Abstract
Ku80 is a subunit of the Ku heterodimer that binds to DNA double-strand break ends as part of the non-homologous end joining (NHEJ) pathway. Ku80 is also involved in homologous recombination (HR) via its interaction with BRCA1. Ku80 is encoded by the XRCC5 gene that contains a variable number tandem repeat (VNTR) insertion in its promoter region. Different VNTR genotypes can alter XRCC5 expression and affect Ku80 production, thereby affecting NHEJ and HR pathways. VNTR polymorphism is associated with multiple types of sporadic cancer. In this study, we investigated its potential association with familial breast cancer at the germline level. Using PCR, PAGE, Sanger sequencing, and statistical analyses, we compared VNTR genotypes in the XRCC5 promoter between healthy individuals and three types of familial breast cancer cases: mutated BRCA1 (BRCA1+), mutated BRCA2 (BRCA2+), and wild-type BRCA1/BRCA2 (BRCAx). We observed significant differences of VNTR genotypes between control and BRCA1+ group (P < 0.0001) and BRCA2+ group (P = 0.0042) but not BRCAx group (P = 0.2185), and the differences were significant between control and cancer-affected BRCA1+ cases (P < 0.0001) and BRCA2+ cases (P = 0.0092) but not cancer-affected BRCAx cases (P = 0.4251). Further analysis indicated that 2R/2R (OR = 1.94, 95%CI = 1.26–2.95, P = 0.0096) and 2R/1R (OR = 1.58, 95%CI = 1.11–2.26, P = 0.0388) were associated with increased risk but 1R/1R (OR = 0.55, 95%CI = 0.35–0.84, P = 0.0196) and 1R/0R (OR = 0, 95%CI = 0–0.29, P = 0.0012) were associated with decreased risk in cancer-affected BRCA1+ group; 2R/1R (OR = 1.94, 95%CI = 1.14–3.32, P = 0.0242) was associated with increased risk in cancer-affected BRCA2+ group. No correlation was observed for the altered risk between cancer-affected or -unaffected carriers and between different age of cancer diagnosis in cancer-affected carriers. The frequently observed VNTR association with in BRCA1+ and BRCA2+ breast cancer group indicates that VNTR polymorphism in the XRCC5 promoter is associated with altered risk of breast cancer in BRCA1+ and BRCA2+ carriers.
Keywords: Ku80, XRCC5, promoter, VNTR, familial breast cancer, BRCA1, BRCA2, association
Introduction
Breast cancer is the major cancer type in women. Up to 20% of breast cancer cases have familial genetic background, with multiple family members across generations affected by the disease (1). The discovery of the germline mutations in BRCA1 and BRCA2 confirmed the presence of genetic predisposition for familial breast cancer (2–4). These genes maintain genome stability in normal cells by repairing double-strand breaks mainly through homologous recombination (HR) pathway; their mutated forms lead to genome instability and increased risk for breast cancer development (5). There are two types of DNA double-strand break repair mechanisms: non-homologous end joining (NHEJ) and HR (6). Deficiency in the HR pathway, mainly caused by BRCA germline mutations, is well known to increase the risk of breast cancer (7); however, it is not equally clear whether deficiency in NHEJ pathway can also increase breast cancer risk (8).
Ku is a heterodimer consisting of Ku80 encoded by XRCC5 and Ku70 encoded by XRCC6. Ku recognizes DNA double-strand break ends to initiate the NHEJ pathway, and Ku can also affect the function of the HR pathway by interacting with BRCA1 (9–13). Deletion of XRCC5 in mice leads to increased chromosomal instability, immune deficiency, growth retardation, and cancer (14, 15). Altered expression of XRCC5 promotes oncogenic phenotypes, including hyper proliferation and resistance to apoptosis, genomic instability, and tumorigenesis (16), and has been observed in various types of sporadic cancer, including bladder, breast, colorectal, skin, esophageal, gastric, head, and neck cancer (17–22).
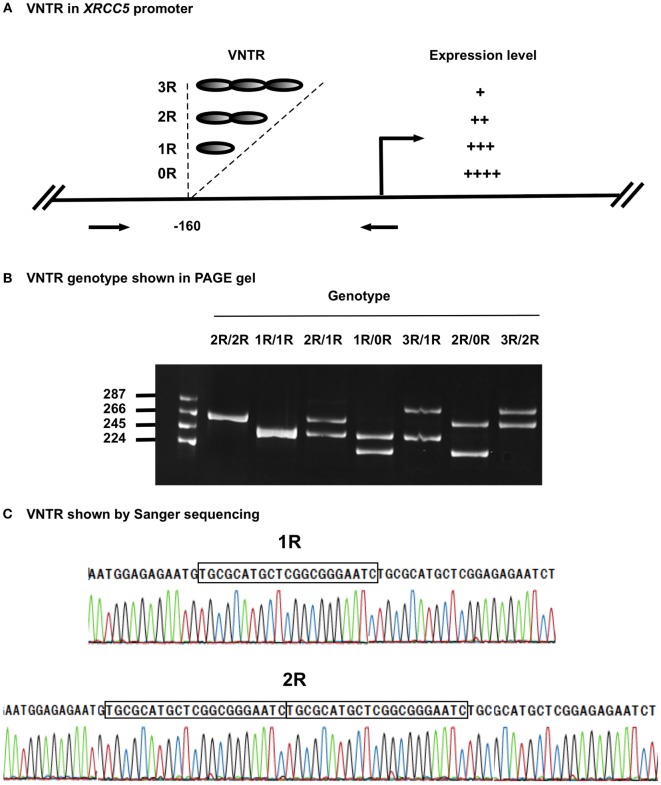
Variable number tandem repeats (VNTRs) are tandem repeat DNA sequences often located in gene regulatory regions that can influence gene expression (23–25). VNTRs follow a Mendelian pattern of inheritance. The XRCC5 promoter contains a VNTR at −160 bp, with a 21-bp repetitive unit (TGCGCATGCTCGGCGGGAATC) hosting a putative Sp1-binding site (26). Studies in Chinese and Iranian populations have demonstrated the presence of VNTR alleles ranging from 0 to 3 21-bp tandem repeats (0R, 1R, 2R, and 3R), with individual genotypes of 0R/0R, 1R/0R, 1R/1R, 2R/0R, 2R/1R, 2R/2R, 3R/0R, 3R/1R, and 3R/2R (22, 23). Experimental data indicate that the number of VNTR repeats is inversely related to XRCC5 expression, with an increase in the number of VNTR repeats linked to decreased XRCC5 expression (27–29) (Figure 1A). VNTR polymorphisms in the XRCC5 promoter are associated with sporadic bladder, gastric, and breast cancer (30–32).
Figure 1.
VNTR in XRCC5 promoter. (A) VNTR types and position in the promoter of XRCC5. The VNTR is located at −160 bp, with 3R, 2R, 1R, and 0R alleles. Arrows refer to PCR primers used to amplify the VNTR region for genotyping. It also shows higher copies of VNTR lead to lower XRCC5 expression (21–23). (B) Size distribution of different VNTR genotypes. PCR products of different genotypes were separated on an 8% PAGE gel. 2R/2R and 1R/1R had single band, other were heterozygotes with two bands, of which 2R/1R, 1R/0R, and 3R/2R had 21-base differences, and 3R/1R and 2R/0R had 42-base differences; (C) Sanger sequencing validation of 1R/1R and 2R/2R genotypes. It shows the 21-base unit (TGCGCATGCTCGGCGGGAATC) in 1R, and 42-base unit in 2R. 3R/3R DNA was not available for sequencing due to its rarity in human population.
Given the transmission pattern of VNTR, the uncertainty regarding the role of NHEJ in familial breast cancer, the presence of VNTR polymorphisms in the XRCC5 promoter, and the association of VNTR polymorphisms with sporadic cancer, we hypothesized that VNTR in the XRCC5 promoter could be involved in familial breast cancer. Therefore, we screened germline VNTR polymorphisms in the XRCC5 promoter in three types of familial breast cancer (BRCA1+, BRCA2+, and BRCAx). The results showed that certain genotypes of VNTR polymorphisms are associated with the risk of familial breast cancer in BRCA1+ and BRCA2+ carriers.
Materials and Methods
Study Population
The familial breast cancer cases used in this study included three subtypes: familial breast cancer with BRCA1 mutation (BRCA1+), familial breast cancer with BRCA2 mutation (BRCA2+), and familial breast cancer without BRCA1 or BRCA2 mutations (BRCAx). Samples were obtained from the Hereditary Cancer Center at Creighton University (Tables S1–S3 in Supplementary Material). Healthy control samples of age- and gender-matched, de-identified Caucasian individuals were obtained from the Nebraska Biobank of the University of Nebraska Medical Center and The Nebraska Medical Center. The use of patient samples for this study was approved by the Institutional Review Board of Creighton University School of Medicine (00-12265) and of the University of Nebraska Medical Center (718-11-EP). Written and informed consent to participate in the study and to publicate the results was obtained from all subjects.
Genotyping VNTR Polymorphisms in the XRCC5 Promoter
PCR amplification, PAGE gel separation, and Sanger sequencing were used to determine VNTR genotype in the XRCC5 promoter of each patient. PCR primer sequences were based on a previously published study (22) with sense primer 5′AGGCGGCTCAAACACCACAC3′ and antisense primer 5′CAAGCGGCAGATAGCGGAAAG3′. The PCR mixture consisted of DNA (20 ng), sense and antisense primers (10 pmol), and GoTaqH DNA polymerase (2 U, Promega). The PCR cycling conditions were 7 min at 95°C; 35 cycles of 30 s at 95°C, 30 s at 62°C, and 45 s at 72°C; and a final extension of 7 min at 72°C. An 8% PAGE gel was used to separate PCR products to determine allele type and genotype in each case (3R allele = 287 bp; 2R allele = 266 bp; 1R allele = 245 bp; and 0R allele = 224 bp). Representative products were isolated from PAGE gels and validated by Sanger sequencing.
VNTR Genotypes in the XRCC5 Promoter of Caucasians
Data from Iranian and Chinese healthy populations showed that VNTR genotypes in the XRCC5 promoter can vary between ethnic groups (27, 28). To determine whether the data from these healthy populations can be used as suitable healthy controls for our study in breast cancer of Caucasian cases, we tested the genotypes of 100 healthy local Caucasian individuals and compared these with the genotypes from 535 Caucasian Iranian and 235 Chinese populations (27). The results showed no significant difference in genotypes between the local and Iranian Caucasian populations (P = 0.3774) with 2R/2R, 2R/1R, and 1R/1R as the major genotypes, but a significant difference was seen in the genotypes between local Caucasian and Chinese populations (P < 0.0001), and Iranian Caucasian and Chinese (P < 0.0001), whose genotypes included 2R/2R, 2R/1R, 2R/0R, 1R/1R, 1R/0R, and 0R/0R (Table 1). The 535 Iranian cases were from a Caucasian population living in the Fars province of Iran (27). Because these Iranian cases and our local cases were of the same ethnicity and there were no significant differences in genotypes between the two groups, the genotypes of the 100 local cases and the 535 Iranian cases were combined to make up the control population for downstream analyses. The combined control group is at Hardy-Weinberg equilibrium (X2 = 4.3485, df = 6, P = 0.6296).
Table 1.
Genotype distribution in three ethnical populations.
| Genotype | Local | Iranian | Chinese |
|---|---|---|---|
| 3R/2R | 0 (0) | 4 (1) | 0 (0) |
| 3R/1R | 1 (1) | 8 (1) | 0 (0) |
| 3R/0R | 1 (1) | 1 (0) | 0 (0) |
| 2R/2R | 16 (16) | 84 (16) | 28 (12) |
| 2R/1R | 50 (50) | 205 (38) | 57 (24) |
| 2R/0R | 5 (5) | 29 (5) | 71 (30) |
| 1R/1R | 22 (22) | 168 (31) | 12 (5) |
| 1R/0R | 5 (5) | 33 (6) | 37 (16) |
| 0R/0R | 0 (0) | 3 (1) | 30 (13) |
| Total | 100 (100) | 535 (100) | 235 (100) |
| P value | Local to Iranian: 0.3774 | ||
| Local to Chinese: <0.0001 | |||
| Iranian to Chinese: <0.0001 | |||
Statistical Analyses
Fisher’s exact test was applied to determine the differences of VNTR polymorphism between the groups of familial breast cancer populations and control population, each type of breast cancer and cancer-affected and -unaffected subgroups within each type of cancer. Both odds ratios and their 95% confidence intervals and P-values were computed by using exact methods to keep consistency (33). Benjamini and Hochberg method was used to control the false positive rate at 0.05 (34). Analyses were performed using SAS® software version 9.4 (SAS Institute Inc., Cary, NC, USA).
Results
Samples Used in the Study
BRCA1+ carrier refers to the women who tested positive for a pathogenic BRCA1 mutation; BRCA2+ refers to the women who tested positive for a pathogenic BRCA2 mutation; and BRCAx refers to the women who tested negative for the mutations in BRCA1, BRCA2, and p53, with two or more first or second degree relatives affected with primary in situ or invasive breast, ovarian, fallopian tube, or peritoneal cancer, and at least one person must have negative test result. Under each group, the cases were further divided into breast cancer (ovarian cancer)-affected and -unaffected carriers. The average ages at breast cancer diagnosis among the groups were 41.4 (BRCA1+), 43.6 (BRCA2+), and 47.7 (BRCAx). The age distributions are consistent with existing data that BRCA1 and BRCA2 mutation carriers tend to suffer cancer at earlier age. Most of the breast cancers were ductal type and ER-positive; all, but one, of the cases of ovarian cancer were invasive at diagnosis (Table 2).
Table 2.
Summary of the BRCA1+, BRCA2+, and BRCAx carriers used in the study*.
| Items | BRCA1+ | BRCA2+ | BRCAx |
|---|---|---|---|
| Unaffected cases | 60 | 29 | 11 |
| Average current age | 56.9 ± 14.4 | 49.1 ± 14.4 | 66.4 ± 15.8 |
| Affected cases | 166 | 69 | 89 |
| Average age at diagnosis | 41.4 ± 10.8 | 43.6 ± 10.3 | 47.7 ± 12.0 |
| Proband | 38 | 15 | 62 |
| Non-proband | 128 | 54 | 27 |
| Breast cancer | 166 | 69 | 89 |
| ER | 22(+)43(−) | 17(+)8(−) | 27(+)6(−) |
| Unknown | 101 | 44 | 56 |
| PR | 17(+)45(−) | 15(+)9(−) | 21(+)9(−) |
| Unknown | 104 | 45 | 59 |
| HER2/neu | 4(+)10(−) | 3(+)4(−) | 6(+)17(−) |
| Unknown | 152 | 62 | 66 |
| Lymph nodes | 38(+)54(−) | 16(+)23(−) | 16(+)16(−) |
| Unknown | 74 | 30 | 57 |
| Left | 56 | 17 | 24 |
| Right | 55 | 25 | 27 |
| Bilateral | 40 | 19 | 8 |
| Unknown | 15 | 8 | 30 |
| Adenocarcinoma not specified | 28 | 7 | 17 |
| Ductual carcinoma | 89 | 43 | 59 |
| Lobular carcinoma | 4 | 4 | 4 |
| Medullary carcinoma | 24 | 3 | |
| Mucoid or colloid carcinoma | 3 | ||
| Unknown | 21 | 12 | 6 |
| Invasive | 148 | 63 | 79 |
| In situ | 5 | 2 | 3 |
| Both invasive and in situ | 6 | 3 | 3 |
| Unknown | 7 | 1 | 4 |
| Ovarian cancer | 21 | 5 | 15 |
| Left | 1 | ||
| Right | 3 | ||
| Bilateral | 4 | 1 | 2 |
| Unknown | 14 | 4 | 12 |
| Fallopian tube | 1 | 1 | |
| Lymph nodes | 7(+)10(−) | 1(+) | 2(+)3(−) |
| Unknown | 4 | 4 | 10 |
| Carcinoma, not specified | 3 | 4 | |
| Clear cell adenocarcinoma | 1 | 1 | |
| Papillary adenocarcinoma | 2 | 2 | 1 |
| Adenocarcinoma (cystadenocarcinoma) | 9 | 1 | 1 |
| Endometrioid adenocarcinoma | 2 | ||
| Serous (cyst)adenocarcinoma | 5 | 1 | 3 |
| Dysgerminoma | 1 | ||
| Unknown | 1 | 5 | |
| Invasive | 20 | 5 | 9 |
| In situ | 1 | 1 | |
| Unknown | 5 |
*Some number in categories may not add up to the total due to incompleteness of tested cases.
VNTR Genotypes in the XRCC5 Promoter
The four VNTR alleles in the XRCC5 promoter consist of three 21-bp (TGCGCATGCTCGGCGGGAATC) tandem repeats (3R), two 21-bp repeats (2R), one 21-bp repeat (1R), or without repeat (0R). The combination of PCR, PAGE, and Sanger sequencing methods provided an effective means to determine VNTR genotypes formed by the four alleles. Figure 1B shows the genotypes of homozygotes (1R/1R and 2R/2R) and heterozygotes (3R/2R, 3R/1R, 2R/1R, 2R/0R, and 1R/0R), and Figure 1C shows the sequences of the 21-bp repeats from the homozygotes (1R/1R and 2R/2R).
VNTR Genotype Distribution, BRCA Predisposition, and Cancer Status
We compared the VNTR genotype distributions in the XRCC5 promoter between three types of familial breast cancer: BRCA1+, BRCA2+, and BRCAx (Tables S1–S3 in Supplementary Material). The results show that the BRCA1+ and BRCA2+ groups differed significantly from the control group (BRCA1+ group: P < 0.0001; BRCA2+ group: P = 0.0042), but no difference was observed between the BRCAx groups and control group (P = 0.1308) (Table 3). To test whether different VNTR genotype distribution exists relating to disease status, the three types of familial breast cancer were divided into breast cancer-affected and breast cancer-unaffected subgroups and further compared each subgroup with the control group. The results show that the differences were only present between the cancer-affected subgroups in both groups of BRCA1+ (cancer-affected: P < 0.0001, cancer-unaffected: P = 0.2216) and BRCA2+ (cancer-affected: P = 0.0092, cancer-unaffected: P = 0.2748), but not in BRCAx (cancer-affected: P = 0.4251, cancer-unaffected: P = 0.5664) (Table 4). These results suggest the presence of association between VNTR genotypes and BRCA1 and BRCA2 mutation carriers affected with breast cancer.
Table 3.
Genotype distribution in three types of familial breast cancer.
| Genotype | Control | Familial breast cancer |
||
|---|---|---|---|---|
| BRCA1+ | BRCA2+ | BRCAx | ||
| Total | 635 (100) | 226 (100) | 98 (100) | 100 (100) |
| 3R/2R | 4 (1) | 0 (0) | 0 (0) | 0 (0) |
| 3R/1R | 9 (1) | 2 (1) | 0 (0) | 0 (0) |
| 3R/0R | 2 (0) | 0 (0) | 0 (0) | 0 (0) |
| 2R/2R | 100 (16) | 61 (27) | 27 (28) | 23 (23) |
| 2R/1R | 255 (40) | 113 (50) | 51 (52) | 48 (48) |
| 2R/0R | 34 (5) | 4 (2) | 1 (1) | 3 (3) |
| 1R/1R | 190 (30) | 45 (20) | 17 (17) | 19 (19) |
| 1R/0R | 38 (6) | 1 (0) | 2 (2) | 7 (7) |
| 0R/0R | 3 (0) | 0 (0) | 0 (0) | 0 (0) |
| P value | <0.0001 | 0.0042 | 0.2185 | |
Table 4.
Genotypes between cancer-affected and unaffected familial breast cancer.
| Genotype | Control |
BRCA1+ |
BRCA2+ |
BRCAx |
|||
|---|---|---|---|---|---|---|---|
| Cancer | No cancer | Cancer | No cancer | Cancer | No cancer | ||
| Total | 635 (100) | 166 (100) | 60 (100) | 69 (100) | 29 (100) | 89 (100) | 11 (100) |
| 3R/2R | 4 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 3R/1R | 9 (1) | 2 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 3R/0R | 2 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 2R/2R | 100 (16) | 44 (27) | 17 (28) | 17 (25) | 10 (34) | 20 (22) | 3 (27) |
| 2R/1R | 255 (40) | 85 (51) | 28 (47) | 39 (57) | 12 (41) | 41 (46) | 6 (64) |
| 2R/0R | 34 (5) | 3 (2) | 1 (2) | 0 (0) | 1 (3) | 3 (3) | 0 (0) |
| 1R/1R | 190 (30) | 32 (19) | 13 (22) | 13 (19) | 4 (14) | 18 (20) | 1 (9) |
| 1R/0R | 38 (6) | 0 (0) | 1 (2) | 0 (0) | 2 (7) | 7 (8) | 0 (0) |
| 0R/0R | 3 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| P value | <0.0001 | 0.2216 | 0.0092 | 0.2748 | 0.4251 | 0.5664 | |
We also compared the genotypes between the affected and unaffected subgroups in each group, and observed no difference in between (Table 5). We also evaluated the relationship between ages at diagnosis and VNTR polymorphism and observed no significant relationship in all three groups (data not shown). Therefore, there is no relationship between age of disease, cancer status, and VNTR polymorphism.
Table 5.
Comparison between affected and unaffected group.
| Genotype | Affected case | Unaffected case | Odds ratio | 95% CI | P-value | Adjusted |
|---|---|---|---|---|---|---|
| BRCA1+ | ||||||
| 2R/2R | 45 | 16 | 0.9299 | 0.46–1.96 | 0.8637 | 0.9422 |
| 2R/1R | 87 | 26 | 1.265 | 0.66–2.42 | 0.5403 | 1 |
| 1R/1R | 32 | 13 | 0.7906 | 0.37–1.79 | 0.5664 | 1 |
| 1R/0R | 0 | 1 | 0 | 0–64.08 | 0.2522 | 1 |
| BRCA2+ | ||||||
| 2R/2R | 17 | 10 | 0.769 | 0.27–2.26 | 0.6281 | 0.9422 |
| 2R/1R | 39 | 12 | 1.8417 | 0.70–4.91 | 0.1904 | 1 |
| 1R/1R | 13 | 4 | 1.3929 | 0.38–6.45 | 0.7707 | 1 |
| 1R/0R | 0 | 2 | 0 | 0–1.44 | 0.0854 | 1 |
| BRCAx | ||||||
| 2R/2R | 20 | 3 | 0.77 | 0.19–3.19 | 0.7118 | 0.9350 |
| 2R/1R | 41 | 6 | 0.71 | 0.20–2.50 | 0.5951 | 1 |
| 1R/1R | 18 | 1 | 2.54 | 0.30–21.12 | 0.6850 | 1 |
| 1R/0R | 7 | 0 | Infinity | 0.23–infinity | 1 | 1 |
Specific Genotypes Associated with Risk of Familial Breast Cancer
Through comparing between control, breast cancer-affected and breast cancer-unaffected groups, we tested odds ratio to identify specific genotypes associated with risk of breast cancer (Table 6). Considering that the 3R and 0R groups contain only few cases in both control and carrier population, we removed 3R/2R, 3R/1R, 3R/0R, 2R/0R, and 0R/0R but focused on the 2R/2R, 2R/1R, 1R/1R, and 1R/0R as they contributed most of the cases. The results showed that
BRCA1+ group. 2R/2R (OR = 1.94, 95%CI = 1.26–2.95, P = 0.0096) and 2R/1R (OR = 1.58, 95%CI = 1.11–2.26, P = 0.0388) were associated with increased risk of breast cancer in cancer-affected BRCA1+ group, and 1R/1R (OR = 0.55, 95%CI = 0.35–0.84, P = 0.0196) and 1R/0R (OR = 0, 95%CI = 0–0.29, P = 0.0012) were associated with the decreased risk in cancer-affected BRCA1+ group;
BRCA2+ group. 2R/1R (OR = 1.94, 95%CI = 1.14–3.32, P = 0.0242) was associated with increased risk in cancer-affected BRCA2+ group. 2R/2R, 1R/1R, and 1R/0R had no association with the risk in cancer-affected BRCA2+ group;
BRCAx group. 2R/2R, 2R/1R, 1R/1R, and 1R/0R had no association with the risk of breast cancer in breast cancer-affected BRCAx group.
Table 6.
Association of VNTR genotypes in XRCC5 promoter with familial breast cancer-affected and -unaffected groups.
| Genotype | Control | Affected case | Odds ratio | 95% CI | P-value | Adjusted | Unaffected case | Odds ratio | 95% CI | P-value | Adjusted |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BRCA1+ | |||||||||||
| 2R/2R | 100 | 45 | 1.94 | 1.26–2.95 | 0.0016 | 0.0096 | 16 | 2.09 | 1.05–3.97 | 0.0221 | 0.1326 |
| 2R/1R | 255 | 87 | 1.58 | 1.11–2.26 | 0.0087 | 0.0388 | 26 | 1.25 | 0.69–2.23 | 0.3190 | 0.4785 |
| 1R/1R | 190 | 32 | 0.55 | 0.35–0.84 | 0.0049 | 0.0196 | 13 | 0.69 | 0.33–1.35 | 0.2906 | 0.4978 |
| 1R/0R | 38 | 0 | 0 | 0–0.29 | 0.0001 | 0.0012 | 1 | 0.28 | 0.01–1.73 | 0.2414 | 0.5794 |
| BRCA2+ | |||||||||||
| 2R/2R | 100 | 17 | 1.75 | 0.97–3.15 | 0.0865 | 0.1038 | 10 | 2.82 | 1.13–6.58 | 0.0174 | 0.2088 |
| 2R/1R | 255 | 39 | 1.94 | 1.14–3.32 | 0.0101 | 0.0242 | 12 | 1.05 | 0.45–2.38 | 1 | 1 |
| 1R/1R | 190 | 13 | 0.54 | 0.27–1.04 | 0.0680 | 0.0907 | 4 | 0.37 | 0.09–1.11 | 0.0632 | 0.1896 |
| 1R/0R | 38 | 0 | 0 | 0–0.73 | 0.0427 | 0.0641 | 2 | 1.16 | 0.13–4.93 | 0.6916 | 0.7545 |
| BRCAx | |||||||||||
| 2R/2R | 100 | 20 | 1.55 | 0.90–2.67 | 0.1101 | 1 | 3 | 2.01 | 0.52–7.69 | 0.3945 | 0.5257 |
| 2R/1R | 255 | 41 | 1.27 | 0.84–1.82 | 0.2882 | 0.3096 | 6 | 1.79 | 0.54–5.92 | 0.3654 | 0.5481 |
| 1R/1R | 190 | 18 | 0.59 | 0.35–1.02 | 0.0583 | 0.3168 | 1 | 0.23 | 0.03–1.84 | 0.1885 | 0.5655 |
| 1R/0R | 38 | 7 | 1.34 | 0.58–3.10 | 0.4913 | 1 | 0 | 0 | 0–4.75 | 1 | 1 |
Discussion
Gene regulatory regions have long been considered a potential source of “missing heritability” in cancer (35, 36). Our study provides evidence showing that VNTR polymorphisms in the XRCC5 promoter is associated with risk of familial breast cancer with BRCA1+ and BRCA2+ predisposition. Data from sporadic breast cancer showed that 2R/1R was not associated (OR = 1.09, 95%CI = 0.78–1.53, P = 0.595), but 0R/0R was associated with the disease (OR = 9.55, 95%CI = 1.19–76.6, P = 0.034) (31). The different results suggest that the association of VNTR polymorphisms in the XRCC5 promoter differs between familial breast cancer and sporadic breast cancer.
For the BRCA1+ and BRCA2+ groups, the results can be explained by synergistic roles between Ku80 and BRCA1/BRCA2 in maintaining genome stability through the NHRJ and HR pathways (37–39). Altered expression of Ku80 can disturb the synergy resulting in increased breast cancer risk in BRCA mutation carriers. The results also suggest that genotype 1R/1R and 1R/0R can reduce the risk of breast cancer in BRCA1+ carriers. Based on current knowledge, it is difficult to relate VNTR polymorphisms to BRCAx familial breast cancer, as genetic predisposition in this heterogeneous group of familial breast cancer remains to be determined. We did not observe a relationship between age at onset of disease, cancer status, and VNTR polymorphism. This could be due to the weaker influence by the VNTR polymorphism compared with that of the BRCA mutation predisposition. Alternatively, it could be due to the limited sample size used in the study, which restricts the statistical power to detect the potential significance. Further studies with larger sample size will help to address the issue.
In summary, our study indicates that 2R/2R and 2R/1R were significantly associated with increased risk, and 1R/1R and 1R/0R were significantly associated with the decreased risk of BRCA1+ breast cancer, whereas 2R/1R was significantly associated with the increased risk of BRCA2+ breast cancer.
Author Contributions
JC and BD performed the genotyping and Sanger sequencing experiments; YK analyzed the genotyping data; JC and JL performed statistical analyses; CS, DB, and HL recruited breast cancer cases and extracted genomic DNA; and SW designed the study, interpreted the data, and drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declared that the research was conducted in the absence of any commercial and financial relationships that could be constructed as a potential conflict of interest.
Acknowledgments
This study was supported by NIH grant R21CA180008 (SW), the Chinese Scholar Council fellowship (JC), the Charles F. and Mary C. Heider Chair in Cancer Research, and by revenue from Nebraska cigarette taxes awarded to Creighton University by the Nebraska Department of Health and Human Services (HL). The Nebraska Biobank of University of Nebraska Medical Center provided the DNA samples from control individuals; the Hereditary Cancer Center of the Creighton University provided breast cancer samples. Written informed consent was obtained from the patient/participant (delete as appropriate) for publication of their individual details and accompanying images in this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fonc.2016.00092
Abbreviations
BRCA1, breast cancer 1, early onset; BRCA2, breast cancer 2, early onset; BRCAx, familial breast cancer with wild type BRCA1 and BRCA2; HR, homologous recombination; NHEJ, non-homologous end joining; PAGE, polyacrylamide gel electrophoresis; VNTR, variable number tandem repeat.
References
- 1.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet (2008) 40:17–22. 10.1038/ng.2007.53 [DOI] [PubMed] [Google Scholar]
- 2.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science (1990) 250:1684–9. 10.1126/science.2270482 [DOI] [PubMed] [Google Scholar]
- 3.Narod SA, Feunteun J, Lynch HT, Watson P, Conway T, Lynch J, et al. Familial breast-ovarian cancer locus on chromosome 17q12-q23. Lancet (1991) 388:82–3. 10.1016/0140-6736(91)90076-2 [DOI] [PubMed] [Google Scholar]
- 4.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science (1994) 266:66–71. 10.1126/science.7545954 [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet (1999) 22:37–43. 10.1038/8743 [DOI] [PubMed] [Google Scholar]
- 6.Kanaar R, Hoeijmakers JH, van Gent DC. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol (1998) 8:483–9. 10.1016/S0962-8924(98)01383-X [DOI] [PubMed] [Google Scholar]
- 7.Rothkamm K, Krüger I, Thompson LH, Löbrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol (2003) 23:5706–15. 10.1128/MCB.23.16.5706-5715.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mérel P, Prieur A, Pfeiffer P, Delattre O. Absence of major defects in non-homologous DNA end joining in human breast cancer cell lines. Oncogene (2002) 21:5654–9. 10.1038/sj.onc.1205742 [DOI] [PubMed] [Google Scholar]
- 9.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature (2000) 404:510–4. 10.1038/35006670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couëdel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, et al. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev (2004) 18:1293–304. 10.1101/gad.1209204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res (2013) 2:130–43. 10.3978/j.issn.2218-676X.2013.04.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev (2001) 15:3237–42. 10.1101/gad.946401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell (2012) 151:1474–87. 10.1016/j.cell.2012.11.054 [DOI] [PubMed] [Google Scholar]
- 14.Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity (1997) 7:653–65. 10.1016/S1074-7613(00)80386-6 [DOI] [PubMed] [Google Scholar]
- 15.Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, et al. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science (1994) 265:1442–5. 10.1126/science.8073286 [DOI] [PubMed] [Google Scholar]
- 16.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol (2004) 24:9305–16. 10.1128/MCB.24.21.9305-9316.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gullo C, Au M, Feng G, Teoh G. The biology of Ku and its potential oncogenic role in cancer. Biochim Biophys Acta (2006) 1765:223–34. 10.1016/j.bbcan.2006.01.001 [DOI] [PubMed] [Google Scholar]
- 18.Lim JW, Kim H, Kim KH. Expression of Ku70 and Ku80 mediated by NF-kappa B and cyclooxygenase-2 is related to proliferation of human gastric cancer cells. J Biol Chem (2002) 277:46093–100. 10.1074/jbc.M206603200 [DOI] [PubMed] [Google Scholar]
- 19.Chang HW, Kim SY, Yi SL, Son SH, Song do Y, Moon SY, et al. Expression of Ku80 correlates with sensitivities to radiation in cancer cell lines of the head and neck. Oral Oncol (2006) 42:979–86. 10.1016/j.oraloncology.2005.12.016 [DOI] [PubMed] [Google Scholar]
- 20.Pucci S, Mazzarelli P, Rabitti C, Giai M, Gallucci M, Flammia G, et al. Tumor specific modulation of KU70/80 DNA binding activity in breast and bladder human tumor biopsies. Oncogene (2001) 20:739–47. 10.1038/sj.onc.1204148 [DOI] [PubMed] [Google Scholar]
- 21.Mazzarelli P, Parrella P, Seripa D, Signori E, Perrone G, Rabitti C, et al. DNA end binding activity and Ku70/80 heterodimer expression in human colorectal tumor. World J Gastroenterol (2005) 11:6694–700. 10.3748/wjg.v11.i42.6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alshareeda AT, Negm OH, Albarakati N, Green AR, Nolan C, Sultana R, et al. Clinicopathological significance of KU70/KU80, a key DNA damage repair protein in breast cancer. Breast Cancer Res Treat (2013) 139:301–10. 10.1007/s10549-013-2542-x [DOI] [PubMed] [Google Scholar]
- 23.Nakamura Y, Koyama K, Matsushima M. VNTR (variable number of tandem repeat) sequences as transcriptional, translational, or functional regulators. J Hum Genet (1998) 43:149–52. 10.1007/s100380050059 [DOI] [PubMed] [Google Scholar]
- 24.Brookes KJ. The VNTR in complex disorders: the forgotten polymorphisms? A functional way forward? Genomics (2013) 101:273–81. 10.1016/j.ygeno.2013.03.003 [DOI] [PubMed] [Google Scholar]
- 25.Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet (2016) 48(1):22–9. 10.1038/ng.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig DL, Chen F, Peterson SR, Nussenzweig A, Li GC, Chen DJ. Ku80 gene expression is Sp1-dependent and sensitive to CpG methylation within a novel cis element. Gene (1997) 199:181–94. 10.1016/S0378-1119(97)00366-1 [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Wang M, Yin S, Fu G, Li C, Chen R, et al. A novel variable number of tandem repeats (VNTR) polymorphism containing Sp1 binding elements in the promoter of XRCC5 is a risk factor for human bladder cancer. Mutat Res (2008) 638:26–36. 10.1016/j.mrfmmm.2007.08.011 [DOI] [PubMed] [Google Scholar]
- 28.Rajaei M, Saadat I, Saadat M. Introducing a novel allele for the polymorphism of variable number of tandem repeats in the promoter region of XRCC5. Biochem Biophys Res Commun (2012) 427:503–5. 10.1016/j.bbrc.2012.09.085 [DOI] [PubMed] [Google Scholar]
- 29.Rajaei M, Saadat I, Saadat M. The novel allele (3R) of the VNTR polymorphism in the XRCC5 promoter region dramatically decreases the gene expression. Biochem Biophys Res Commun (2013) 430:640–1. 10.1016/j.bbrc.2012.11.099 [DOI] [PubMed] [Google Scholar]
- 30.Saadat M, Pashaei S, Amerizade F. Susceptibility to gastric cancer and polymorphisms of insertion/deletion at the intron 3 of the XRCC4 and VNTR at the promoter region of the XRCC5. Pathol Oncol Res (2015) 21:689–93. 10.1007/s12253-014-9875-6 [DOI] [PubMed] [Google Scholar]
- 31.Monsees GM, Kraft P, Chanock SJ, Hunter DJ, Han J. Comprehensive screen of genetic variation in DNA repair pathway genes and postmenopausal breast cancer risk. Breast Cancer Res Treat (2011) 125:207–14. 10.1007/s10549-010-0947-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajaei M, Saadat I, Omidvari S, Saadat M. Association between polymorphisms at promoters of XRCC5 and XRCC6 genes and risk of breast cancer. Med Oncol (2014) 31:885. 10.1007/s12032-014-0885-8 [DOI] [PubMed] [Google Scholar]
- 33.Agresti A. Categorical Data Analysis. 3rd ed Hoboken, NJ: John Willey & Sons; (2013). [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol (1995) 57:289–300. [Google Scholar]
- 35.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science (2012) 337:1190–5. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet (2014) 46:1160–5. 10.1038/ng.3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei L, Lan L, Hong Z, Yasui A, Ishioka C, Chiba N. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol Cell Biol (2008) 28:7380–93. 10.1128/MCB.01075-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang G, Plo I, Wang T, Rahman M, Cho JH, Yang E, et al. BRCA1-Ku80 protein interaction enhances end-joining fidelity of chromosomal double-strand breaks in the G1 phase of the cell cycle. J Biol Chem (2013) 288:8966–76. 10.1074/jbc.M112.412650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA (2015) 313:1347–61. 10.1001/jama.2014.5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.