Abstract
The present study analyzes the lack of culturability of different non-Saccharomyces strains due to interaction with Saccharomyces cerevisiae during alcoholic fermentation. Interaction was followed in mixed fermentations with 1:1 inoculation of S. cerevisiae and ten non-Saccharomyces strains. Starmerella bacillaris, and Torulaspora delbrueckii indicated longer coexistence in mixed fermentations compared with Hanseniaspora uvarum and Metschnikowia pulcherrima. Strain differences in culturability and nutrient consumption (glucose, alanine, ammonium, arginine, or glutamine) were found within each species in mixed fermentation with S. cerevisiae. The interaction was further analyzed using cell-free supernatant from S. cerevisiae and synthetic media mimicking both single fermentations with S. cerevisiae and using mixed fermentations with the corresponding non-Saccharomyces species. Cell-free S. cerevisiae supernatants induced faster culturability loss than synthetic media corresponding to the same fermentation stage. This demonstrated that some metabolites produced by S. cerevisiae played the main role in the decreased culturability of the other non-Saccharomyces yeasts. However, changes in the concentrations of main metabolites had also an effect. Culturability differences were observed among species and strains in culture assays and thus showed distinct tolerance to S. cerevisiae metabolites and fermentation environment. Viability kit and recovery analyses on non-culturable cells verified the existence of viable but not-culturable status. These findings are discussed in the context of interaction between non-Saccharomyces and S. cerevisiae.
Keywords: contact-dependent interaction, culturability loss, excreted compounds, viable but not-culturable (VBNC), wine
Introduction
Spontaneous wine fermentation is driven by a succession of different yeast species. A great variety of non-Saccharomyces yeast species originate from grape berries and survive during the early stages of fermentation, such as species from the genera Candida, Hanseniaspora, Lachancea, Metschnikowia, Pichia, and Torulaspora (Fleet, 2003). Some species such as Starmerella bacillaris and Hanseniaspora uvarum grow to a high density (105–107 cells/mL) and dominate other non-Saccharomyces species (Wang et al., 2015b). There is a growing interest regarding the impact of non-Saccharomyces yeasts on the final wines, and some species are now used as fermentation starters (Jolly et al., 2014). However, fermentative Saccharomyces cerevisiae soon replaces non-Saccharomyces species to become the main or the only species present in the late stages of fermentation. Non-Saccharomyces strains are assumed to “die off” because these cells gradually lose their ability to form colonies on growth media, i.e., they lose the capacity to grow. The culturability loss of non-Saccharomyces strains has drawn widespread attention in recent years due to new findings that mention the role of excreted compounds in the interaction between Saccharomyces and non-Saccharomyces yeasts (Ciani and Comitini, 2015; Liu et al., 2015; Albergaria and Arneborg, 2016). Moreover, in a work by Branco et al. (2015), it was shown that viable but not-culturable (VBNC) status was related to interaction through excreted compounds. Therefore, as more non-conventional wine yeasts have been explored as wine starters in mixed fermentation with S. cerevisiae (Masneuf-Pomarede et al., 2016), studies on culturability loss of different non-Saccharomyces strains will help in understanding their final impact on wine quality.
The culturability loss of non-Saccharomyces strains at the late stages of alcoholic fermentation is a complicated phenomenon due to the multitude of factors involved. It is conventionally regarded to be related to their insufficient adaptability to environmental changes in fermentations, such as nitrogen limitation (Monteiro and Bisson, 1991), low oxygen availability (Holm Hansen et al., 2001) and inhibition of increased ethanol (Fleet, 2003), as well as extrinsic factors such as SO2(Ribéreau-Gayon et al., 2006). However, Nissen et al. (2003) proposed that S. cerevisiae S101 adopted a contact-dependent mechanism to induce the culturability loss of some non-Saccharomyces strains (Lachancea thermotolerans and Torulaspora delbrueckii). Subsequently, the contact-dependent mechanism was confirmed by studies using the same S. cerevisiae strain (Nissen et al., 2004; Renault et al., 2013; Kemsawasd et al., 2015a). However, it was found that S. cerevisiae CCMI 885 excreted toxic compounds, which inhibited the growth of Hanseniaspora guilliermondii and H. uvarum, demonstrating the interaction of these species through excreted antimicrobial compounds (Pérez-Nevado et al., 2006). Recent studies further elucidated that S. cerevisiae CCMI 885 produced antimicrobial peptides, which altered intracellular pH, membrane permeability and culturability of non-Saccharomyces strains (Albergaria et al., 2010; Branco et al., 2014, 2015). Interestingly, in the work of Wang et al. (2015c), not only the excreted products from S. cerevisiae NSa but also the synthetic media, induce a lack of culturability of H. uvarum. However, the synthetic must was weaker at inducing a lack of culturability of H. uvarum than S. cerevisiae supernatant, which included the same media plus the yeast metabolites. Thus, the role of environmental changes should be taken into consideration when studying the interaction between different yeasts.
Until now, studies on culturability loss of non-Saccharomyces yeasts have mainly focused on several potential wine starters: H. guilliermondii, H. uvarum, Kluyveromyces marxianus, L. thermotolerans, and T. delbrueckii (reviewed in Albergaria and Arneborg, 2016). However, few studies have focused on the culturability differences among strains. According to Branco et al. (2014), different D. bruxellensis strains showed strain-specific sensitivity toward antimicrobial peptides excreted by S. cerevisiae. The differences between contact-dependent mechanisms and interactions through extracellular compounds were ascribed to the S. cerevisiae strains used (Kemsawasd et al., 2015a). Therefore, more yeast species and strains should be considered to gain a better understanding of the interaction between S. cerevisiae and non-Saccharomyces yeasts.
This study was aimed at (i) investigating the strain and species differences in culturability loss, (ii) analyzing the interaction mechanisms that exist in different strains, and (iii) determining the viable status of non-culturable cells. We investigated the interaction between S. cerevisiae NSa (the same strain used in our former work, Wang et al., 2015c) and 10 non-Saccharomyces strains from different sources belonging to H. uvarum, S. bacillaris, M. pulcherrima, and T. delbrueckii to analyze the interactions in mixed fermentation between S. cerevisiae and each individual strain. Through the use of three types of media (supernatants from S. cerevisiae fermentation, synthetic media mimicking S. cerevisiae fermentation and mixed fermentation), the performance of each non-Saccharomyces strain was compared and studied. Synthetic must was used to rule out other effects and to define the media to mimic the must at different stages of fermentation. Recovery analysis and viability assays were also conducted to evaluate the status of non-culturable cells.
Materials and methods
Yeasts strains and culture conditions
Eleven yeast strains were used in this study, containing H. uvarum CECT13130, NSb and CECT1444T, S. bacillaris NSc, NSd and CECT11046, M. pulcherrima Mp com and Mp 51, T. delbrueckii Td com and CECT13135, and S. cerevisiae NSa. These strains were obtained from different collections: CECT13130, NSa, NSb, NSc, NSd, Mp 51, CECT13135 and NSa were natural isolates from our collection (Wang et al., 2014). CECT1444 and CECT11046 were from Spanish Type Culture Collection. Mp com (Flavia) and Td com (Biodiva) were commercial strains from Lallemand Inc. (Canada).
The species identity of all strains was determined by 5.8S-ITS-RFLP analysis (Esteve-Zarzoso et al., 1999) and sequence analysis of the D1/D2 domain of 26S rDNA (Kurtzman and Robnett, 1998). Yeasts were grown overnight in YPD medium (1% yeast extract, 2% peptone, and 2% glucose, w/v, pH 6.2) at 28°C before use.
Alcoholic fermentations, sampling, and setting culturability
Synthetic must (100 g/L fructose, 100 g/L glucose, 290 mg N/L amino nitrogen, and 120 mg N/L ammonium nitrogen, pH 3.3) was prepared according to Andorrà et al. (2012). 350 mL of synthetic must was added to a 500 mL screw cap bottle, inoculated with 106 cells/mL of each yeast strain and kept at 25°C in a shaker at a speed of 120 rpm. Fermentations were performed in the presence of air because the caps were not screwed tightly on the bottles. Each of the mixed fermentations was inoculated with one non-Saccharomyces strain and S. cerevisiae NSa. As a comparison, a single S. cerevisiae fermentation was carried out with the NSa strain. Fermentations were conducted in duplicate; when an interaction was observed, the fermentations were repeated in another duplicate and thus four replicas were used to set the interaction analysis.
Samples were taken every day to follow sugar and nitrogen consumption, ethanol production and yeast population dynamics until the end of fermentation. Concentrations of ethanol, fructose and glucose were tested using an enzymatic kit from Roche Diagnostics (Germany). The level of individual amino acids and ammonium was analyzed by HPLC according to Andorrà et al. (2012). Yeast populations in all samples were quantified using a microscope and plating after appropriate dilution in sterile water. YPD agar medium was used to calculate the total number of yeast cells present, and lysine agar medium (Oxoid LTD., England) was used for quantification of non-Saccharomyces strains.
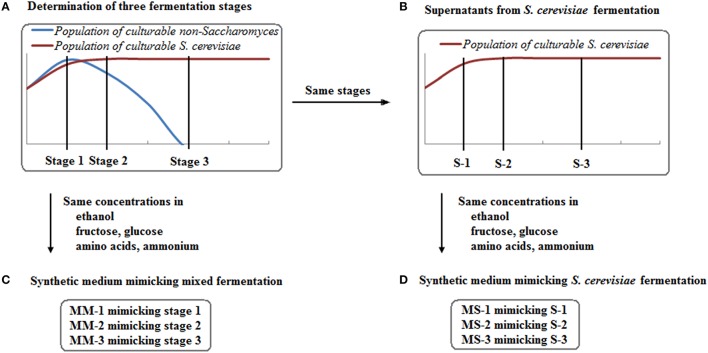
Three stages were set up for each species depending on the culturability of the non-Saccharomyces species in mixed fermentations (Figure 1A): 1. When culturable populations reached the highest level; 2. When culturable populations started to decrease; 3. When no colonies grew on plates, or at the end of fermentation for some strains if colonies were still seen on plates. The concentration of main chemical components (ethanol, fructose, glucose, individual amino acids, and ammonium), fermentation time and non-Saccharomyces strain culturability at these stages were listed to mimic the conditions of each fermentation stage where the interaction between non-Saccharomyces strains and S. cerevisiae was set (Figure 1 and Table 1). Data from these stages at mixed and single fermentations were used for the interaction assays in next step.
Figure 1.
Experimental design for setting up three fermentation stages (A) and preparing three types of synthetic media (B–D).
Table 1.
Fermentation stages, population size and chemical characteristics of the media at different single and mixed fermentation stages.
| Species | Fermentation time (h) | Culturable non-saccharomyces (cfu/mL) | Ethanol (v/v) | Fructose (g/L) | Glucose (g/L) | Total assimilable nitrogen (mg N/L) | Names of synthetic media |
|---|---|---|---|---|---|---|---|
| H. uvarum | 24 | 2.6 ± 1.3 × 107 | 1.6 ± 0.2 | 74.6 ± 4.3 | 59.0 ± 4.1 | 66.8 ± 26.7 | MM-1 |
| 24 | – | 1.8 ± 0.0 | 76.7 ± 0.0 | 52.3 ± 0.0 | 5.0 ± 0.0 | MS-1 | |
| 48 | 2.0 ± 1.1 × 107 | 3.1 ± 0.6 | 49.8 ± 2.1 | 25.1 ± 6.1 | 6.6 ± 3.3 | MM-2 | |
| 48 | – | 4.2 ± 0.1 | 45.2 ± 1.6 | 13.7 ± 1.7 | 1.2 ± 0.0 | MS-2 | |
| 96 | nd | 9.4 ± 0.5 | 12.7 ± 5.0 | 0.9 ± 0.7 | nd | MM-3 | |
| 96 | – | 10.2 ± 0.4 | 4.1 ± 2.9 | nd | nd | MS-3 | |
| M. pulcherrima | 24 | 8.1 ± 3.3 × 106 | 2.2 ± 0.6 | 89.6 ± 4.1 | 53.1 ± 7.2 | 87.4 ± 54.1 | MM-1 |
| 24 | – | 1.8 ± 0.0 | 76.7 ± 0.0 | 52.3 ± 0.0 | 5.0 ± 0.0 | MS-1 | |
| 48 | 3.2 ± 6.8 × 106 | 6.7 ± 0.9 | 56.1 ± 4.9 | 23.6 ± 5.7 | 2.5 ± 1.2 | MM-2 | |
| 48 | – | 4.2 ± 0.1 | 45.2 ± 1.6 | 13.7 ± 1.7 | 1.2 ± 0.0 | MS-2 | |
| 96 | nd | 10.2 ± 0.5 | 15.9 ± 6.0 | 0.7 ± 0.8 | nd | MM-3 | |
| 96 | – | 10.2 ± 0.4 | 4.1 ± 2.9 | nd | nd | MS-3 | |
| S. bacillaris | 24 | 4.2 ± 3.7 × 107 | 1.5 ± 0.2 | 68.6 ± 3.0 | 50.7 ± 2.2 | 18.3 ± 4.7 | MM-1 |
| 24 | – | 1.8 ± 0.0 | 76.7 ± 0.0 | 52.3 ± 0.0 | 5.0 ± 0.0 | MS-1 | |
| 96 | 8.8 ± 8.3 × 106 | 9.9 ± 0.5 | 5.1 ± 1.9 | nd | nd | MM-2 | |
| 96 | – | 10.2 ± 0.4 | 4.1 ± 2.9 | nd | nd | MS-2 | |
| 120 | 4.7 ± 8.2 × 101 | 11.6 ± 0.1 | 0.1 ± 0.1 | nd | nd | MM-3 | |
| 120 | – | 11.5 ± 0.4 | 0.1 ± 0.1 | nd | nd | MS-3 | |
| T. delbrueckii | 24 | 1.9 ± 0.7 × 107 | 3.1 ± 0.1 | 90.8 ± 3.4 | 48.4 ± 0.9 | 20.1 ± 13.9 | MM-1 |
| 24 | – | 1.8 ± 0.0 | 76.7 ± 0.0 | 52.3 ± 0.0 | 5.0 ± 0.0 | MS-1 | |
| 96 | 1.1 ± 1.9 × 106 | 10.4 ± 0.4 | 19.8 ± 5.1 | 1.1 ± 1.9 | nd | MM-2 | |
| 96 | – | 10.2 ± 0.4 | 4.1 ± 2.9 | nd | nd | MS-2 | |
| 144 | 1.8 ± 2.4 × 106 | 11.7 ± 0.6 | 1.5 ± 1.9 | nd | nd | MM-3 | |
| 120 | – | 11.5 ± 0.4 | 0.1 ± 0.1 | nd | nd | MS-3 |
This chemical composition was used to define the media mimicking the three selected fermentation stages. All values are the average of different strains within the same species. “MM” means synthetic media with main metabolites (ethanol, fructose, glucose and nitrogen) mimicking mixed fermentations, whereas “MS” is named after synthetic media with main metabolites mimicking S. cerevisiae fermentation. The Arabic numbers 1, 2, and 3 refer to the three stages selected in fermentations. “–” refers to the absence of a non-Saccharomyces population as derived from single S. cerevisiae fermentations, whereas “nd” means not detected. The total assimilable nitrogen is the sum of nitrogen from assimilable amino acids and ammonium. Only amino acid concentrations higher than 0.9 mg N/L are considered, and the concentrations are shown in Supplementary Table 1.
Culture assays using different synthetic media
To further understand the culturability of non-Saccharomyces strains in mixed fermentation, three types of synthetic media were prepared (Figure 1): supernatant from S. cerevisiae fermentation (S), synthetic medium mimicking S. cerevisiae fermentation (MS) and synthetic medium mimicking mixed fermentation (MM). S was collected from S. cerevisiae fermentation (Figure 1B), centrifuged and filtered using a 0.22 μm Whatman syringe filter (GE Healthcare Life Science, Germany). S was spread onto YPD agar plates to confirm the absence of S. cerevisiae cells. As a comparison, MS was prepared with metabolites (ethanol, fructose, glucose, individual amino acid, and ammonium) mimicking S, with the absence of S. cerevisiae excreted compounds (Figure 1D). By performing culture assays using S and MS, the effect of main fermentation metabolites (the same for S and MS) and other putative S. cerevisiae metabolites (only in S) could be observed. Considering the possible differences of the main metabolites produced by S. cerevisiae fermentation and mixed fermentation, MM was prepared with corresponding components mimicking the mixed fermentation (Figure 1C). Moreover, no micronutrients or vitamins were added to MS and MM due to fast consumption at the beginning of alcoholic fermentation. All of the synthetic media were prepared for the three fermentation stages selected in 2.2 and were named with Arabic numbers to differentiate these stages (Figure 1 and Table 1).
Single fermentations of each non-Saccharomyces strain were then performed to provide adapted cells as described in Wang et al. (2015c). These adapted cells were incubated in YPD to ensure viability and incubated in synthetic media to check their culturability on plates. Culture assays were conducted at 25°C in duplicate with a shaking speed of 120 rpm; when a culturability decrease was observed, the culture assays were repeated in another duplicate and thus four replicas were used to follow the culturability changes of non-Saccharomyces yeasts. Samples were taken at 24, 48, and 120 h to quantify yeast cells using a microscope and YPD plating after appropriate dilution in sterile water. Cells losing culturability in synthetic media were collected for the following recovery analysis and viability assay.
Recovery analysis and viability assay of non-culturable cells from synthetic media
To test the viability of non-culturable cells from synthetic media, two approaches were used. Membrane integrity was analyzed by using the LIVE/DEAD® BactLight™ Bacterial Viability kit (Molecular Probes Inc., USA). In this assay, yeast cells were stained and observed using a fluorescence microscope equipped with filter system I3 and N2.1 (Leica DM 4000B) as in Hierro et al. (2006). The capacity to grow in rich liquid media was analyzed by incubating the cells in fresh YPD medium. Cells that could be recovered were considered to be viable but not culturable in synthetic media. Cells that could not be recovered after two consecutive 48 h incubations in fresh YPD medium were analyzed again by the LIVE/DEAD® BactLight™ Bacterial Viability kit.
Statistical analysis
One-way ANOVA by IBM SPSS Statistics 23 was used to calculate the value of significance for the variation analysis, and included a post-hoc Tukey test when needed. The consumption ratio (% of the total) of nutrients was used directly for the analysis of variation.
Results
Culturable population and metabolic characteristics of non-saccharomyces strains during alcoholic fermentation
Overall, both S. cerevisiae and non-Saccharomyces strains reached the maximum population number of 107–108 cells/mL 24 h after inoculation, and this size was maintained during mixed and single fermentations. Culturability of non-Saccharomyces strains decreased in all mixed fermentations. This decrease varied not only among different yeast species but also among some strains within the same species (Figure 2). Culturable H. uvarum increased to 107–108 cfu/mL at 24 h and began to decrease at 48 h. No colonies were formed on lysine plates for CECT1444 after 72 h and for CECT13130 and NSb after 96 h. Similar to H. uvarum, M. pulcherrima grew to 107 cfu/mL and quickly started to decrease. At 96 h, no colonies of Mp com were recovered on lysine plates, and for Mp 51, no colonies were recovered after 48 h. Culturable S. bacillaris maintained a population size of 107–108 cfu/mL until 96 h at which time the population started to decline. After 120 h, 10 to 100 cfu/mL of NSd and CECT11046 were recovered, however no colonies of NSc grew from lysine plates. Finally, T. delbrueckii strains reached approximately 107 cfu/mL and were maintained up to 48 h. After 48 h, CECT13135 started to decline, and no colonies were recovered after 144 h. The other T. delbrueckii strain, Td com, showed a slow decrease in culturable population, and at 144 h, 106 cfu/mL of Td com were still culturable. Based on the culturability of non-Saccharomyces strains in mixed fermentations, three stages were set up for each species. The fermentation times and main metabolites of these three fermentation stages are shown in Table 1. As a comparison, the same stage in fermentation of S. cerevisiae is also listed.
Figure 2.
Culturable population of non-Saccharomyces in mixed fermentations with S. cerevisiae. Culturable S. cerevisiae populations were shown in orange line using the same line type as the non-Saccharomyces co-inoculated. (A) H. uvarum (B) M. pulcherrima (C) S. bacillaris (D) T. delbrueckii.
Despite the different culturability of non-Saccharomyces strains in mixed fermentations, no obvious variations in fermentation length were observed, and all fermentations finished after 120 or 144 h. Similar to S. cerevisiae fermentation, all mixed fermentations consumed glucose faster than fructose, and the final ethanol concentration reached 11–12% V (Table 1). However, analysis of the consumption of these main metabolites after 24 h revealed strain-dependent differences. As shown in Table 2, the strains that lost culturability faster were those that consumed some metabolites faster in the mixed fermentations: (i) Within H. uvarum, mixed fermentations inoculated with CECT1444 consumed glucose, ammonium and arginine faster than the other two strains; (ii) For M. pulcherrima, mixed fermentations with Mp 51 metabolized fructose, glucose, alanine, ammonium, arginine and glutamine faster than mixed fermentations with Mp com; (iii) Arginine was consumed faster during mixed fermentations with S. bacillaris NSc than the other two strains; (iv) More arginine was consumed during mixed fermentations with T. delbrueckii CECT13135 than Td com.
Table 2.
Consumption ratio of glucose, alanine, ammonium, arginine, and glutamine at 24 h of fermentation.
| Non-Saccharomyces strains | % of the total | Time of culturability loss | ||||||
|---|---|---|---|---|---|---|---|---|
| Fructose | Glucose | Alanine | Ammonium | Arginine | Glutamine | |||
| sc | 23.3 | 47.7 | 100.0 | 100.0 | 100.0 | 100.0 | nd | |
| sc+hu | CECT13130 | 27.2 | 37.6* | 90.4* | 68.1* | 65.1* | 100.0 | 96 h |
| NSb | 23.3 | 39.2* | 93.1* | 63.4* | 66.7* | 100.0 | 96 h | |
| CECT1444 | 25.7 | 46.2# | 91.6* | 100.0# | 71.9*# | 100.0 | 72 h | |
| sc+mp | Mp com | 6.9*# | 40.7*# | 13.4*# | 69.1*# | 53.7*# | 88.4*# | 96 h |
| Mp 51 | 13.9* | 53.2* | 96.7 | 91.2* | 67.8* | 100.0 | 48 h | |
| sc+sb | NSc | 35.1* | 51.6 | 100.0 | 100.0 | 91.4*# | 100.0 | 120 h |
| NSd | 30.4* | 48.5 | 96.9 | 100.0 | 82.7* | 100.0 | nd | |
| CECT11046 | 28.8* | 47.7 | 98.3 | 100.0 | 84.1* | 100.0 | nd | |
| sc+td | Td com | 10.8* | 51.6 | 99.4 | 100.0 | 75.9*# | 100.0 | nd |
| CECT13135 | 7.7* | 51.6 | 99.9 | 100.0 | 93.5* | 100.0 | 144 h | |
sc means S. cerevisiae single fermentation, and mixed fermentations are presented as sc+hu (S. cerevisiae + H. uvarum), sc+mp (S. cerevisiae + M. pulcherrima), sc+sb (S. cerevisiae + S. bacillaris), sc+td (S. cerevisiae + T. delbrueckii). nd means not detected.
significance ≤ 0.05 with respect control (sc) by one-way ANOVA.
significantly different from the other strains of the same species as determined by a post-hoc Tukey test.
The influence of excreted compounds from S. cerevisiae and media composition on the culturability of non-saccharomyces strains
To further elucidate the culturability of non-Saccharomyces strains and the interaction with S. cerevisiae during mixed fermentation, we performed culture assays using S (supernatant from S. cerevisiae fermentation), MS (synthetic media mimicking S. cerevisiae fermentation), and MM (synthetic media mimicking mixed fermentation) based on the three stages of fermentation (Table 1). Although the non-Saccharomyces strains maintained a population size of 107–108 cells/mL for 120 h, as determined by cell counting under a microscope, not all strains were culturable during the 5-day period. The culturability was dependent on the media used, as well as the yeast species and strain.
No effect on culturability was seen in any media in fermentation stage 1 (S-1, MS-1, MM-1), which corresponded to the fermentation stage where culturable populations of non-Saccharomyces strains were the highest (generally between 107 and 108 cfu/ml). However, in the media from fermentation stage 2 (only S-2), and in the media from fermentation stage 3 (all the media), a decrease in culturable populations was observed (Figures 3–6), although the extent of the decrease was species- and strain-dependent.
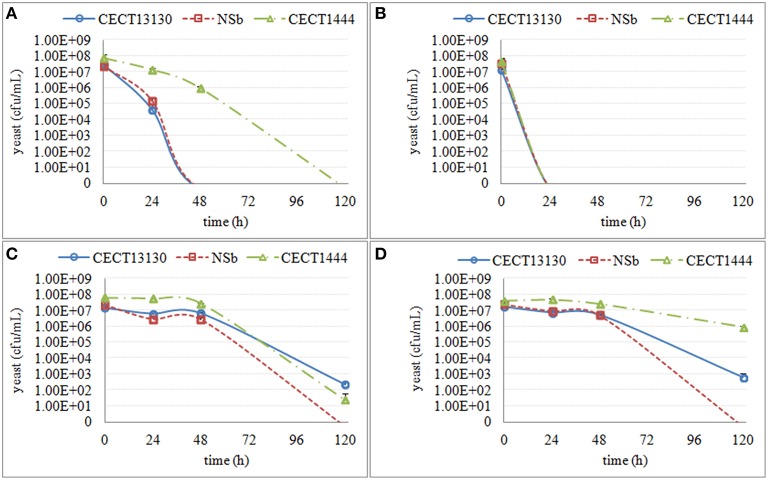
Figure 3.
The culturable population of three H. uvarum strains (CECT13130, NSb and CECT1444) grown in different synthetic media for 120 h. (A) the growth in supernatant from the second stage of S. cerevisiae fermentation S-2 (B) the growth in supernatant from the third stage of S. cerevisiae fermentation S-3 (C) the growth in synthetic media MS-3 (D) the growth in synthetic media MM-3.
Within H. uvarum strains, the decrease of culturability was seen at 24 h in S-2 and S-3 and at 120 h in MS-3 and MM-3 (Figure 3). H. uvarum strains lost culturability in both S-2 and S-3; however, the decrease in culturability in S-2 occurred more slowly than in S-3. Further, among the three strains, CECT1444 showed a much slower decrease in culturability in S-2. The media (MS-3 and MM-3) also affected the culturability, but to a lesser extent, and they were evident only at 120 h. There was also a strain difference, such that NSb was more affected than the other two strains.
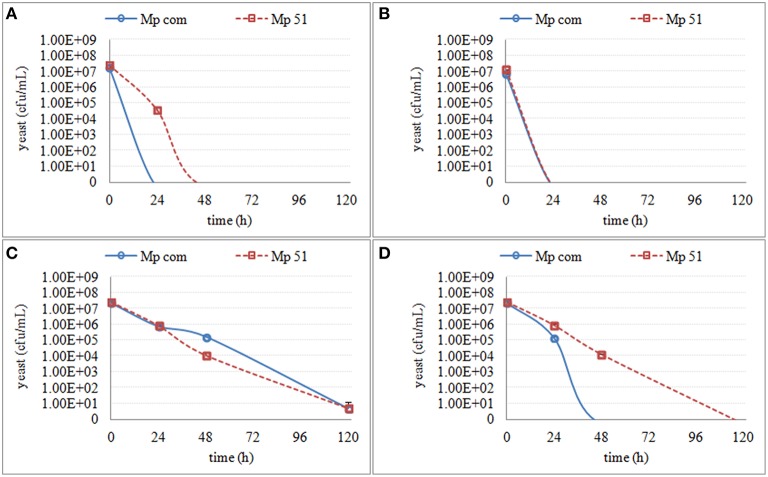
Regarding the M. pulcherrima strains, a slow decrease in culturability was observed in all media mimicking the fermentation stages (MS-2, MM-2, MS-3, and MM-3), whereas a sharp decrease was seen in the S. cerevisiae supernatants (S-2 and S-3). Mp com and Mp51 showed different culturability in both S-2 and MS-3 (Figure 4), with Mp com exhibiting higher sensitivity.
Figure 4.
The culturable population of two M. pulcherrima strains (Mp com and Mp 51) grown in different synthetic media for 120 h. (A) the growth in supernatant from the second stage of S. cerevisiae fermentation S-2 (B) the growth in supernatant from the third stage of S. cerevisiae fermentation S-3 (C) the growth in synthetic media MS-3 (D) the growth in synthetic media MM-3.
The culturability of three S. bacillaris strains was less affected by different synthetic media, as some colonies were recovered (Figure 5). All of the strains showed a decrease in culturability during all studied periods (120 h), with no relevant differences between strains. Only in S-3 was a difference in sensitivity observed with strain NSc, which showed much lower culturability than the other two strains.
Figure 5.
The culturable population of three S. bacillaris strains (NSc, NSd and CECT11046) grown in different synthetic media for 120 h. (A) the growth in supernatant from the second stage of S. cerevisiae fermentation S-2 (B) the growth in supernatant from the third stage of S. cerevisiae fermentation S-3 (C) the growth in synthetic media MS-3 (D) the growth in synthetic media MM-3.
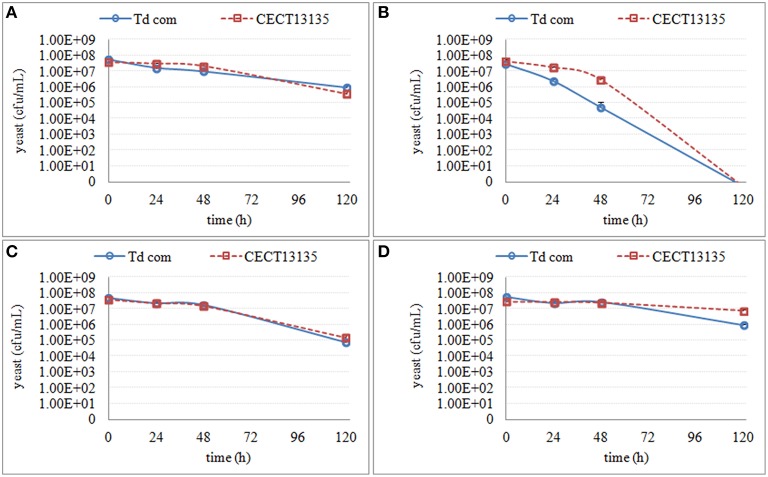
Similar to S. bacillaris, the effect of synthetic media on the culturability of the two T. delbrueckii strains was limited (Figure 6). However, when the cells were cultured in S-3, no colonies were recovered after 120 h. Instead, only a small decrease of culturability was observed in MS-3 and S-2.
Figure 6.
The culturable population of two T. delbrueckii strains (Td com and CECT13135) grown in different synthetic media for 120 h. (A) the growth in supernatant from the second stage of S. cerevisiae fermentation S-2 (B) the growth in supernatant from the third stage of S. cerevisiae fermentation S-3 (C) the growth in synthetic media MS-3 (D) the growth in synthetic media MM-3.
Although the decrease of culturability varied among different species, S-3 consistently showed the most obvious effect compared with other synthetic media. For the two species more affected (H. uvarum and M. pulcherrima), a more obvious effect was shown in S-2 than in MS-3 or MM-3. Thus, it is likely that some substances secreted from S. cerevisiae played a principal role in the interaction between S. cerevisiae and non-Saccharomyces strains, and that changes in the media (ethanol, nitrogen and sugar) also mediated the interaction.
The viability of the non-culturable cells
To improve the understanding of the lack of culturability of the non-Saccharomyces strains, all of the samples (34 cases in total) that did not grow on plates were tested using two different methods: membrane integrity using the LIVE/DEAD viability kit and recovery by suspension in liquid YPD with agitation.
On one hand, these non-culturable cells were immediately analyzed using the LIVE/DEAD viability kit (Supplementary Figure 1). The results showed that live fluorescent cells were only found three times: non-culturable cells of the Mp com strain at 24 h in S-2 yielded 0.13% of live fluorescent cells, at 48 h in MM-3 8.17% and the Mp 51 strain at 120 h in MM-2 2.84 %. All other non-culturable cells yielded dead fluorescent cells.
On the other hand, all non-culturable cells were evaluated by recovery analysis. The non-culturable cells with live fluorescent were recovered when incubated in liquid YPD medium, whereas some of the non-culturable cells with dead fluorescent could also be recovered. The latter cases were found seven times involving three non-Saccharomyces species. For example, after 120 h exposure to mimicking media MS-3 and MM-3, H. uvarum NSb, as well as H. uvarum CECT1444 after S-2 and the two T. delbrueckii strains after S-3, appeared dead in the fluorescence analysis but could be recovered. In the case of H. uvarum CECT13130, this observation was seen only in the early stages of exposure (48 h) to the media. Likewise, Mp com could be recovered after 120 h exposure to mimicking media MM-2 but could not be recovered from MM-3. Both cases indicate the existence of an intermediate, transient step before the cells are completely dead.
The LIVE/DEAD viability kit was again used to check the cells that could not be recovered by consecutive incubation in liquid YPD medium. All cells that could not be recovered yielded only dead fluorescent.
Discussion
The culturability loss of non-Saccharomyces strains during late stages of alcoholic fermentation has been well documented (Fleet, 2003). However, despite recent advances, the cellular mechanism underlying culturability loss is still a matter of discussion (Ciani and Comitini, 2015; Liu et al., 2015; Albergaria and Arneborg, 2016). In a previous study (Wang et al., 2015c), we investigated how S. cerevisiae NSa interacted with H. uvarum NSb by the use of a compartmented dialysis system, cell-free supernatant and mimicking synthetic media. Due to the absence of a contact-dependent mechanism in S. cerevisiae NSa, in the present study we decided to focus on the effects of compounds secreted by S. cerevisiae NSa, and the changes in main metabolites (ethanol, glucose, fructose, amino nitrogen and ammonium nitrogen).
Our results indicated that cell-free supernatant from S. cerevisiae fermentation influenced cellular culturability much more than mimicking synthetic media at the same fermentation stage (same chemical composition for major metabolites). Therefore, as mentioned in Wang et al. (2015c), some putative S. cerevisiae metabolites played a main role in the interaction between S. cerevisiae NSa and other non-Saccharomyces strains. A faster culturability loss was induced by S. cerevisiae supernatant at stage 3 than the initial two stages, which demonstrated the possible accumulation, or higher effect, of the S. cerevisiae secreted compounds as fermentation proceeded. Studies from Pérez-Nevado et al. (2006) and Williams et al. (2015) further related the accumulation to the amount of sugar consumed by S. cerevisiae. Likewise, antimicrobial peptides identified by Branco et al. (2014) were derived from a glycolytic enzyme, showing a probable link with sugar metabolism of S. cerevisiae. More research is still required to illustrate how sugar consumption regulates the secretion of antimicrobial peptides or other putative metabolites.
Moreover, according to our previous report (Wang et al., 2015c), as fermentation proceeds, the changes of the main metabolites also decreased the culturability of the cells, and the present results indeed showed that synthetic media at stage 3 caused a decrease in culturability. However, this effect occurs more slowly in the sensitive species and strains, and in our culture assays, complete culturability loss was mostly found after 48 or 120 h. This indicates that the changes in the main metabolites play a role in the interaction between S. cerevisiae NSa and other non-Saccharomyces strains, and vice versa. Because not all of the species were equally affected, it also showed the capacity of some non-Saccharomyces strains to withstand a harsh environment (ethanol higher than 10% vol, glucose lower than 1 g/L, fructose lower than 16 g/L and no available nitrogen).
The interaction between S. cerevisiae and non-Saccharomyces strains also relied on the participating yeast species. In our mixed fermentations, cells of S. bacillaris and T. delbrueckii could coexist longer with S. cerevisiae than H. uvarum and M. pulcherrima. Other studies proposed that oxygen availability, glucose uptake rate and nitrogen source might contribute to the longer co-existence (Holm Hansen et al., 2001; Nissen et al., 2004; Andorrà et al., 2012; Taillandier et al., 2014). We indeed found that mixed fermentation inoculated with S. bacillaris or T. delbrueckii present a consumption rate of glucose, alanine, ammonium and arginine more similar to single fermentations with S. cerevisiae than those mixed fermentations inoculated with H. uvarum and M. pulcherrima. However, the relation between species tolerance and consumption of some nutrients still needs further investigation.
As expected, strain differences within each species were observed in mixed fermentation, culture assays and recovery analyses. First, strains decrease their culturability to a different extent during mixed fermentation. Second, when incubated in the same synthetic media in culture assays, strains showed different culturability or tolerance to a harsh environment. Third, non-culturable cells from the same synthetic media showed different recovery abilities depending on the strain. The strain difference, to some extent, increased the complexity of interaction analysis between S. cerevisiae and non-Saccharomyces strains. In our case, the hypothesis of “strain tolerance to hard environment” cannot simply explain the strain differences in mixed fermentations. H. uvarum CECT1444 exhibited a slow culturability decrease in S-2, MM-2 and MM-3 as compared to the other two strains and thus was regarded as a strain that is highly tolerant to harsh environments. However, despite being a highly tolerant strain, in mixed fermentation, the culturability decreased even earlier than the other two H. uvarum strains. When we analyzed the metabolites at 24 h of mixed fermentation inoculated with H. uvarum CECT1444, a faster consumption of glucose, ammonium and arginine was detected. Andorrà et al. (2012) and Kemsawasd et al. (2015b) reported the influence of nitrogen consumption on yeast growth and fermentation performance. Nevertheless, further research should be undertaken to elucidate this effect, which was also observed in strains Mp 51, S. bacillaris NSc and T. delbrueckii CECT13135 compared with other strains within the same species.
Another important finding was the appearance of non-culturable cells when incubated with synthetic media, yielding more than 90 % of cells with “dead” fluorescence by viability analysis but that could be recovered by incubation in YPD medium. However, when these cells were incubated longer in the synthetic media (24 h more), all showed dead fluorescence and could no longer be recovered in YPD medium. This phenomenon demonstrated the existence of VBNC status of at least H. uvarum, M. pulcherrima, and T. delbrueckii during alcoholic fermentation. As hypothesized by Branco et al. (2015), VBNC status could be understood as a transition status of yeast from culturable cells to dead cells, involving sub-lethally and severely injured cells. During this transition process, the ability to form colonies is the first lost vital activity and progressive changes in the permeability of cell membrane occur as found in this study, however the DNA or RNA remains stable (Andorrà et al., 2010; Wang et al., 2014, 2015a). Branco et al. (2015) measured how antimicrobial peptides secreted by S. cerevisiae affected cell viability and reported that injured cells had a similar pH as the external pH, whereas cells without compromised membranes (impermeable to propidium iodide) maintained a higher pH. Further research is still required to determine how the interactions between S. cerevisiae and non-Saccharomyces impacts physiological status and metabolic capacity of cells in different status.
In conclusion, we investigated the interaction between one S. cerevisiae strain and ten non-Saccharomyces strains during alcoholic fermentation. We demonstrated that the decrease of culturability was mainly caused by metabolites secreted by S. cerevisiae, although the change of the composition in main metabolites in the media also played a role. We also found that culturability loss of non-Saccharomyces yeasts was not only species-dependent but also strain-dependent. The finding of VBNC status and strain differences in culturability is meaningful to the exploration of Saccharomyces-non-Saccharomyces interactions. The understanding of these interactions is relevant for the development of non-Saccharomyces strains as starters in wine production.
Author contributions
Conceived and designed the experiments: AM, BE. Performed the experiments: CW. Generated and analyzed the data: CW, AM, BE. Wrote the paper: CW, AM, BE.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The present work was supported by the Wildwine EU Project (grant agreement 315065). CW is the recipient of a grant from China Scholarship Council (State Scholarship Fund No. 201206300028), and we thank the guidance from Rosa Pastor for HPLC analysis.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00502
References
- Albergaria H., Arneborg N. (2016). Dominance of Saccharomyces cerevisiae in alcoholic fermentation process: role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 100, 2035–2046. 10.1007/s00253-015-7255-0 [DOI] [PubMed] [Google Scholar]
- Albergaria H., Francisco D., Gori K., Arneborg N., Gírio F. M. (2010). Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 86, 965–972. 10.1007/s00253-009-2409-6 [DOI] [PubMed] [Google Scholar]
- Andorrà I., Berradre M., Mas A., Esteve-Zarzoso B., Guillamón J. M. (2012). Effect of mixed culture fermentations on yeast populations and aroma profile. Food Sci. Technol. 49, 8–13. 10.1016/j.lwt.2012.04.008 [DOI] [Google Scholar]
- Andorrà I., Esteve-Zarzoso B., Guillamón J. M., Mas A. (2010). Determination of viable wine yeast using DNA binding dyes and quantitative PCR. Int. J. Food Microbiol. 144, 257–262. 10.1016/j.ijfoodmicro.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Branco P., Francisco D., Chambon C., Hébraud M., Arneborg N., Almeida M. G., et al. (2014). Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl. Microbiol. Biotechnol. 98, 843–853. 10.1007/s00253-013-5411-y [DOI] [PubMed] [Google Scholar]
- Branco P., Viana T., Albergaria H., Arneborg N. (2015). Antimicrobial peptides (AMPs) produced by Saccharomyces cerevisiae induce alterations in the intracellular pH, membrane permeability and culturability of Hanseniaspora guilliermondii cells. Int. J. Food Microbiol. 205, 112–118. 10.1016/j.ijfoodmicro.2015.04.015 [DOI] [PubMed] [Google Scholar]
- Ciani M., Comitini F. (2015). Yeast interactions in multi-starter wine fermentation. Curr. Opin. Food Sci. 1, 1–6. 10.1016/j.cofs.2014.07.001 [DOI] [Google Scholar]
- Esteve-Zarzoso B., Belloch C., Uruburu F., Querol A. (1999). Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 49, 329–337. 10.1099/00207713-49-1-329 [DOI] [PubMed] [Google Scholar]
- Fleet G. H. (2003). Yeast interactions and wine flavour. Int. J. Food Microbiol. 86, 11–22. 10.1016/S0168-1605(03)00245-9 [DOI] [PubMed] [Google Scholar]
- Hierro N., Esteve-Zarzoso B., González Á., Mas A., Guillamón J. M. (2006). Real-Time quantitative PCR (QPCR) and reverse transcription-QPCR for detection and enumeration of total yeast in wine. Appl. Environ. Microb. 72, 7148–7155. 10.1128/AEM.00388-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm Hansen E., Nissen P., Sommer P., Nielsen J. C., Arneborg N. (2001). The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J. Appl. Microbiol. 91, 541–547. 10.1046/j.1365-2672.2001.01426.x [DOI] [PubMed] [Google Scholar]
- Jolly N. P., Varela C., Pretorius I. S. (2014). Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. 10.1111/1567-1364.12111 [DOI] [PubMed] [Google Scholar]
- Kemsawasd V., Branco P., Almeida M. G., Caldeira J., Albergaria H., Arneborg N. (2015a). Cell-to-cell contact and antimicrobial peptides play a combined role in the death of Lachanchea thermotolerans during mixed-culture alcoholic fermentation with Saccharomyces cerevisiae. FEMS Microbiol. Lett. 362:fnv103. 10.1093/femsle/fnv103 [DOI] [PubMed] [Google Scholar]
- Kemsawasd V., Viana T., Ardö Y., Arneborg N. (2015b). Influence of nitrogen sources on growth and fermentation performance of different wine yeast species during alcoholic fermentation. Appl. Microbiol. Biotechnol. 99, 10191–10207. 10.1007/s00253-015-6835-3 [DOI] [PubMed] [Google Scholar]
- Kurtzman C. P., Robnett C. J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73, 331–371. 10.1023/A:1001761008817 [DOI] [PubMed] [Google Scholar]
- Liu Y., Rousseaux S., Tourdot-Maréchal R., Sadoudi M., Gougeon R., Schmitt-Kopplin P., et al. (2015). Wine microbiome, a dynamic world of microbial interactions. Crit. Rev. Food Sci. Nutr. [Epub ahead of print]. 10.1080/10408398.2014.983591. [DOI] [PubMed] [Google Scholar]
- Masneuf-Pomarede I., Bely M., Marullo P., Albertin W. (2016). The genetics of non-conventional wine yeasts: current knowledge and future challenges. Front Microbiol. 6:1563. 10.3389/fmicb.2015.01563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro F. F., Bisson L. F. (1991). Biological assay of nitrogen content of grape juice and prediction of sluggish fermentations. Am. J. Enol. Viticult. 42, 47–57 [Google Scholar]
- Nissen P., Nielsen D., Arneborg N. (2003). Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell-cell contact-mediated mechanism. Yeast 20, 331–341. 10.1002/yea.965 [DOI] [PubMed] [Google Scholar]
- Nissen P., Nielsen D., Arneborg N. (2004). The relative glucose uptake abilities of non-Saccharomyces yeasts play a role in their coexistence with Saccharomyces cerevisiae in mixed cultures. Appl. Microbiol. Biotechnol. 64, 543–550. 10.1007/s00253-003-1487-0 [DOI] [PubMed] [Google Scholar]
- Pérez-Nevado F., Albergaria H., Hogg T., Girio F. (2006). Cellular death of two non-Saccharomcyes wine-related yeasts during mixed fermentations with Saccharomcyes cerevisiae. Int. J. Food Microbiol. 108, 336–345. 10.1016/j.ijfoodmicro.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Renault P. E., Albertin W., Bely M. (2013). An innovative tool reveals interaction mechanisms among yeast populations under oenological conditions. Appl. Microbiol. Biotechnol. 97, 4105–4119. 10.1007/s00253-012-4660-5 [DOI] [PubMed] [Google Scholar]
- Ribéreau-Gayon P., Dubourdieu D., Donèche B., Lonvaud A. (2006). Handbook of Enology. Chichester, IL: John Wiley & Sons, Ltd. [Google Scholar]
- Taillandier P., Lai Q. P., Julien-Ortiz A., Brandam C. (2014). Interactions between Torulaspora delbrueckii and Saccharomyces cerevisiae in wine fermentation: influence of inoculation and nitrogen content. World J. Microb. Biot. 30, 1959–1967. 10.1007/s11274-014-1618-z [DOI] [PubMed] [Google Scholar]
- Wang C., Esteve-Zarzoso B., Cocolin L., Mas A., Rantsiou K. (2015a). Viable and culturable populations of Saccharomyces cerevisiae, Hanseniaspora uvarum and Starmerella bacillaris (synonym Candida zemplinina) during Barbera must fermentation. Food Res. Int. 78, 195–200. 10.1016/j.foodres.2015.10.014 [DOI] [PubMed] [Google Scholar]
- Wang C., Esteve-Zarzoso B., Mas A. (2014). Monitoring of Saccharomyces cerevisiae, Hanseniaspora uvarum, and Starmerella bacillaris (synonym Candida zemplinina) populations during alcoholic fermentation by fluorescence in situ hybridization. Int. J. Food Microbiol. 191, 1–9. 10.1016/j.ijfoodmicro.2014.08.014 [DOI] [PubMed] [Google Scholar]
- Wang C., García-Fernández D., Mas A., Esteve-Zarzoso B. (2015b). Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front Microbiol. 6:1156. 10.3389/fmicb.2015.01156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Mas A., Esteve-Zarzoso B. (2015c). Interaction between Hanseniaspora uvarum and Saccharomyces cerevisiae during alcoholic fermentation. Int. J. Food Microbiol. 206, 67–74. 10.1016/j.ijfoodmicro.2015.04.022 [DOI] [PubMed] [Google Scholar]
- Williams K. M., Liu P., Fay J. C. (2015). Evolution of ecological dominance of yeast species in high-sugar environments. Evolution 69, 2079–2093. 10.1111/evo.12707 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.