Abstract
Epidermal growth factor receptor (EGFR) has been used as the target in drug design for cancer treatment including the liver cancer. Men and women have different levels of EGFR expression during the life. The whole genome expression profiles of livers of recombinant inbred (RI) strains derived from C57BL/6J (B6) X DBA/2J (D2) were used to compare three major molecular aspects of Egfr gene: the relative expression levels, gene network and eQTLs that regulate the expression of Egfr between female and male mice. Our data suggest that there is a significant difference in the expression levels in the liver between female and male mice. Several important genes in the gene network of Egfr are differentially expressed between female and male mice. The regulatory elements for the expression levels of Egfr between female and male mice are also different. In summary, our data reveals an important sex difference in the Egfr pathways in the liver of the mice. These data may have substantial impact on drug development and dosage determinant for women and men in the clinical trials.
Keywords: Cancer, Egfr, liver, Mice, Pathway, Sex.
Introduction
Epidermal growth factor receptor (Egfr) is involved in tissue development and in a variety of cellular functions, such as cell proliferation, differentiation, motility, and survival 1. Most importantly, it has a significant role in cancer development 2. It is highly expressed in liver and also regulated by estrogen 3. Therefore, it is important to understand its molecular pathways in liver and similarities and differences between men and women.
There are considerable reports on the role of Egfr in the cancer development. Recently, Lanaya et al. 4 reported that the presence of Egfr-positive liver macrophages in Hepatocellular carcinoma patients is associated with poor survival. Accompanied is the fact that men and women have different levels of Egfr during life 5-6. Mouse models have also suggest that accelerates hepatocellular carcinoma progression in a sex-dependent manner in a mouse model of hepatocarcinogenesis 7-8. These data indicated that gender difference may affect the cancer development, cancer treatment, and cancer survival.
Several drugs for the treatment of hepatocarcinomas have been developed by targeting the Egfr. For example, erlotinib, an epidermal growth factor receptor and tyrosine kinase inhibitor, is a targeted drug that was approved for the treatment of hepatocellular carcinomas 9. Lapatinib is an inhibitor of both epidermal growth factor receptor and HER2/NEU, whereas both of which are implicated in hepatocarcinogenesis 10. While the drugs based on anti Egfr are in development and in clinical trials, its gender specificity should have been thoroughly investigated. In order to obtain meaningful data, the study of sex-specific molecular pathways needs samples of populations of both sexes of the same age, genomic background, and environment. It is extremely difficult to collect such a population from humans. Therefore, utilizations of animal models have been as the alternative approach for the study of the gender difference of humans 11.
The purpose of this work is to systematically investigate the sex differences of Egfr using high quality data from the liver of a mouse population of recombinant inbred (RI) strains derived from C57BL/6J (B6) X DBA/2J (D2) 12-13.
Materials and Methods
Data sets for analysis of gene expression profiles
UNC Agilent G4121A Liver Database (Jul04) was used for this analysis (http://www.genenetwork.org/webqtl/main.py). The data set includes gene expression in livers of naive mice of both sexes from C57BL/6J, DBA/2J, B6D2F1, and 37 BXD strains using Agilent G4121A 12-13 . The data sets at GeneNetwork are publically available and they were validated using sex specific probe sets such as Xist and Dby. In general, sex-balanced samples (one array from male cases, one array from female cases) from RI strains derived from C57BL/6J (B6) X DBA/2J (D2) and parental strains were used. While they are collected at the similar age, performed with the same microarray platform, and grown in the same environment, most of them are from different RI strains. However, these RI strains are all derived from the same two progenitors. Also, the gene expression profiles of both female and male of two progenitors are generated separately. Additional details regarding animals, microarrays, data acquisition, processing, and analyses can be found at WebQTL.org (http://webqtl.org/dbdoc/LV_G_0106_B.html).
Collections of data on expression levels of Egfr and associated genes
For the expression data of Egfr and its associated genes we collected the expression data of Egfr axis from two sets of data, the male and the female sets. We used the Actin beta as controls for the Egfr. When multiple probes were presented for the gene, the probe with the highest expression level was chosen for the analysis while the others were used as reference.
The association of the expression levels of Egfr between female and male mouse populations
We conducted the association analysis using the method reported previously 14. R values were compared between the association of female and male mice. We followed the standard criteria for the strong, correlation, and none correlation. When R value is equal or more than 0.7 or -0.7, we regard the correlation as strong positive or negative. When R value is between 0.35 and 0.69 or -0.35 and -0.69, the correlation exists but not strong. Any R values between 0 and 0.35 or 0 and -0.35 will be treated as none correlation 14. The gene networks were constructed using tools in the GeneNetwork. We constructed the gene network based on the Network Graph. Network graph is often used to visualize multiple sets of interactions. It consists of nodes and edge connecting nodes. In addition, the network graph can represent the strength of the interaction by the thickness of the edge—that is, the higher the number of interactions between the two nodes, the thicker the edge becomes.
Mapping transcriptomic loci (eQTL) that regulates the expression level of Egfr in female and male mice
Transcriptome mapping with GeneNetwork was conducted to identify the chromosomal regions that affect the expression of Egfr in female and male mice. After we obtained the transcriptome maps, we compared them to see whether there is a difference between maps of female and male mice and among different tissues. We focused on the loci that have LRS score equal to or higher than the significant levels. If we did not find the loci at the significant levels, we then looked at the loci at the suggestive levels 14-15.
For the loci showing sex difference, we examined the candidate genes that regulate the Egfr expression level. We focused on the genes under the peak region of the eQTL, which usually is the chromosome region with LRS scores at the significant or suggestive levels 14.
After we identified the genes in the chromosome region under the peak region of eQTL, we searched the literature report to see whether any of these genes have been reported to regulate or functionally associate with Egfr. We used the PGMapper to conduct such a search 16. It is a software tool for automatically matching defined functional terms to genes from a defined genome region by combining the mapping information from the Ensembl database and gene function information from the OMIM and PubMed databases.
Additional Statistical Analysis
The top 50 associated genes in each dataset on the basis of Pearson correlation were used for plotting Network Graphs in GeneNetwork. T-test was performed to test the levels of gene expression levels between the male and female mice. P value of 0.05 was regarded as the largest value of indication of significant difference between the two populations.
Results
Expression levels of Egfr between female and male in the Liver
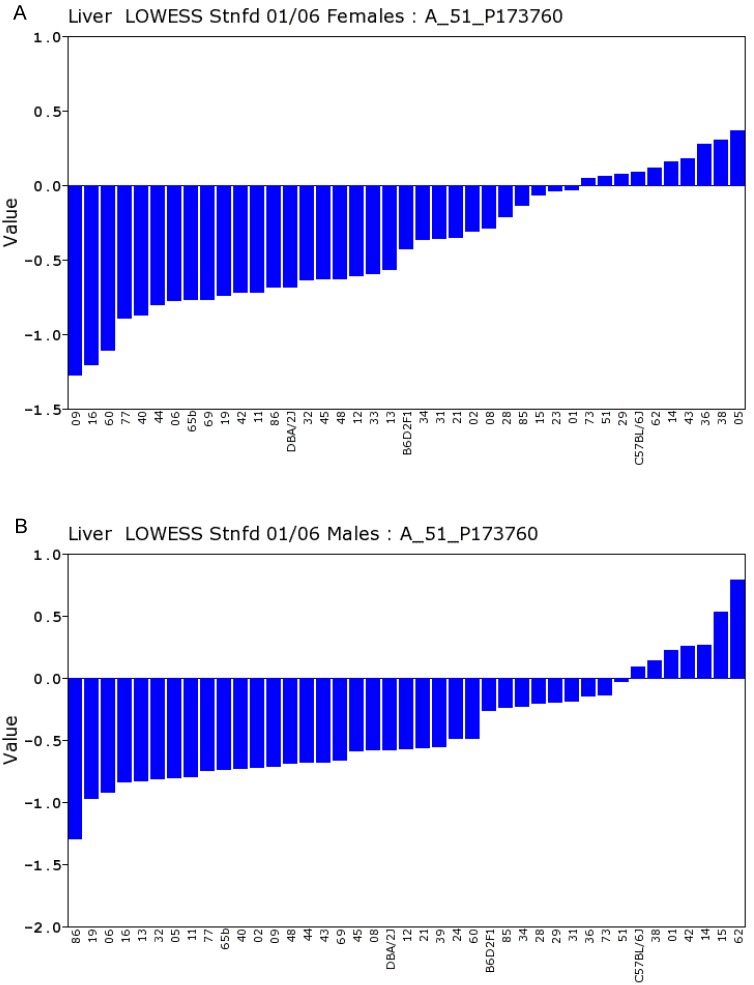
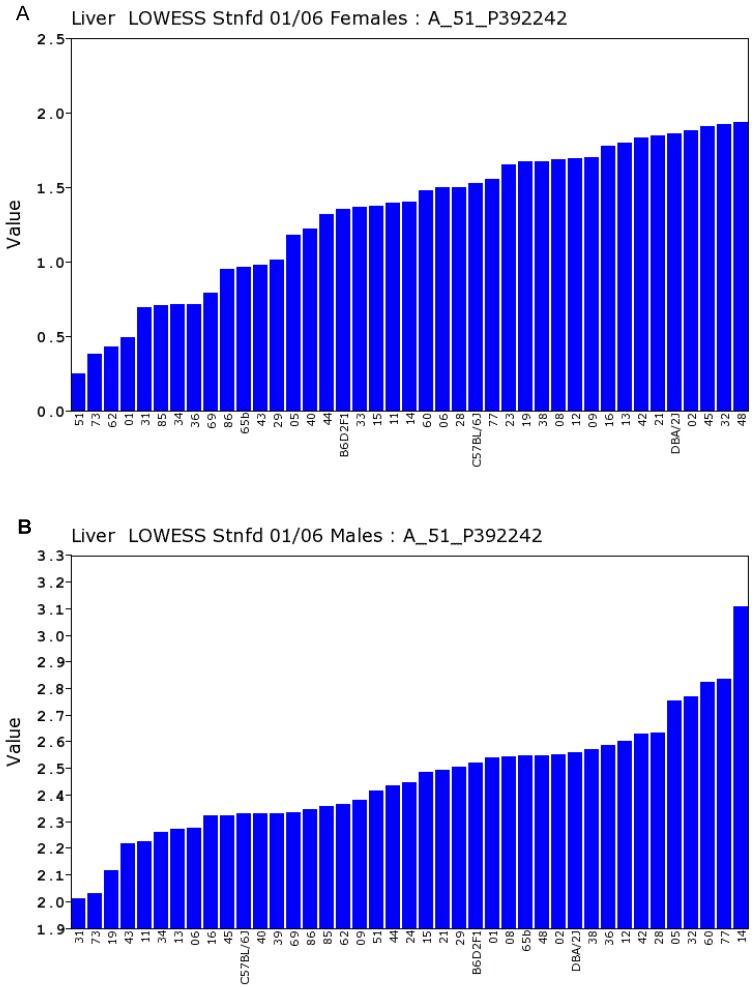
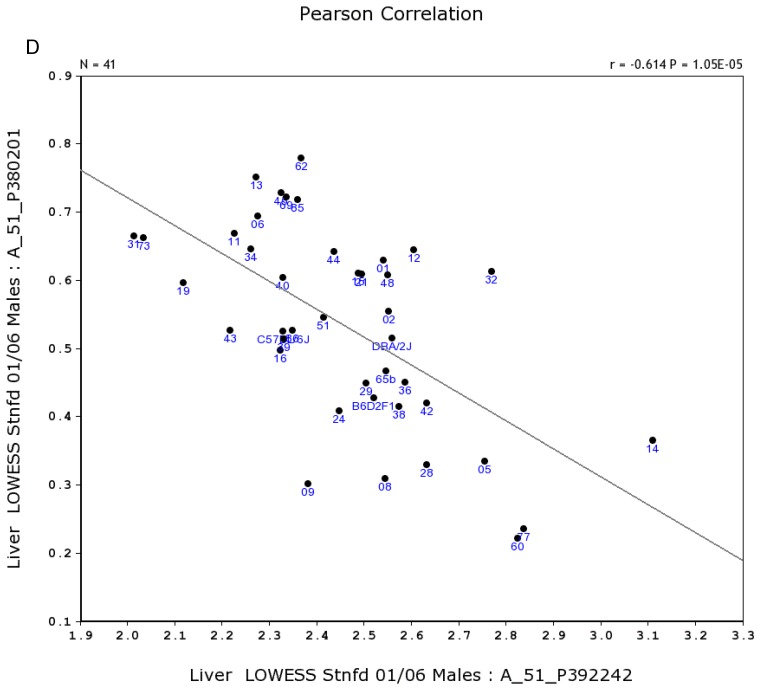
We conducted statistical analysis using data of gene expression of livers from a total of 39 strains, which have both the data from Actin β (beta) and Egfr in both sexes. There is one probe for Actin β on the Agilent G4121A chip. Figure 1A and 1B showed the expression levels of Actin β in female and male mice of different strains. For Actin B, the P value for T test between female and male is 0.88. The R value from the correlation analysis is 0.60. These data indicated that there is a high correlation in the expression levels of Actin β between male and female mice. Similarly, we obtained one probe for Egfr on the UNC Agilent G4121A chip. Figure 2A and 2B showed the expression levels of Egfr in the female and male mice of different strains. We also conducted the same statistical analyses on the levels of Egfr expression between the male and female mice. For Egfr, the P value for T test between the female and male is 1.07488E-17. The R value from the correlation analysis is 0.31. Therefore, there seems a difference in the expression levels between the male and female mice. Thus, there is no difference in the expression level of Actin β between the female and male populations while there is a significant sex difference in the Egfr expression levels. Therefore, further analyses were conducted as shown in the next several sections.
Figure 1.
Expression levels of Actin β in the liver between the female and male mice in the BXD strains. Numbers on Y-axis are for the relative levels of the expression of Actin β. The information of strains and names of standard inbred strains are listed under the X-axis. A. The expression levels of Actin β in the liver of the female mice. B. The expression levels of Actin β in the liver of the male mice.
Figure 2.
Expression levels of Egfr in the liver between the female and male mice in the BXD strains. Numbers on Y-axis are for the relative levels of the expression of Egfr. The information of strains and names of standard inbred strains are listed under the X-axis. A. The expression levels of Egfr in the liver of the female mice. B. The expression levels of Egfr in the liver of the male mice.
Gene network of Egfr between female and male in moue liver
Using the data on the expression level of probe named A_51_P392242, the probe for Egfr and whole genome expression profiles in the livers of female RI strains, we identified the top 50 probes of genes with their expression levels mostly correlated to that of Egfr (Supplementary Table S1).
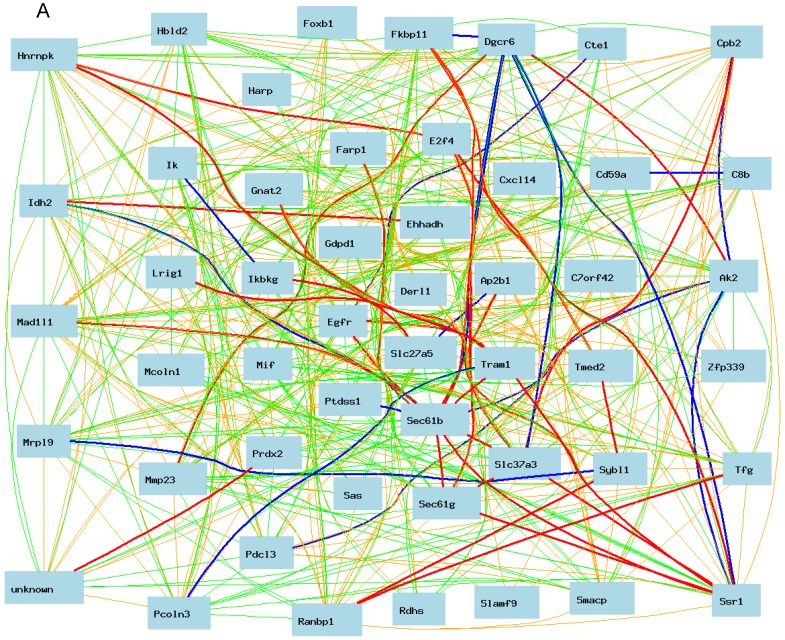
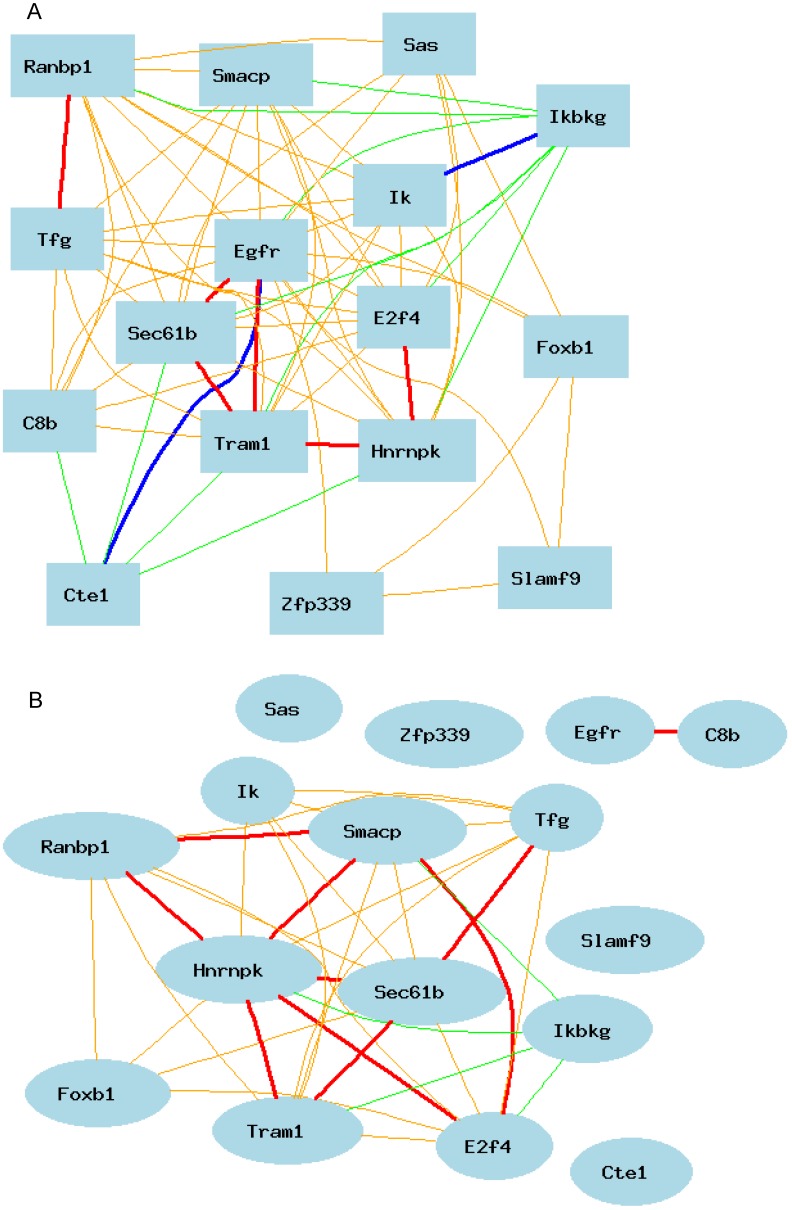
As shown in Figure 3A, Egfr is positively and negatively correlated to several genes in the female mice. The expression of mouse Egfr is strongly positively correlated to translocating chain-associating membrane protein 1 (Tram1), RIKEN cDNA 1190006C12 gene (for Sec61b); Both Tram1 and Sec61b then have strong connections to multiple genes. Egfr is strongly negatively correlated to dedicator of cytosolic acyl-CoA thioesterase 1 (Cte1). Cte1 has negative impact on several other genes.
Figure 3.
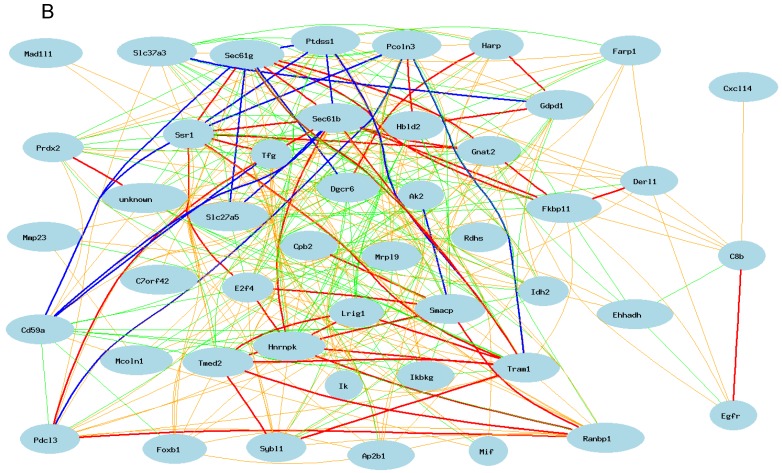
Gene network of top 50 genes that are closely associated to Egfr in the male mouse liver and their network in the female liver. The 50 nodes in the graph below show the selected traits. Only nodes with the edges are displayed. The graph's canvas is 40.0 by 40.0 cm; the node labels are drawn with a 18.0 point font, and the edge labels are drawn with a 18.0 point font. A. Gene network in the female liver. The 684 edges between the nodes, filtered from the 1225 total edges and drawn as curves, show Pearson correlation coefficients greater than 0.5 or less than -0.5. B. Gen network in the liver of the male mice. The 366 edges between the nodes, filtered from the 1225 total edges and drawn as curves, show Pearson correlation coefficients greater than 0.5 or less than -0.5.
We then constructed a gene network in the liver of the male mice using the same probes from female gene expression profiles of liver. As shown in Figure 3B, Egfr has even a stronger connection to C8b in female than that in male mice. It has no significant influence on the expression levels of Tram1, Cte1, and Sec61b in female. In addition, in male mice, Egfr has strong positive influence on the expression levels of E2F transcription factor 4 (E2f4); Forkhead box B1 (Foxb1); heterogeneous nuclear ribonucleoprotein K (Hnrnpk); IK cytokine (Ik); RAN binding protein 1 (Ranbp1); RIKEN cDNA 2700085A14 gene (Sas); CD2 antigen family, member 10 (Slamf9); small acidic protein (Smacp); Trk-fused gene (Tfg); and zinc finger protein 339 (Zfp339). In female there is no strong association between Egfr and these genes.
The expression levels of key genes correlate with Egfr differently between female and male in the moue liver
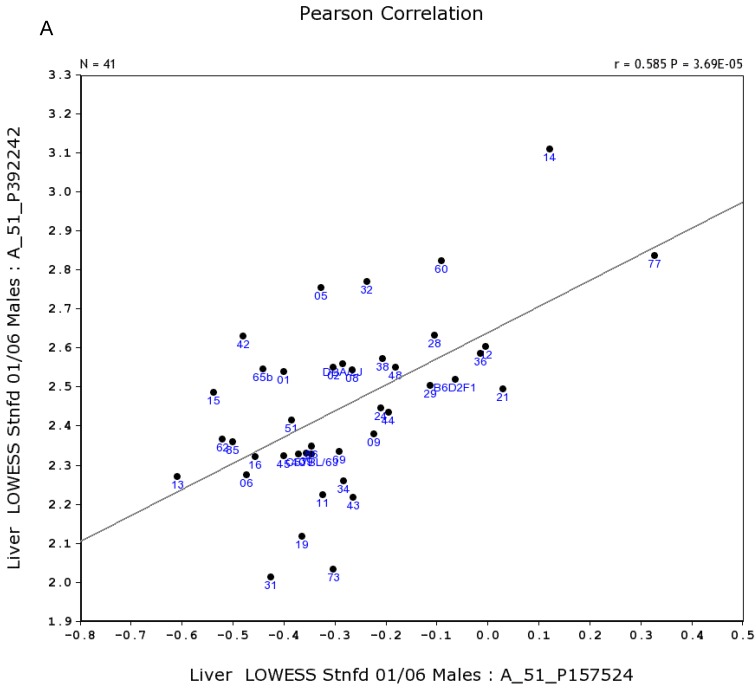
Based on these differences, we constructed second level of gene network between male and female mice. Figure 4A shows the positive and negative connections of these 15 genes to Egfr. The expression level of Egfr is connected to the expression level of each of these genes in the male liver. In female, as shown in Figure 4B, the expression of Egfr is not correlated with the expression level of any except the C8b.
Figure 4.
Genes positively and negatively connected to the expression of Egfr in the mouse liver. The 16 nodes in the graph below show the selected traits. All the nodes are displayed. The graph's canvas is 40.0 by 40.0 cm; the node labels are drawn with a 16.0 point font, and the edge labels are drawn with a 16.0 point font. A. Genes positively and negatively connected to the expression of Egfr in the male liver. The 69 edges between the nodes, filtered from the 120 total edges and drawn as curves, show Pearson correlation coefficients greater than 0.5 or less than -0.5. B. Gene network based on the expression level in the female liver for genes that are positively and negatively connected to the expression of Egfr. The 35 edges between the nodes, filtered from the 120 total edges and drawn as curves, show Pearson correlation coefficients greater than 0.5 or less than -0.5.
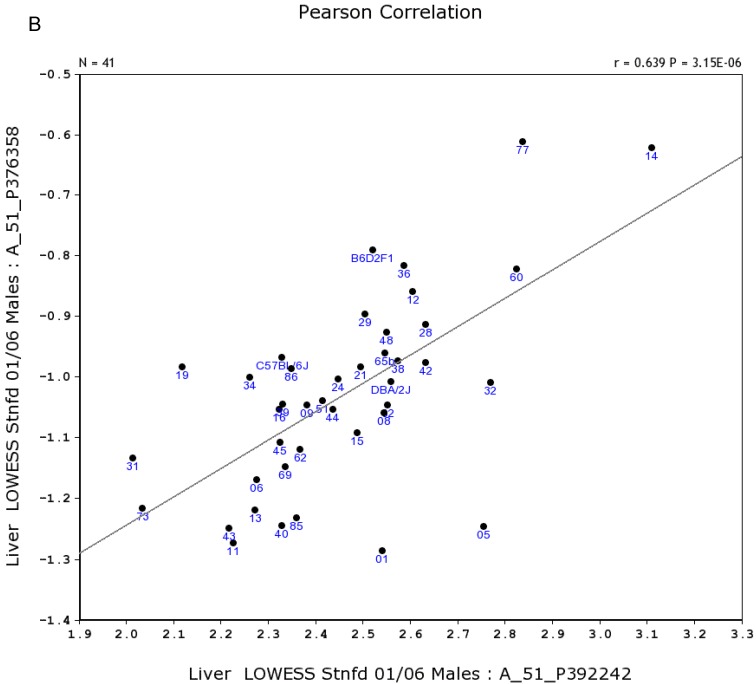
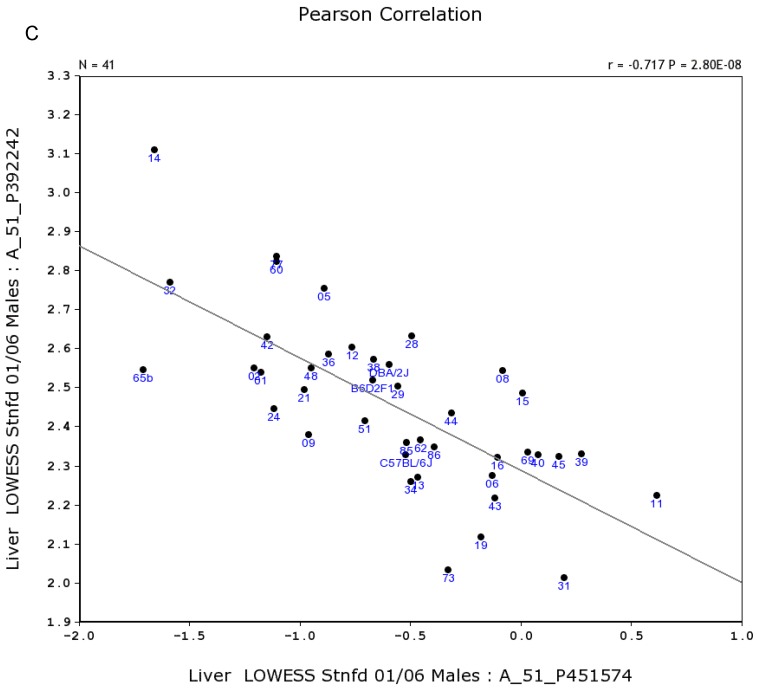
We further examined the correlation between the expression level of Egfr and several genes in the male and female mice. Specifically, in the male, the expression levels of Egfr is strongly positively correlated to the expression levels of E2f4 (Figure 5A) and Hnrnpk (Figure 5B) among the RI strains, while in the female there is no such a strong connection. In the male, the expression level of Egfr is strongly negatively connected to the expression of Cte1 (Figure 5C) and inhibitor of kappaB kinase gamma (Ikbkg) (Figure 5D) while in the female there is no such a connection. These data confirm a significant difference in the Egfr pathways between male and female mice.
Figure 5.
Correlation between the expression levels of Egfr and four key genes in the male mice. A. Egfr is strongly positively correlated to the expression of E2f4. B. Egfr is strongly positively correlated to the expression of Hnrnpk. C. Egfr is strongly negatively correlated to the expression of Cte1. D. Egfr is strongly negatively correlated to the expression of Ikbkg.
Gene regulation of the expression of Egfr in the female and male liver
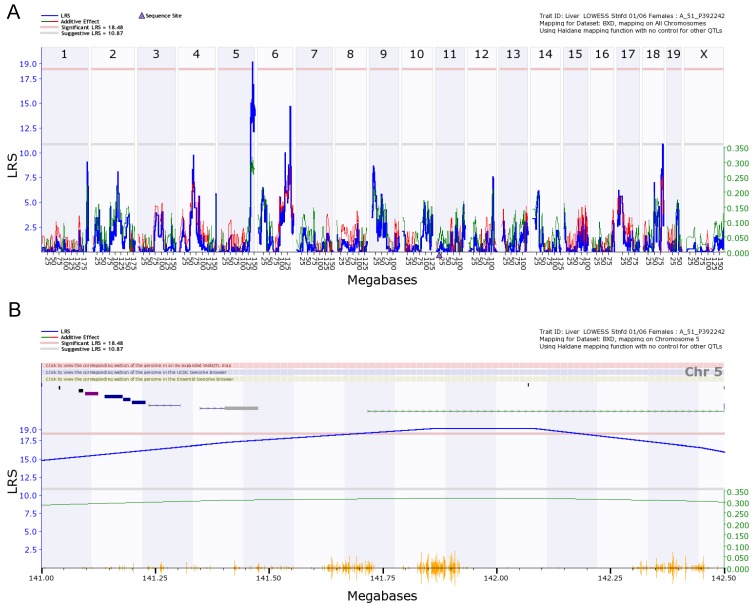
We finally examined the genetic factors that regulate the expression of Egfr in the livers of female and male mice. With 5000 permutation test, we detected eQTLs for the regulation of Egfr from female liver on chromosome 5 and 6 (Figure 6A). The eQTL on chromosome 5 is significant. The significant threshold is 18.48, and the suggestive threshold is 10.87. The eQTL on chromosome 5 reached the level of 19. We did not detect an eQTL on either chromosome 5 or 6 from the data of male liver. The eQTL for male mice are on chromosome 7 and 8, but all suggestive. Thus, the level of Egfr expression is differently regulated between male and female mice.
Figure 6.
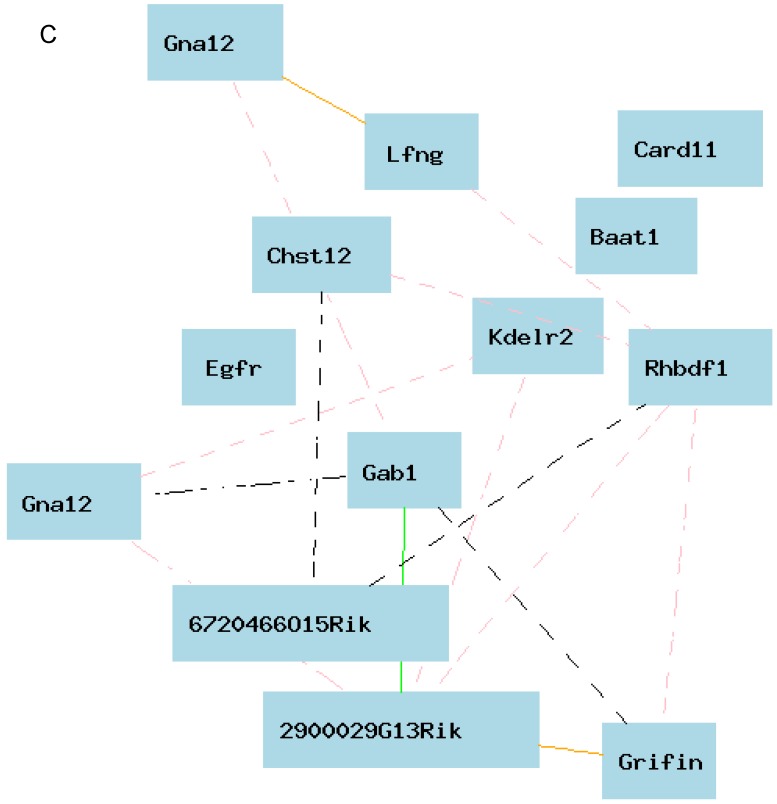
eQTL map of Egfr based on the whole genome expression profiles of the liver of the female population. A. EQTL map of whole genome showing the loci on chromosome 5, 6, and 18. B. The peak region of eQTL on chromosome 5. C. Gene network of 12 candidate genes of eQTL on chromosome 5 and Egfr derived from the liver of the female mice. The 13 nodes in the graph below show the selected traits. All the nodes are displayed. The 15 edges between the nodes, filtered from the 78 total edges and drawn as curves, show Pearson correlation coefficients greater than 0.3 or less than -0.3. The graph's canvas is 40.0 by 40.0 cm; the node labels are drawn with a 16.0 point font, and the edge labels are drawn with a 16.0 point font.
Figure 6B shows the peak region of the eQTL on chromosome 5. The region between 141.00 and 142.50 contains 12 genetic elements, including 10 known genes and two probes for other genetic components (Table 1). In order to explore the possibility of any of these 12 genes as the causal gene, we investigated two aspects of these genes. We first examined the correlations between the expression levels of these genes and that of Egfr. Gene network indicated that there is no connections between the expression level in any of these 12 genes and that of Egfr (Figure 6C). If we concentrated on the very top of peak region of the eQTL on chromosome 5, which is located between 141.5 and 142.5 Mbp (Figure 5B), we obtained only 3 genetic elements as candidate genes. These candidates are sidekick homolog 1 (Sdk1), RIKEN cDNA 3200001G23 gene (3200001G23Rik), and KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 (Kdelr2).
Table 1.
Information of candidate genes for eQTL of expression level of Egfr in male mice.
| Symbol | Mb Start | Length (Kb) | SNP (count) | SNP Density | Avg.Expr | Gene Description |
|---|---|---|---|---|---|---|
| Chst12 | 140.9816 | 19.63 | 2 | 0.101885 | 1.287 | Carbohydrate sulfotransferase 12 |
| Grifin | 141.0391 | 1.879 | 0 | 0 | 0.681 | Galectin-related inter-fiber proteinprovided |
| Lfng | 141.0833 | 8.205 | 0 | 0 | 0.077 | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| Ttyh3 | 141.0966 | 28.385 | 1 | 0.03523 | - | Tweety homolog 3 (Drosophi... |
| Iqce | 141.1395 | 38.874 | 7 | 0.180069 | -- | IQ motif containing E |
| AA881470 | 141.181 | 14.31 | 2 | 0.139762 | -- | EST AA881470 |
| Amz1 | 141.2001 | 29.186 | 4 | 0.137052 | -- | Archaelysin family metallopeptidase 1 |
| Gna12 | 141.2359 | 70.488 | 21 | 0.297923 | 0.691 | Guanine nucleotide binding protein, alpha 12 |
| Card11 | 141.349 | 127.598 | 24 | 0.188091 | -- | Caspase recruitment domain family, member 11 |
| Sdk1 | 141.7175 | 972.258 | 605 | 0.622263 | -- | Sidekick homolog 1 (chicken)... |
| 3200001G23Rik | 142.0698 | 1.197 | 0 | 0 | -- | RIKEN cDNA 3200001G23 gene |
| Kdelr2 | 142.5 | 0.05 | 0 | 0 | 1.070 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
We next examined the polymorphisms of these three candidate genes between strains of B6 and D2 based on the Jackson laboratory database of Mouse Genome Informatics (http://www.informatics.jax.org/). We searched all the polymorphisms within each gene and 2 kb upstream and downstream of the gene. There are 23 polymorphic sites in Kdelr2 but none of them are different between D2 and B6. There are three polymorphisms in 3200001G23Rik (transcript for unknown gene) between D2 and B6. They are all located in the intron regions. There are 309 polymorphisms in Sdk1, all of which are SNPs. Most of these SNPs are within the intron while 4 of these are in the coding region. Three of these are synonymous while one is nonsynonymous polymorphism. The ID of the one nonsynonymous polymorphism is rs29564987. It is C/T missense polymorphism, where D2 codes for a “C” and B6 codes for a “T”. Accordingly, the Sdk1 is the most favorite candidate gene for the regulation of expression of Egfr in the liver of the mouse.
Discussion
Our data showed that there are significant differences in the molecular pathways of Egfr in the liver between female and male mice. The data agree with the fact that the Egfr is regulated by estrogen 3, 17. Our data further indicated that the sex differences exist not only in the molecular pathways in which Egfr regulates and interacts with other genes but also in the regulation mechanisms of Egfr by other genes. On the other hand, our eQTL analysis did not map the loci for Egfr onto the position where the estrogen locates. Future studies may reveal whether this is because that in the liver the estrogen does not play a major role for the sex differential expression. The significant difference in the molecular pathways of Egfr between the female and male mice in these strains has laid a foundation for this important study.
Most of the sex differentially regulated genes in this study have not been investigated for liver cancer. However, a notable feature of these genes is their critical roles in the pathways of gender specific cancers. For example, E2f4 have been reported to regulate liver cancer cell survival and risk for breast cancer 18-20; Hnrnpk has been linked to the neuroendocrine differentiation signaling in prostate cancer cells 21 as well as to the development of breast cancer 22. In brain cancer the expression of Hnrnpk has been linked to the Egfr 23. The increased transcription level of Ranbp1 has been linked to the decreases taxol sensitivity in RKO colon carcinoma cells 24. Remarkably, Ikbkg has been linked to several cancers and its cytokine network has been the target for drug development for breast cancer 25. Furthermore, Ikbkg and Egfr have been reported to upregulate PKM2 expression and promote tumorigenesis 26. Understanding the gender differences of their functions in the cancer genetics and responses to drug treatment will greatly enhance the future drug development and therapeutic application.
Remarkably, the regulation of the Egfr expression level between female and male mice showed much difference. In the male, there was no significant loci for the regulation of the expression level of Egfr, while in the female, there were at least two loci being identified. According to our data, Sdk1 is the most favorite candidate for the regulation of expression level of Egfr in the female mice. Although some researches on the molecular pathways of Sdk1 have been done, it has not been directly linked to cancer. It is possible that it may play a secondary role in the cancer development by regulation of expression level of Egfr differentially between female and male. On the other hand, the data may indicate that there is a complicated regulation of Egfr expression between the female and the male, in some cases, there may be different in binding sites in the regulation of Egfr expression. Although the differential expression levels of Egfr between the female and the male mice apparently is not regulated by estrogen, the difference in the gene network of Egfr between the female and the male are mainly caused by genes in the estrogen pathway. For example, E2f4 and Hnrnpk are well known by their interactions with estrogen. In addition, Lasarte et al. reported that estradiol downregulates NF-κb translocation by Ikbkg transcriptional repression in dendritic cells 27. Furthermore, the relation between estrogen and Sdk1 has not been studied. There is a possibility that Sdk1 has a role in the estrogen pathway. Nevertheless, further study is necessary before any conclusion is reached.
In humans, although there is no intensive investigation on the gender difference of anti EGFR therapy for the liver cancer, there are few reports showing the potential gender difference. For example, there is apparently a gender difference in hazard ratio (HR) in patients in a phase III, randomized, double-blind, placebo-controlled trial of sorafenib plus erlotinib 9 in patients with advanced hepatocellular carcinoma 28. While the HR of overall survival (OS) in the female patients is 1.135 (0.708-1.821), the male patients is 0.926 (0.762-1.126). Brivanib is an ATP-competitive inhibitor against VEGFR2, moderate potency against VEGFR-1 and FGFR-1. In a study using brivanib as adjuvant therapy to transarterial chemoembolization in the patients with hepatocellular carcinoma, Kaplan-Meier's curves for OS in the female patients is 1.71 (0.78-3.74), but in the Male patients is 0.79 (0.56-1.11) 29. The evidences and the data from animal studies again emphasize the importance of gender difference not only in disease incidence but also in response to drug treatment and overall survival in the liver cancer patients.
One limitation of this study is that our data is at one time point. Future studies at different age stages may lead to different results and comprehensive understanding of the sex difference in the molecular pathways of Egfr axis. In addition, our study is based on the RI strains that were derived from two mouse strains. Mouse strain specific pathways have been known 30-31. Nevertheless, the two parental strains, C57BL/6J and DBA/2J, are the strains that have been widely used in the biomedical research. We feel confident on the reliability and accuracy of the data. The raw data in this study were from multiple homozygous RI strains 12-13. Gene expression profiles from each strain were generated from multiple mice. Mice of both sexes were from the same strain with the same genomic background except that they differed by sex. All the animals were kept in the same animal facility with the same environment. The none-difference of the expression levels of Actin B between female and male mice in these strains demonstrated the reliability of the data.
We are aware that notable differences between the mouse models and humans have been found 32. The differentially expressed Egfr between the female and male mice may not be differentially expressed between men and women in humans. However, our data will serve as a reminder that it is important to keep in mind on the gender difference in the drug development targeting EGFR and its family members.
Supplementary materials
Supplementary Tables.
Acknowledgments
This work was partially supported by the Center of Genomics and Bioinformatics and Center of Connective Tissue Research at the University of Tennessee Health Science Center; The collaborative project between the first hospital of Qiqihare city and the University of Tennessee health Science Center.
Author contributions
WG and HC designed the experiments. LW, JX and WG analyzed the data, constructed the gene networks, and mapped the eQTLs. HC and WG conceived and managed the project. LW, JX and WG contributed to the manuscript preparation. All authors contribute to the manuscript finalization.
References
- 1.Voldborg BR, Damstrup L, Spang-Thomsen M, Poulsen HS. Epidermal growth factor receptor (Egfr) and Egfr mutations, function and possible role in clinical trials. Ann Oncol. 1997;8(12):1197–206. doi: 10.1023/a:1008209720526. [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y. The Egfr family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(Suppl 4):S3–8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 3.Filardo EJ. Epidermal growth factor receptor (Egfr ) transactivation by estrogen via the G-protein-coupled receptor, GPR30: a novel signaling pathway with potential significance for breast cancer. J Steroid Biochem Mol Biol. 2002;80(2):231–8. doi: 10.1016/s0960-0760(01)00190-x. [DOI] [PubMed] [Google Scholar]
- 4.Lanaya H, Natarajan A, Komposch K, Li L, Amberg N, Chen L, Wculek SK, Hammer M, Zenz R, Peck-Radosavljevic M, Sieghart W, Trauner M, Wang H, Sibilia M. Egfr has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat Cell Biol. 2014;16(10):972–81. doi: 10.1038/ncb3031. 1-7. doi: 10.1038/ncb3031. Epub 2014 Aug 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malamitsi-Puchner A, Tziotis J, Tsonou A, Protonotariou E, Sarandakou A, Creatsas G. Changes in serum levels of vascular endothelial growth factor in males and females throughout life. J Soc Gynecol Investig. 2000;7:309–312. [PubMed] [Google Scholar]
- 6.Baserga M, Bares AL, Hale MA, Callaway CW, McKnight RA, Lane PH, Lane RH. Uteroplacental insufficiency affects kidney VEGF expression in a model of IUGR with compensatory glomerular hypertrophy and hypertension. Early Hum Dev. 2009;85:361–367. doi: 10.1016/j.earlhumdev.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandon-Warner E, Walling TL, Schrum LW. et al. Chronic ethanol feeding accelerates hepatocellular carcinoma progression in a sex-dependent manner in a mouse model of hepatocarcinogenesis. Alcohol Clin Exp Res. 2012;36(4):641–653. doi: 10.1111/j.1530-0277.2011.01660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JK, Abe K, Bridges AS, Patel NJ. et al. Sex-dependent disposition of acetaminophen sulfate and glucuronide in the in situ perfused mouse liver. Drug Metab Dispos. 2009;37(9):1916–1921. doi: 10.1124/dmd.109.026815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu CH, Kang YK, Yang TS, Shun CT, Shao YY, Su WC, Sandoval-Tan J, Chiou TJ, Jin K, Hsu C, Cheng AL. Bevacizumab with erlotinib as first-line therapy in Asian patients with advanced hepatocellular carcinoma: a multicenter phase II study. Oncology. 2013;85(1):44–52. doi: 10.1159/000350841. [DOI] [PubMed] [Google Scholar]
- 10.Bekaii-Saab T, Markowitz J, Prescott N, Sadee W, Heerema N, Wei L, Dai Z, Papp A, Campbell A, Culler K, Balint C, O'Neil B, Lee RM, Zalupski M, Dancey J, Chen H, Grever M, Eng C, Villalona-Calero M. A multi-institutional phase II study of the efficacy and tolerability of lapatinib in patients with advanced hepatocellular carcinomas. Clin Cancer Res. 2009;15(18):5895–5901. doi: 10.1158/1078-0432.CCR-09-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiao Y, Chen H, Yan J, Wang L, Huang Y, Liu X, Williams RW, Lu L, Wang Y, Gu W. Genome-wide gene expression profiles in antioxidant pathways and their potential sex differences and connections to vitamin C in mice. Int J Mol Sci. 2013;14(5):10042–10062. doi: 10.3390/ijms140510042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatti DM, Zhao N, Chesler EJ, Bradford BU, Shabalin AA, Yordanova R, Lu L, Rusyn I. Sex-specific gene expression in the BXD mouse liver. Physiol Genomics. 2010;42(3):456–468. doi: 10.1152/physiolgenomics.00110.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gatti D, Maki A, Chesler EJ, Kirova R, Kosyk O, Lu L, Manly KF, Williams RW, Perkins A, Langston MA, Threadgill DW, Rusyn I. Genome-level analysis of genetic regulation of liver gene expression networks. Hepatology. 2007;46(2):548–57. doi: 10.1002/hep.21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Liu H, Jiao Y, Wang E, Clark SH, Postlethwaite AE, Gu W, Chen H. Differences between Mice and Humans in Regulation and the Molecular Network of Collagen, Type III, Alpha-1 at the Gene Expression Level: Obstacles that Translational Research Must Overcome. Int J Mol Sci. 2015;16(7):15031–56. doi: 10.3390/ijms160715031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Jiao Y, Sun S, Jarrett HW, Sun D, Gu W. Gene network of a phosphoglycerate mutase in muscle wasting in mice. Cell Biol Int. 2015;39(6):666–77. doi: 10.1002/cbin.10437. [DOI] [PubMed] [Google Scholar]
- 16.Xiong Q, Qiu Y, Gu W. PGMapper: a web-based tool linking phenotype to genes. Bioinformatics. 2008;24(7):1011–3. doi: 10.1093/bioinformatics/btn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aizen J, Thomas P. Role of Pgrmc1 in estrogen maintenance of meiotic arrest in zebrafish oocytes through Gper/Egfr. J Endocrinol. 2015;225(1):59–68. doi: 10.1530/JOE-14-0576. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez JL, Sandoval J, Serviddio G, Sastre J, Morante M, Perrelli MG, Martínez-Chantar ML, Viña J, Viña JR, Mato JM, Avila MA, Franco L, López-Rodas G, Torres L. Id2 leaves the chromatin of the E2F4-p130-controlled c-myc promoter during hepatocyte priming for liver regeneration. Biochem J. 2006;398(3):431–7. doi: 10.1042/BJ20060380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrer-Vicens I, Riffo-Campos ÁL, Zaragozá R, García C, López-Rodas G, Viña JR, Torres L, García-Trevijano ER. In vivo genome-wide binding of Id2 to E2F4 target genes as part of a reversible program in mice liver. Cell Mol Life Sci. 2014;71(18):3583–97. doi: 10.1007/s00018-014-1588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González-Hernández A, Henríquez-Hernández LA, Cabrera de León A, Rodríguez-Pérez Mdel C, Murias-Rosales A, Domínguez-Coello S, Brito-Díaz B, Almeida-González D, Aguirre-Jaime A, Díaz-Chico BN. Microsatellite polymorphisms in the EGFR, NOTCH4 and E2F4 genes and their association with breast cancer risk. Int J Biol Markers. 2012;27(3):e219–26. doi: 10.5301/JBM.2012.9583. doi: 10.5301/JBM.2012.9583. [DOI] [PubMed] [Google Scholar]
- 21.Ciarlo M, Benelli R, Barbieri O, Minghelli S, Barboro P, Balbi C, Ferrari N. Regulation of neuroendocrine differentiation by AKT/hnRNPK/AR/β-catenin signaling in prostate cancer cells. Int J Cancer. 2012;131(3):582–90. doi: 10.1002/ijc.26402. [DOI] [PubMed] [Google Scholar]
- 22.Hamrita B, Chahed K, Kabbage M, Guillier CL, Trimeche M, Chaïeb A, Chouchane L. Identification of tumor antigens that elicit a humoral immune response in breast cancer patients' sera by serological proteome analysis (SERPA) Clin Chim Acta. 2008;393(2):95–102. doi: 10.1016/j.cca.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B, Zhu S, Gu Y, Villa GR, Akhavan D, Nathanson D, Gini B, Mareninov S, Li R, Camacho CE, Kurdistani SK, Eskin A, Nelson SF, Yong WH, Cavenee WK, Cloughesy TF, Christofk HR, Black DL, Mischel PS. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17(6):1000–8. doi: 10.1016/j.cmet.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amato R, Scumaci D, D'Antona L, Iuliano R, Menniti M, Di Sanzo M, Faniello MC, Colao E, Malatesta P, Zingone A, Agosti V, Costanzo FS, Mileo AM, Paggi MG, Lang F, Cuda G, Lavia P, Perrotti N. Sgk1 enhances RANBP1 transcript levels and decreases taxol sensitivity in RKO colon carcinoma cells. Oncogene. 2013;32(38):4572–8. doi: 10.1038/onc.2012.470. [DOI] [PubMed] [Google Scholar]
- 25.Barbie TU, Alexe G, Aref AR, Li S, Zhu Z, Zhang X, Imamura Y, Thai TC, Huang Y, Bowden M, Herndon J, Cohoon TJ, Fleming T, Tamayo P, Mesirov JP, Ogino S, Wong KK, Ellis MJ, Hahn WC, Barbie DA, Gillanders WE. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J Clin Invest. 2014;124(12):5411–23. doi: 10.1172/JCI75661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang L, You MJ, Koh MY, Cote G, Aldape K, Li Y, Verma IM, Chiao PJ, Lu Z. EGFR-induced and PKCε monoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol Cell. 2012;48(5):771–84. doi: 10.1016/j.molcel.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasarte S1, Elsner D, Sanchez-Elsner T, Fernandez-Pineda A, López-Fernández LA, Corbí AL, Muñoz-Fernandez MA, Relloso M. Estradiol downregulates NF-κb translocation by Ikbkg transcriptional repression in dendritic cells. Genes Immun. 2013;14(7):462–9. doi: 10.1038/gene.2013.35. doi: 10.1038/gene.2013.35. Epub 2013 Jul 25. [DOI] [PubMed] [Google Scholar]
- 28.Zhu AX, Kudo M, Assenat E. et al. Effect of everolimus on survival in advanced hepatocellular carcinoma after failure of sorafenib, the EVOLVE-1 randomized clinical trial. JAMA. 2014;312(1):57–67. doi: 10.1001/jama.2014.7189. [DOI] [PubMed] [Google Scholar]
- 29.Kudo M, Han G, Finn RS. et al. Brivanib as adjuvant therapy to transarterial chemoembolization in patients with hepatocellular carcinoma, A randomized phase III trial. Hepatology. 2014;60(5):1697–1707. doi: 10.1002/hep.27290. [DOI] [PubMed] [Google Scholar]
- 30.Mo Y, Chen J, Schlueter C.F, Hoyle G.W. Differential susceptibility of inbred mouse strains to chlorine-induced airway fibrosis. Am J Physiol Lung Cell Mol Physiol. 2013;304:L92–L102. doi: 10.1152/ajplung.00272.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haston C.K., Cory S., Lafontaine L., Dorion G., Hallett M.T. Strain-dependent pulmonary gene expression profiles of a cystic fibrosis mouse model. Physiol Genomics. 2006;25:336–345. doi: 10.1152/physiolgenomics.00208.2005. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Liu H, Jiao Y, Wang E, Clark SH, Postlethwaite AE, Gu W, Chen H. Differences between Mice and Humans in Regulation and the Molecular Network of Collagen, Type III, Alpha-1 at the Gene Expression Level: Obstacles that Translational Research Must Overcome. Int J Mol Sci. 2015;16(7):15031–56. doi: 10.3390/ijms160715031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables.