C for 30 min. Subsequently, lysates were reacted at 4°C for 2 hr with indicated antibodies and immunoprecipitated with Protein A-Sepharose beads for another 1 hr. Immunoprecipitates were collected by brief centrifugation, washed extensively with 0.1% NP-40 buffer and analyzed by immunoblotting as described previously [13].
Cell lines stably expressing shRNA against USP9X and LacZ: Recombinant retroviruses were generated as described previously [1]. Briefly, vectors encoding silencing shRNA against USP9X or Lac-Z were transfected into Plat-GP cells. Retroviruses released into the culture supernatant were then used to infect HeLa cells that express the corresponding silencing shRNA, and selected with 1,000 µg/ml G418 (Nacalai Tesque).
RESULTS
Identification of proteins that interact with ICP0: To screen ICP0-interacting partners more efficiently, we engineered a recombinant HSV-1 virus, YK322, in which both copies of ICP0 are fused to a TEV protease cleavage site, Myc and Flag epitopes (Fig. 1). We then purified ICP0 complexes from YK322-infected cells by tandem affinity purification. Finally, proteins co-purified with ICP0 were identified by mass spectrometry. This strategy identified 4 viral and 50 host cell proteins that appeared to form complexes with ICP0. These included proteins that were previously reported to interact with ICP0, such as UL50, UL39, ICP27, VP22, USP7, ribosomal protein S18, hyperpolarization-activated cyclic nucleotide-gated potassium channel 3 and acyl-coenzyme A thioesterase 8 [6]. Of the putative ICP0-binding proteins identified, we focused on USP9X.
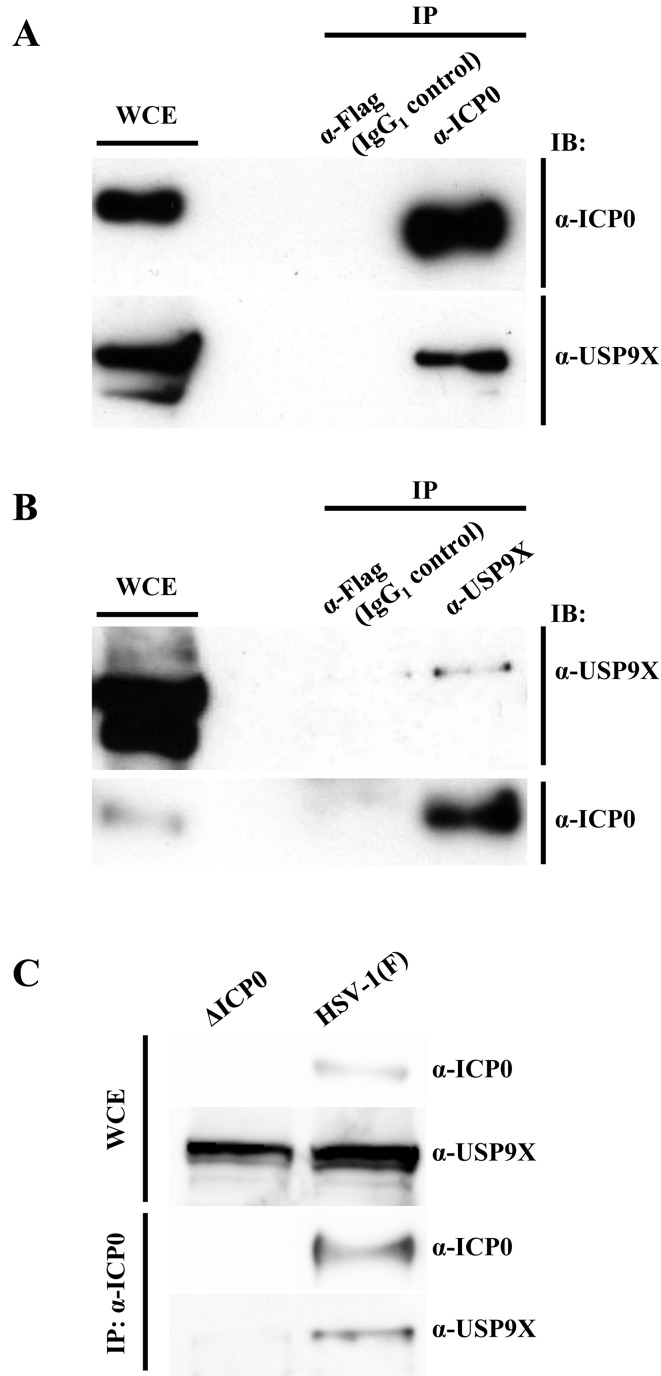
To confirm the interaction between USP9X and ICP0, we performed co-immunoprecipitation in HEp-2 cells infected with wild-type HSV-1 (F) at MOI 5. Lysates were obtained 8 hr postinfection, immunoprecipitated with anti-ICP0 or anti-Flag antibody, and immunoprecipitates were analyzed by immunoblotting with anti-USP9X and anti-ICP0 antibodies. As shown in Fig. 3A, antibodies against ICP0 precipitated both ICP0 and endogenous USP9X, but antibodies against the Flag epitope, which were used as an IgG1 control, did not. Conversely, anti-USP9X antibody precipitated both USP9X and ICP0 from Hep-2 cells infected with wild-type HSV-1 (F) (Fig. 3B). In addition, anti-ICP0 antibody co-precipitated ICP0 and endogenous USP9X from HeLa cells infected with wild-type HSV-1 (F), but not from HeLa cells infected with R7910 (ΔICP0) (Fig. 3C). Collectively, these results indicated that ICP0 interacted with USP9X in HSV-1-infected cells.
Fig. 3.
Co-immunoprecipitation of USP9X with ICP0 in infected cells. (A-B) HEp-2 cells infected for 8 hr with wild-type HSV-1 (F) at MOI 5 were harvested (WCE, whole-cell extract) and immunoprecipitated (IP) with anti-Flag, anti-ICP0 (A) or anti-USP9X (B) antibody. Immunoprecipitates were analyzed by immunoblotting (IB) with indicated antibodies. (C) HeLa cells were infected for 8 hr with wild-type HSV-1 (F) or R7910 (ΔICP0) at MOI 5, harvested and immunoprecipitated with anti-ICP0 antibody. Immunoprecipitates were analyzed by immunoblotting with indicated antibodies.
Effect of ICP0 on USP9X expression: ICP0 is a viral E3 ubiquitin ligase that drives the proteasomal degradation of several cellular targets, including USP7, a reported ICP0 binding partner [5, 8]. To examine the effect of ICP0 on USP9X abundance in HSV-1 infection, HeLa cells mock-infected or infected for 8 hr with wild-type HSV-1 (F) or R7910 (ΔICP0) at MOI 5 analyzed by immunoblotting with anti-ICP0 and anti-USP9X antibodies. As shown in Fig. 4, USP9X accumulated to similar levels in all cells. Consistent results were also obtained in Fig. 3C. Thus, the data suggested that USP9X is not a direct substrate of ICP0.
Fig. 4.
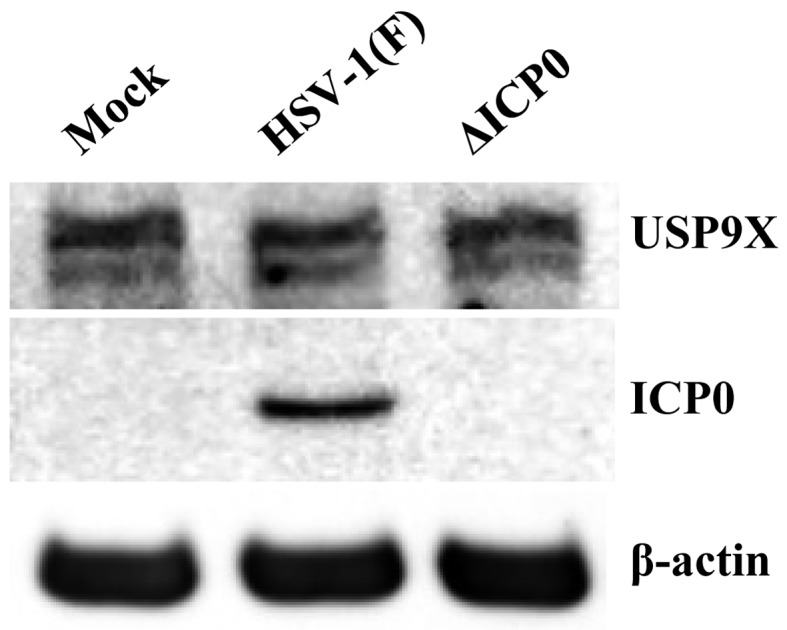
Effect of ICP0 on USP9X expression. HeLa cells infected with wild-type HSV-1 (F) or R7910 (ΔICP0) at MOI 5 were harvested after 8 hr and analyzed by immunoblotting with indicated antibodies.
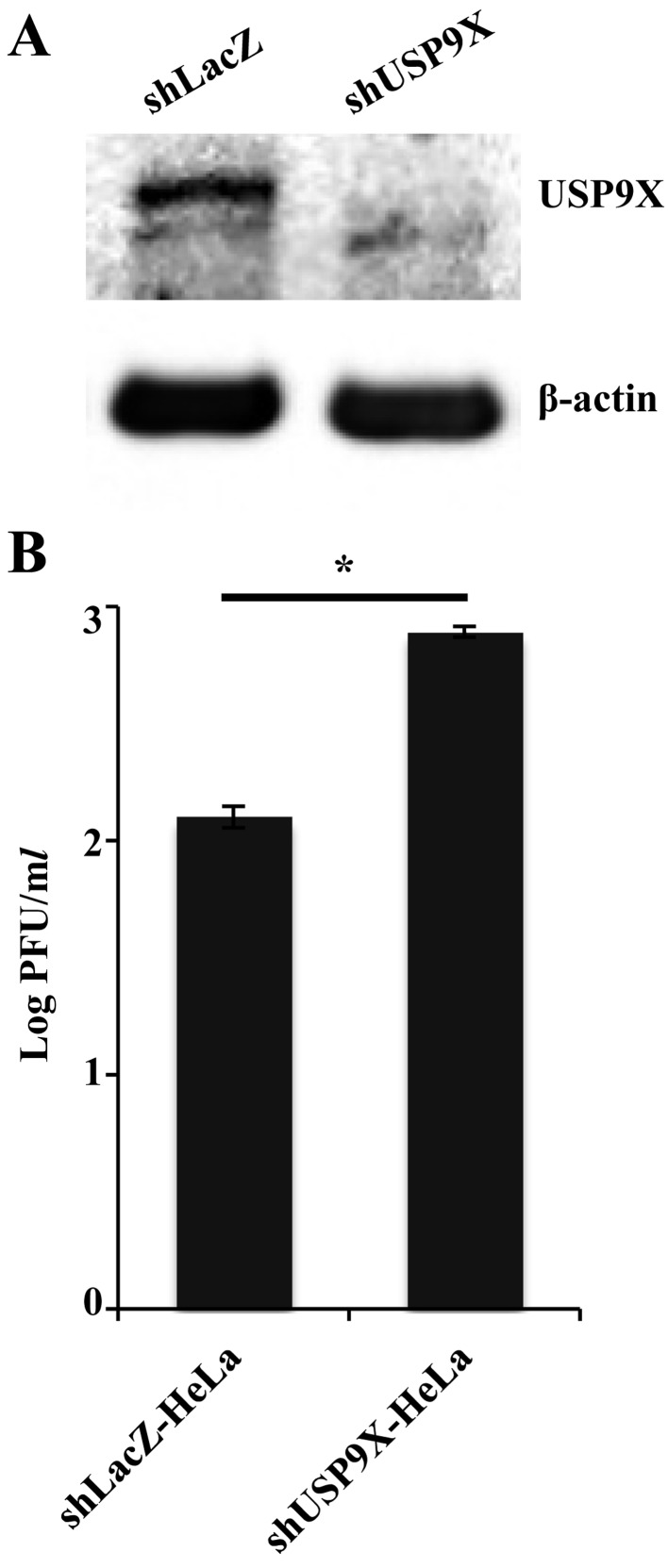
Effect of USP9X on HSV-1 replication: To investigate the role (s) of USP9X in HSV-1 replication, we generated HeLa cell lines stably expressing silencing shRNA against USP9X, as well as a control cell line expressing shRNA against LacZ. Expression of USP9X was considerably diminished in cells expressing USP9X shRNA than in cells with LacZ shRNA (Fig. 5A). These cells were also infected with wild-type HSV-1 (F) at MOI 0.01, and viral titers 48 hr postinfection were measured. As shown in Fig. 5B, the titer of progeny virus in cells expressing USP9X shRNA was significantly higher than in cells expressing LacZ shRNA. These results indicated that USP9X is required to effectively suppress HSV-1 replication.
Fig. 5.
Effect of USP9X on HSV-1 replication. (A) HeLa cells expressing silencing shRNA against LacZ and USP9X were analyzed by immunoblotting with antibodies against USP9X and β-actin. (B) These cells were infected with wild-type HSV-1 (F) at MOI 0.01. Total virus preparations from the media and infected cells were obtained 48 hr postinfection and assayed on U-2 OS cells. Data are mean ± standard error from three independent experiments. *P<0.05 by two-tailed Student’s t-test.
Effect of USP9X on expression of HSV-1 ICP0: As USP9X depletion increased HSV-1 replication, we examined the effect of USP9X depletion on expression of HSV-1 proteins. Cells expressing shRNA against LacZ or USP9X were infected with wild-type HSV-1 (F) at MOI 5 and immunoblotted 8 hr postinfection with antibodies against the viral proteins ICP4, ICP0 and UL12. As shown in Fig. 6, ICP0 was more abundant in cells with depleted USP9X than in control cells, even though expression of ICP4 and UL12 was not affected. These results suggested that USP9X downregulates expression of viral ICP0.
Fig. 6.
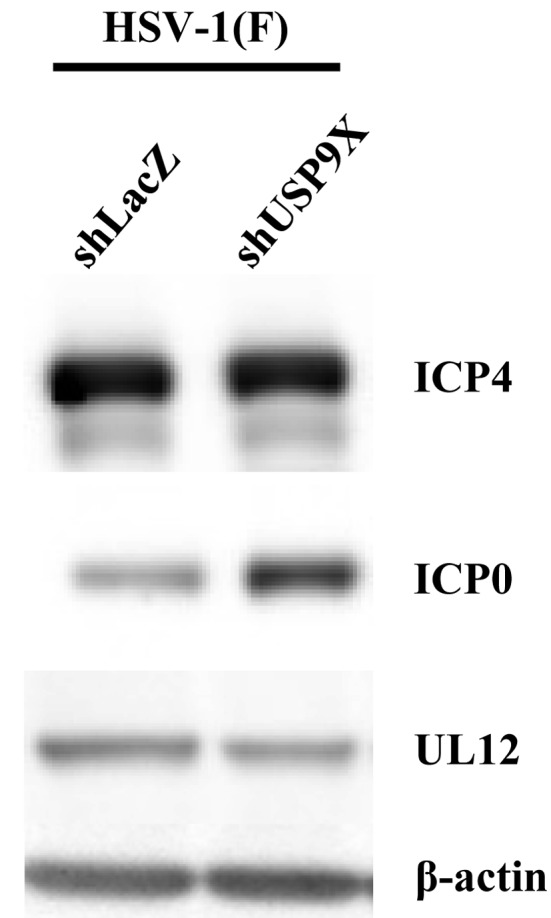
Effect of USP9X on expression of HSV-1 proteins. HeLa cells expressing silencing shRNA against LacZ and USP9X were infected for 8 hr with wild-type HSV-1 (F) at MOI 5, harvested and analyzed by immunoblotting with indicated antibodies.
DISCUSSION
We demonstrate for the first time that cellular USP9X interacts with viral ICP0. USP9X is a substrate-specific deubiquitylating enzyme that stabilizes substrates by removing mono-ubiquitin moieties and a wide array of ubiquitin chains, including K48, K63 and K29 linkages [18]. USP9X was reported to interact with at least 35 proteins, many of which are substrates [2, 7, 22, 23], and also to mediate protein trafficking and endocytosis [18]. In addition, the enzyme regulates polarity, cell death and neural development, and is implicated in neurodegenerative disease [18]. Notably, ICP0 expression was downregulated by USP9X, in contrast to other USP9X-interacting proteins. Accordingly, we found that USP9X depletion increased viral replication, in agreement with a previous report [4] demonstrating that ICP0 enhances HSV-1 replication. These observations suggested that USP9X negatively regulates ICP0 expression to inhibit viral replication, although we cannot exclude the possibility that USP9X may suppress HSV-1 replication through other pathways. However, the mechanism by which USP9X downregulates ICP0 expression is unknown at present, although it is possible that USP9X deubiquitylates and stabilizes an unknown ICP0-interacting factor that downregulates ICP0.
On the other hand, it is conceivable that HSV-1 replication is precisely regulated according to conditions, and the inhibitory effects of USP9X may be essential for optimal HSV-1 replication. In agreement with this hypothesis, accumulating evidence suggests that ICP0 degrades not only inhibitors of HSV-1 replication, such as antiviral proteins of the intrinsic and innate immunity, but also factors that enhance viral replication. For example, ICP0 was reported to interact with and degrade USP7, which is a deubiquitylating enzyme like USP9X, but one that promotes HSV-1 replication by reversing ICP0 self-ubiquitination [5, 8]. In addition, ICP0 was shown to bind and degrade TRIM27, a protein that promotes HSV-1 replication as well [6]. Thus, HSV-1 appears to have evolved multiple mechanisms to upregulate, downregulate and optimize its own replication as needed, and these mechanisms may be critical for viral infection in vivo.
Acknowledgments
We thank Tomoko Ando for excellent technical assistance. This study was supported by the Funding Program for Next Generation World-Leading Researchers, by Grants for Scientific Research from the Japan Society for the Promotion of Science (JSPS), by grants for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports and Technology (MEXT) of Japan, by a contract research fund for the Program of Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from MEXT and the Japan Agency for Medical Research and Development (AMED), and by grants from the Takeda Science Foundation and the Mitsubishi Foundation.
REFERENCES
- 1.Arii J., Goto H., Suenaga T., Oyama M., Kozuka-Hata H., Imai T., Minowa A., Akashi H., Arase H., Kawaoka Y., Kawaguchi Y.2010. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature 467: 859–862. doi: 10.1038/nature09420 [DOI] [PubMed] [Google Scholar]
- 2.Azakir B. A., Angers A.2009. Reciprocal regulation of the ubiquitin ligase itch and the epidermal growth factor receptor signaling. Cell. Signal. 21: 1326–1336. doi: 10.1016/j.cellsig.2009.03.020 [DOI] [PubMed] [Google Scholar]
- 3.Boutell C., Everett R. D.2013. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J. Gen. Virol. 94: 465–481. doi: 10.1099/vir.0.048900-0 [DOI] [PubMed] [Google Scholar]
- 4.Burris C. A., de Silva S., Narrow W. C., Casey A. E., Lotta L. T., Jr, Federoff H. J., Bowers W. J.2008. Hexamethylene bisacetamide leads to reduced helper virus-free HSV-1 amplicon expression titers via suppression of ICP0. J. Gene Med. 10: 152–164. doi: 10.1002/jgm.1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canning M., Boutell C., Parkinson J., Everett R. D.2004. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 279: 38160–38168. doi: 10.1074/jbc.M402885200 [DOI] [PubMed] [Google Scholar]
- 6.Conwell S. E., White A. E., Harper J. W., Knipe D. M.2015. Identification of TRIM27 as a novel degradation target of herpes simplex virus 1 ICP0. J. Virol. 89: 220–229. doi: 10.1128/JVI.02635-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M. J., Soligo S., Morsut L., Inui M., Moro S., Modena N., Argenton F., Newfeld S. J., Piccolo S.2009. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136: 123–135. doi: 10.1016/j.cell.2008.10.051 [DOI] [PubMed] [Google Scholar]
- 8.Everett R. D., Meredith M., Orr A.1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73: 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii H., Kato A., Mugitani M., Kashima Y., Oyama M., Kozuka-Hata H., Arii J., Kawaguchi Y.2014. The Ul12 protein of herpes simplex virus 1 is regulated by tyrosine phosphorylation. J. Virol. 88: 10624–10634. doi: 10.1128/JVI.01634-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagglund R., Roizman B.2004. Role of Icp0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78: 2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haraguchi T., Ozaki Y., Iba H.2009. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 37: e43. doi: 10.1093/nar/gkp040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato A., Arii J., Shiratori I., Akashi H., Arase H., Kawaguchi Y.2009. Herpes simplex virus 1 protein kinase Us3 phosphorylates viral envelope glycoprotein B and regulates its expression on the cell surface. J. Virol. 83: 250–261. doi: 10.1128/JVI.01451-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi Y., Van Sant C., Roizman B.1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71: 7328–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi Y., Van Sant C., Roizman B.1998. Eukaryotic elongation factor 1delta is hyperphosphorylated by the protein kinase encoded by the U(L)13 gene of herpes simplex virus 1. J. Virol. 72: 1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H.2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31: 1007–1014. doi: 10.1016/S0301-472X(03)00260-1 [DOI] [PubMed] [Google Scholar]
- 16.Lanfranca M. P., Mostafa H. H., Davido D. J.2014. HSV-1 ICP0: An E3 ubiquitin ligase that counteracts host intrinsic and innate immunity. Cells 3: 438–454. doi: 10.3390/cells3020438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maruzuru Y., Fujii H., Oyama M., Kozuka-Hata H., Kato A., Kawaguchi Y.2013. Roles of P53 in herpes simplex virus 1 replication. J. Virol. 87: 9323–9332. doi: 10.1128/JVI.01581-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murtaza M., Jolly L. A., Gecz J., Wood S. A.2015. La FAM fatale: USP9X in development and disease. Cell. Mol. Life Sci. 72: 2075–2089. doi: 10.1007/s00018-015-1851-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roizman B., Knipe D. M., Whitley R.2013. Herpes simplex viruses. pp. 1823–1897. In: Fields Virology. 6th ed., Lippincott Williams and Wilkins, Philadelphia. [Google Scholar]
- 20.Sugimoto K., Uema M., Sagara H., Tanaka M., Sata T., Hashimoto Y., Kawaguchi Y.2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82: 5198–5211. doi: 10.1128/JVI.02681-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka M., Kagawa H., Yamanashi Y., Sata T., Kawaguchi Y.2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77: 1382–1391. doi: 10.1128/JVI.77.2.1382-1391.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Théard D., Labarrade F., Partisani M., Milanini J., Sakagami H., Fon E. A., Wood S. A., Franco M., Luton F.2010. USP9x-mediated deubiquitination of EFA6 regulates de novo tight junction assembly. EMBO J. 29: 1499–1509. doi: 10.1038/emboj.2010.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vong Q. P., Cao K., Li H. Y., Iglesias P. A., Zheng Y.2005. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science 310: 1499–1504. doi: 10.1126/science.1120160 [DOI] [PubMed] [Google Scholar]
- 24.Yao F., Schaffer P. A.1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69: 6249–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]