Abstract
Background: Long non-coding RNAs (lncRNAs) have been reported to participate in various diseases. Hirschsprung disease (HSCR) is a common digestive disease in the new born. However, the relationship between lncRNAs and HSCR remains unclarified.
Methods: We used qRT-PCR to detect the relative expression of LOC101926975 in 80 pairs of HSCR bowel tissues and matched normal bowel tissues. CCK-8 assay, transwell assay and flow cytometry were then used to evaluate the function in vitro by knocking down the LOC101926975 in SK-N-BE(2) cells. Receiver operating characteristic (ROC) curve was used to evaluate the potential diagnostic value of LOC101926975.
Results: LOC101926975 was significantly downregulated in HSCR tissues with excellent correlation with FGF1. Dysregulation of LOC101926975 suppressed cell proliferation and induced G0/G1 arrest without impact on cell apoptosis or migration. Meanwhile, the AUC of LOC101926975 was 0.900 which presented great diagnostic value.
Conclusions: Our study firstly investigates the potential function of LOC101926975 in HSCR and infers that LOC101926975 can distinguish HSCR from the normal ones.
Keywords: HSCR, LncRNA, Molecular diagnosis
Introduction
Hirschsprung disease (HSCR) is recognized as a rare congenital gut disease with the incidence of 1/5000 in newborn 1, which is caused by the impaired colonization of the developing bowel by the neural crest cells (NCCs). Any factors that affect NCCs proliferation and migration may induce HSCR 2. RET and EDNRB are still the main genes verified to be related to the disease 3. However, the exact underlying mechanism needs further exploration.
Long non-coding RNAs (lncRNAs) have been verified to regulate various biological processes at transcriptional, post-transcriptional and translational levels 4-6. LncRNAs are a new class of non-coding RNAs which are generally defined as transcripts longer than 200nt in length without protein-coding capacity 7. Recent studies have revealed that HOTTIP can decrease the cell proliferation and migration in HSCR by regulating the expression of HOXA13 8. However, the role of lncRNAs in HSCR is still largely unknown.
Our previous work has demonstrated the expression profile of lncRNAs in HSCR (data not shown). One of them is LOC101926975, which is significantly differentially expressed between HSCR cases and control samples. LOC101926975 is located on chromosome 5 (142745600-142760993) with the neighbor gene named FGF1. Thus, we aimed to explore the expression pattern and function of LOC101926975 in HSCR.
Material and methods
Patients
This study was approved by the Institutional Ethics Committee of Nanjing Medical University and written informed consent was obtained from each subject. A total of 80 pairs of HSCR and matched control tissues were collected from Nanjing Children's Hospital between 2009 and 2015. The normal colon tissues were obtained from patients admitted to the hospital that were proven to be without HSCR or other enteric neural malformations. HSCR diagnosis was confirmed by pathological analysis after surgery.
Cell lines and siRNA transfection
The SK-N-BE(2) cell was obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in DMEM/F12 medium supplemented with 10% FBS (Hyclone, UT, US), 100U/ml penicillin and 100mg/ml streptomycin at 37 oC with 5% CO2. For siRNA transfection, cells were seeded in the six-wells overnight and then incubated with the specific LOC101926975 siRNA (100nM) and control siRNA (100nM) using Lipofectamine 2000 Reagent (Invitrogen, CA, USA). All the siRNAs were offered by the GenePharma (Shanghai, China). The sequence of the specific LOC101926975 siRNA was 5'-GACUGUAGUUCUGAGCUUUTT-3'. The sequence of scrambled siRNA was 5'-UUCUCCGAAGGUGUCACGUTT-3'. The processed cells were harvested for following experiments after 48h.
Flow cytometry analysis
We used flow cytometry to evaluate the cell cycle and apoptosis. Cells were collected after 48h transfection. Transfected cells were detected by BD Biosciences FACS Calibur Flow Cytometry (BD Biosciences, NJ, US). For apoptosis assay, Annexin V-FITC/Propidium Iodide Kit (KeyGen Biotech, Nanjing, China) was used to stain the harvested cells. Experiments were performed in triplicate independently.
Cell proliferation assay
The CCK-8 Cell Proliferation Kit (Beyotime, Nantong, China) was used to measure the cell viability according to the guidelines. Experiments were performed in triplicate independently.
Migration assay
The capacity of cell migration was measured using Transwell migration chambers (8 μm pore size, Millipore Corporation, Billerica, MA). The single-cell suspension of 1 x 105 transfected cells in 100 µl of serum-free medium was added to the upper chamber. The bottom well contained 600ul DMEM/F12 medium with 10% FBS. After incubation for 24 h, the cells were fixed with methanol, stained with crystal violet staining solution (Beyotime, Nantong, China). The number of invasive tumor cells was counted using Image-pro Plus 6.0. Experiments were performed in triplicate independently.
RNA extraction and qRT-PCR
Total RNAs were isolated from HSCR and healthy bowel tissues using Trizol reagent (Life Technologies, CA, US) according to the manufacture's instructions. The qRT-PCR was performed with the SYBR (Takara, Tokyo, Japan) by the ABI7900HT. GAPDH was used as internal control. The relative expression of RNA was calculated by the 2-△CT method. The primer sequences were listed as follows: GAPDH: 5'-GTCAACGGATTTGGTCTGTATT-3' (forward), 5'-AGTCTTCTGGGTGGCAGTGAT-3' (reverse); FGF1: 5'-CTGAGTGTGGGAGTGCAGAG-3' (forward), 5'-GACCCCAAAGCCTCTGCTTA-3' (reverse); LOC101926975: 5'-AACCCAGTGTTCAAAACCCCA-3' (forward), 5'-GCAGGGGAAAATACCAGGGAA-3' (reverse).
Data analysis
Date analysis were performed by using SPSS 17.0 software (SPSS, Chicago, IL) and presented by Graphpad software (GraphPad Software, Inc., CA, US). Data of the relative expression level of RNA in human tissue samples were presented as a box plot of the median and range of log-transformed expression level accessed by Wilcoxon rank-sum test. The data for the experiments in vitro that were repeated three times, were plotted as mean ± SEM via double-sided Student's t-test. Receiver operating characteristic (ROC) curve was used to evaluate the diagnostic value. p < 0.05 was considered statistically significant.
Results
LOC101926975 is down-regulated in HSCR
A total of 160 colon tissues containing 80 HSCR cases and 80 matched controls were collected in this study. There is no statistically difference between two groups in ages, sex and body weight as shown in Table 1.
Table 1.
Clinical features of study population
| Variable | Control(n=80) | HSCR(n=80) | P |
|---|---|---|---|
| Age(days,mean,SE) | 128.70(7.04) | 117.10(6.32) | 0.21* |
| Weight(kg,mean,SE) | 5.59(0.14) | 5.29(0.12) | 0.12* |
| Sex(%) | |||
| Male | 49(61.25) | 60(75.00) | 0.06^ |
| Female | 31(38.75) | 20(25.00) |
*Student's t-test
^Two-sided chi-squared test
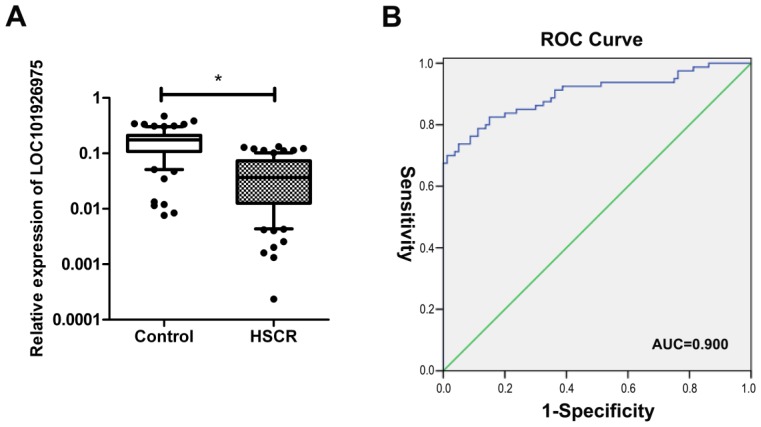
As shown in Fig 1A, the expression of LOC101926975 was significantly reduced in HSCR compared with the corresponding control tissues. Numerous studies have shown lncRNAs also can act as biomarkers of diseases. Thus, we used ROC curve to assess the capacity of LOC101926975 distinguishing HSCR from normal tissues (Fig 1B). The area under the ROC curve was 0.900 with the cut off value of 0.1162 and 0.1288. The result shows that LOC101926975 has the potential diagnostic value.
Figure 1.
Expression of LOC101926975 in HSCR. A. LOC101926975 was significantly downregulated in HSCR tissues compared control samples. B. Receiver Operating Characteristic (ROC) curve for the LOC101926975 to distinguish HSCR cases from controls. * indicates significant difference (p<0.05)
LOC101926975 knockdown inhibits cell proliferation and causes G1 arrest
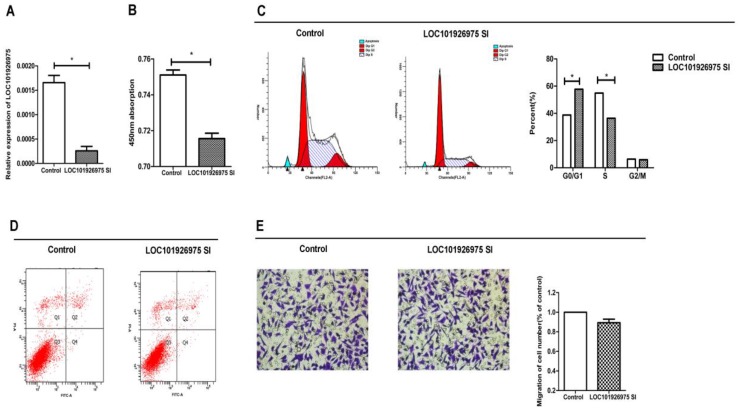
To investigate the function of LOC101926975 in vitro, we used short interfering RNAs (siRNAs) to reduce the expression of LOC101926975 in SK-N-BE(2) cells. The siRNA could effectively reduce the LOC101926975 expression level (Fig 2A). The phenotype changes induced by LOC101926975 knockdown indicated that the low expression of LOC101926975 significantly suppressed the cell proliferation compared with the control cells (Fig 2B). Meanwhile, flow cytometry analysis revealed that LOC101926975 downregulation blocked the G0/G1 to S phase transition (Fig 2C). However, no influence was found on cell migration and apoptosis with the siRNA treatment (Fig 2D, E).
Figure 2.
Function of LOC101926975 in vitro. A. LOC101926975 was effectively knocked down in SK-N-BE(2) cells. Downregulation of LOC101926975 suppressed cell proliferation (B) and caused cell cycle arrest (C) without impact on cell apoptosis (D) or cell migration. Pictures were captured under a light microscope with the magnification, x20 (E). * indicates significant difference (p<0.05)
LOC101926975 may regulate the expression of FGF1
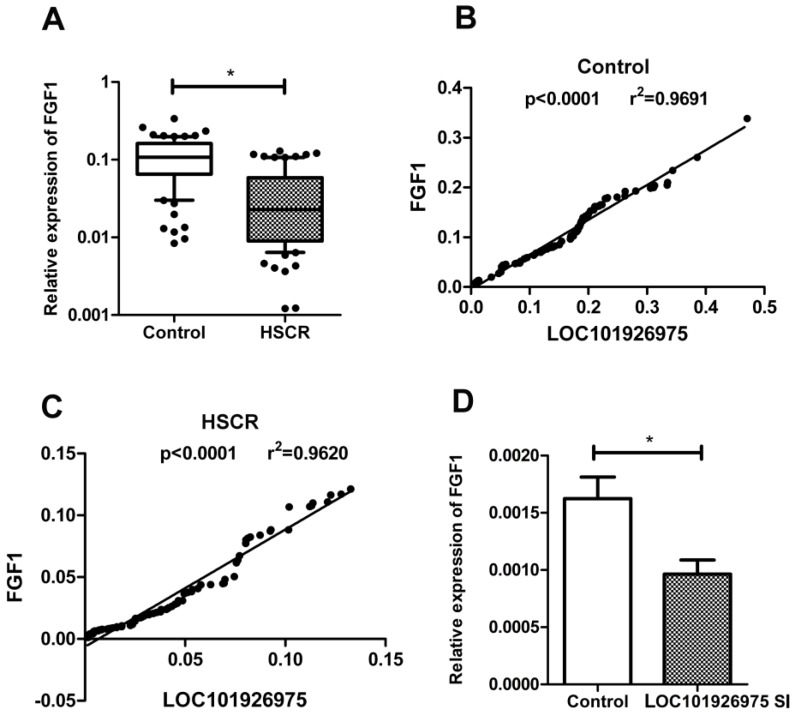
To explore the potential mechanism of LOC101926975 regulating biological process, we focused on FGF1 due to its near location on chromosome. FGF1 is a member of the fibroblast growth factor family, which plays key roles in cell proliferation and embryonic development 9. We found that the expression of FGF1 was also low in HSCR cases (Fig 3A). The correlation analysis showed that the association between FGF1 and LOC101926975 was evident in both controls and cases with the r value of 0.9844 and 0.9804 respectively and p value <0.0001 (Fig 3B, C). And the expression of FGF1 in LOC101926975 knockdown cells was lower than the control according to the results of qRT-PCR (Fig 3D). All above, hinted that LOC101926975 might regulate the expression of FGF1 and thus participated in HSCR.
Figure 3.
Relationship between FGF1 and LOC101926975. The expression of FGF1 was lower in HSCR tissues (A) and was correlated with the expression of LOC101926975 in control samples (B), HSCR tissues (C) and cells (D). * indicates significant difference (p<0.05)
Discussion
HSCR characterized by the absence of enteric neurons in the distal gut is one of the most common digestive diseases in the newborn. The main clinical symptoms are abdominal distension and constipation. Untreated HSCR is a fatal disease especially with enterocolitis 10. Roughly estimated initial costs for neonatal with HSCR is $100,000 in the United States 11. However, we still cannot interpret clearly the genetic factors or environmental underpinnings of HSCR less to say clinical application of replacing enteric nervous system 12. Thus, it is important to explore the pathogenesis of HSCR.
LncRNAs have been demonstrated to play key roles in numerous biological processes and diseases 13-15. In this study, we investigate the functional performance of LOC101926975 in HSCR. LOC101926975 is significantly downregulated in HSCR tissues with FGF1 which is near to this lncRNA on chromosome. Results in vitro show that LOC101926975 impacts cell proliferation and cell cycle without influencing cell migration or apoptosis. FGF1 is a well characterized member of fibroblast growth factor family. Dysregulation of FGF1 is involved in cell proliferation, migration, cell arrest and apoptosis by interacting with FGF receptors 16-19. LncRNAs have been reported to affect the expression of neighboring genes positively or negatively namely cis regulation5. And several known lncRNAs such as Xist and Air can regulate nearby or and distantly located genes by interacting with histone modification complexes 20, 21. Therefore, we wonder that if LOC101926975 can regulate FGF1 expression. And we find that the expression of FGF1 is correlated with LOC101926975 in both cells and population samples. However, there is no difference in migration or apoptosis when FGF1 is knocked down in our study. It seems inconsistent with previous studies, which hints LOC101926975 may also affect other genes in addition to FGF1. And further validation is needed to confirm the specific regulation mechanism of LOC101926975 for FGF1.
LncRNAs also can act as biomarkers in numerous diseases especially cancers22. For instance, RP11-160H22.5, XLOC_014172 and LOC149086 are related to hepatocellular carcinoma 23. Traditional biomarkers are mostly blood-based, which may influence the stability and sensitivities of results 24. In this study, we attempt to evaluate the diagnostic value of LOC101926975 in tissue samples. And LOC101926975 is significantly downregulated in HSCR tissues with the AUC of 0.900, which implies that LOC101926975 can effectively distinguish HSCR cases from control samples.
In conclusion, we demonstrate that LOC101926975 expression is downregulated in HSCR tissues. Dysregulation of LOC101926975 can impact cell proliferation as well as cell cycle and serve as biomarker for HSCR. However, further study is still needed to confirm the result and explain the molecular mechanisms.
Acknowledgments
We thank Dr. Jie Zhang, HuanChen and Changgui Lu (Nanjing Children's Hospital Affiliated to Nanjing Medical University) for sample collection. This study was supported by Natural Science Foundation of China (NSFC 81370473), Natural Science Foundation of China (NSFC 81400574), Natural Science Foundation of China (NSFC 81570467), Natural Science Foundation of Jiangsu Province of China (BK20131388), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). Competing Interests: the authors have no competing interests.
Abbreviations
- HSCR
Hirschsprung disease
- LncRNA
Long non-coding RNA
- ROC
Receiver operating characteristic
- NCCs
neural crest cells.
References
- 1.Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S. et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- 2.Lake JI, Heuckeroth RO. Enteric nervous system development: migration, differentiation, and disease. Am J Physiol Gastrointest Liver Physiol. 2013;305:G1–24. doi: 10.1152/ajpgi.00452.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace AS, Anderson RB. Genetic interactions and modifier genes in Hirschsprung's disease. World J Gastroenterol. 2011;17:4937–44. doi: 10.3748/wjg.v17.i45.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–40. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Xie H, Zhu D, Xu C, Zhu H, Chen P, Li H. et al. Long none coding RNA HOTTIP/HOXA13 act as synergistic role by decreasing cell migration and proliferation in Hirschsprung disease. Biochem Biophys Res Commun. 2015;463:569–74. doi: 10.1016/j.bbrc.2015.05.096. [DOI] [PubMed] [Google Scholar]
- 9.Raju R, Palapetta SM, Sandhya VK, Sahu A, Alipoor A, Balakrishnan L. et al. A Network Map of FGF-1/FGFR Signaling System. J Signal Transduct. 2014;2014:962962. doi: 10.1155/2014/962962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Momoh JT. Hirschsprung's disease: problems of diagnosis and treatment. Ann Trop Paediatr. 1982;2:31–5. doi: 10.1080/02724936.1982.11748220. [DOI] [PubMed] [Google Scholar]
- 11.Shinall MC Jr, Koehler E, Shyr Y, Lovvorn HN 3rd. Comparing cost and complications of primary and staged surgical repair of neonatally diagnosed Hirschsprung's disease. J Pediatr Surg. 2008;43:2220–5. doi: 10.1016/j.jpedsurg.2008.08.048. [DOI] [PubMed] [Google Scholar]
- 12.Hotta R, Natarajan D, Thapar N. Potential of cell therapy to treat pediatric motility disorders. Semin Pediatr Surg. 2009;18:263–73. doi: 10.1053/j.sempedsurg.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Ellis BC, Molloy PL, Graham LD. CRNDE: A Long Non-Coding RNA Involved in CanceR, Neurobiology, and DEvelopment. Front Genet. 2012;3:270. doi: 10.3389/fgene.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J. et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53:1005–19. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang M, Zhai X, Xia B, Wang Y, Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol; 2015. [DOI] [PubMed] [Google Scholar]
- 16.Hossain WA, Morest DK. Fibroblast growth factors (FGF-1, FGF-2) promote migration and neurite growth of mouse cochlear ganglion cells in vitro: immunohistochemistry and antibody perturbation. J Neurosci Res. 2000;62:40–55. doi: 10.1002/1097-4547(20001001)62:1<40::AID-JNR5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Murphy M, Drago J, Bartlett PF. Fibroblast growth factor stimulates the proliferation and differentiation of neural precursor cells in vitro. J Neurosci Res. 1990;25:463–75. doi: 10.1002/jnr.490250404. [DOI] [PubMed] [Google Scholar]
- 18.Cassina P, Pehar M, Vargas MR, Castellanos R, Barbeito AG, Estevez AG. et al. Astrocyte activation by fibroblast growth factor-1 and motor neuron apoptosis: implications for amyotrophic lateral sclerosis. J Neurochem. 2005;93:38–46. doi: 10.1111/j.1471-4159.2004.02984.x. [DOI] [PubMed] [Google Scholar]
- 19.Browaeys-Poly E, Perdereau D, Lescuyer A, Burnol AF, Cailliau K. Akt interaction with PLC(gamma) regulates the G(2)/M transition triggered by FGF receptors from MDA-MB-231 breast cancer cells. Anticancer Res. 2009;29:4965–9. [PubMed] [Google Scholar]
- 20.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K. et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R. et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Shao Y, Zhu M, Li Q, Yang F, Lu X, Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol; 2015. [DOI] [PubMed] [Google Scholar]
- 23.Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6:4505–15. doi: 10.18632/oncotarget.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu HS, Xiao HS. MicroRNAs as potential biomarkers for gastric cancer. World J Gastroenterol. 2014;20:12007–17. doi: 10.3748/wjg.v20.i34.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]