Highlights
-
•
Pregnancy vitamin D and cord blood DNA methylation compared in 1416 infants.
-
•
Associations tested in two large prospective cohort studies, MoBa and ALSPAC.
-
•
Similarly for offspring health outcomes and vitamin D, no convincing associations.
Keywords: DNA methylation, Epigenetics, Vitamin D, 2-Hydroxyvitamin D, Maternal vitamin D, Methylome, Offspring
Abstract
The aim of the study was to investigate whether maternal mid-pregnancy 25-hydroxyvitamin D concentrations are associated with cord blood DNA methylation.
DNA methylation was assessed using the Illumina HumanMethylation450 BeadChip, and maternal plasma 25-hydroxyvitamin D was measured in 819 mothers/newborn pairs participating in the Norwegian Mother and Child Cohort (MoBa) and 597 mothers/newborn pairs participating in the Avon Longitudinal Study of Parents and Children (ALSPAC).
Across 473,731CpG DNA methylation sites in cord blood DNA, none were strongly associated with maternal 25-hydroxyvitamin D after adjusting for multiple tests (false discovery rate (FDR) > 0.5; 473,731 tests). A meta-analysis of the results from both cohorts, using the Fisher method for combining p-values, also did not strengthen findings (FDR > 0.2). Further exploration of a set of CpG sites in the proximity of four a priori defined candidate genes (CYP24A1, CYP27B1, CYP27A1 and CYP2R1) did not result in any associations with FDR < 0.05 (56 tests). In this large genome wide assessment of the potential influence of maternal vitamin D status on DNA methylation, we did not find any convincing associations in 1416 newborns. If true associations do exist, their identification might require much larger consortium studies, expanded genomic coverage, investigation of alternative cell types or measurements of 25-hydroxyvitamin D at different gestational time points.
1. Introduction
Vitamin D is a precursor of the steroid hormone 1,25-dihydroxyvitamin D (1,25(OH)2D), with important roles in calcium and bone metabolism as well as other biological processes. A number of different tissues express the vitamin D receptor (VDR), which acts as a transcriptional factor after binding of 1,25(OH)2D and heterodimerisation with retinoic X receptor (RXR). The major circulating form and indicator of vitamin D status, 25-hydroxyvitamin D (25(OH)D), is supplied by 25-hydroxylation of vitamin D produced in the skin upon UVB radiation or vitamin D from the diet [1]. 25(OH)D is activated in a second hydroxylation step, by 1α-hydroxylase (encoded by CYP27B1), primarily in the kidneys, but also in other tissues expressing CYP27B1 including lymphocytes [1], [2].
Low 25(OH)D levels have been associated with a range of adverse conditions, from pregnancy outcomes to childhood illnesses and chronic disease including osteoporosis, cancer and cardiovascular disease in adulthood [3], although randomized controlled trials of vitamin D supplements do not support causality for extra-skeletal outcomes [4], [5].
Vitamin D metabolism changes during pregnancy, suggesting importance for the mother and fetus [6]. For instance, while circulating 1,25(OH)2D is normally tightly controlled by renal hydroxylation of 25(OH)D, levels increase during pregnancy. In addition to increased renal expression of CYP27B1, this may also partly result from placental expression of CYP27B1 combined with reduced activity of CYP24A1, which catalyzes the first step of the catabolism of 1,25(OH)2D.
Lower maternal 25(OH)D during pregnancy has been associated with a number of adverse perinatal outcomes, such as low birth weight and preterm birth and also later health outcomes in the offspring such as bone health [7], wheezing and atopic disorders [8], and autoimmune disorders like type 1 diabetes [9]. Although a number of studies have reported inverse associations between maternal vitamin D status and postnatal health outcomes, systematic reviews show that there is still substantial heterogeneity between studies in terms of methodology and results, and few or no randomized trials have been performed [6], [8], [10], [11]. Although two recent randomized controlled trials observed suggestive reductions in the incidence of asthma and recurrent wheezing following vitamin D supplementation during pregnancy [12], [13], the primary endpoints were not statistically significant. The evidence for causality of these associations therefore remains largely inconclusive.
In addition to a need for large randomized studies in this field, there is also a need to explore potential mechanisms involved in the hypothesized links between maternal vitamin D status and offspring health. It is well established that maternal 25(OH)D during pregnancy is correlated with cord blood 25(OH)D, but it is possible that some of the observed associations with long term health outcomes may be mediated by fetal programming mechanisms such as DNA methylation in the fetal genome [14]. The activated vitamin D receptor has a large number of potential target genes, identified both experimentally using in vitro models and in silico by identification of vitamin D responsive elements [15]. Potential target genes included some well-established candidates, such as CYP27B1 and CYP24A1, and a large number of yet unconfirmed genes.
A few smaller studies have investigated DNA methylation at some candidate loci in relation to vitamin D [16]. One recent study examined the association between vitamin D deficiency and genome wide DNA methylation in African children [17]. Another study explored epigenetic regulation of vitamin D converting enzymes [18], while a third found a relationship between methylation of the genes CYP24R1 and CYP27A1 and variations in circulating 25(OH)D levels [19]. Epigenome wide studies, in general, are gaining in popularity with the use of the Illumina HumanMethylation450 BeadChip. We are not aware of any published studies on the association between maternal 25(OH)D and genome wide DNA methylation in cord blood. We assessed this association using the Illumina HumanMethylation450 assay in 1416 newborns from two large pregnancy cohort studies: The Norwegian Mother and Child Cohort Study (MoBa) [20] and the Avon Longitudinal Study of Parents and Children (ALSPAC) from the UK [21].
2. Materials and methods
2.1. The Norwegian Mother and Child Cohort Study (MoBa)
Study population and sample acquisition. The Norwegian Mother and Child Cohort Study (MoBa) is a population-based pregnancy cohort administered by the Norwegian Institute of Public Health (NIPH) [20], [22], [23].
Pregnant women were recruited between 1999 and 2008 from 50 of the 52 hospitals in Norway when attending the routine ultrasound examination at approximately 18 weeks of gestation (98% coverage of all pregnant women). The overall participation rate for MoBa was 41% [20]. Blood samples were drawn from the mother and blood from the umbilical cord vein was collected with a syringe. The handling and quality assurance of the biological material has been thoroughly described previously [24].
The present study is based on data from MoBa version VI (108,863 children in total) with linkage to the Medical Birth Registry of Norway (MBR). Participants constituted two subgroups. The first included a sample among those born between July 2002 and December 2003 with completed questionnaires at 18–22 weeks gestation (n = 17,005 eligible children). The second subgroup included children born between July 2002 and July 2004 with completed questionnaires up to 36 months who were classified as having asthma at 36 months. From these two groups, there were 819 children with maternal 25-hydroxyvitamin D levels and cord blood DNA methylation profiles [25]. The study was approved by the Norwegian Data Inspectorate and the Regional Ethics Committee for Medical Research.
25-Hydroxyvitamin D levels. Maternal plasma levels of 25-hydroxyvitamin D3 and 25-hydroxyvitamin D2 were analyzed at Bevital laboratories (www.Bevital.no), using a liquid chromatography-tandem mass spectrometry method (LC–MS/MS) [25]. The within day coefficient of variance for 25-hydroxyvitamin D2 was 4.3–4.5%, while the between day coefficient of variance was 4.6–7.7%. The within day coefficient of variance for 25-hydroxyvitamin D3 was 4.4–5.3%, while the between day coefficient of variance was 7.3–8.2%. The contribution of 25-hydroxyvitamin D2 in the study sample was negligible, and the sum of 25-hydroxyvitamin D3 and -D2, termed 25(OH)D, was used in the analysis.
DNA methylation profile generation. Cord blood DNA methylation was assayed using the Illumina Infinium HumanMethylation450 BeadChip (www.illumina.com), which was designed to conduct epigenome-wide association studies (EWAS) and includes 485,512 methylation sites per sample at single-nucleotide resolution. This chip covers 99% of RefSeq genes, with an average of 17CpG sites per gene region across the promoter, 5′UTR, first exon, gene body, and 3′UTR. In addition, the chip covers 96% of CpG islands, with additional coverage of island shores. DNA methylation levels of CpG sites is detected using bisulfite-converted genomic DNA (gDNA), where unmethylated (Un) cytosine bases are converted to uracil, while methylated (Me) cytosine remains unchanged. Bisulfite conversion was performed using the EZ-96 DNA Methylation kit (Zymo Research Corporation, Irvine, CA) according to manufacturer instructions. Illumina reports an average Beta value for the methylation level of the interrogated sites based on the following formula: βij = Max(Meij, 0)/(Max (Meij, 0) + Max (Unij, 0) + 100), for each person j and each of the 485,512 CpG sites i. Batch effects in these analyses were avoided as all samples were analyzed on the same day, by the same individual, using the same instrument. As previously described, chip, chip set and plate were not appreciable sources of variability [26], [27], so they were not included as covariates in regression models. BMIQ was performed on the methylation data to assimilate type I and II probes [28], [29]. More information regarding quality control of the MoBa cohort dataset can be found elsewhere [26], [27].
Statistical analyses. MM type robust linear regression [30] was carried out with CpG values (0 ≤ β ≤ 1) as outcome and plasma 25(OH)D as the explanatory variable. MM type regression is very robust to outliers (breakdown point when 50% incorrect observations compared to 1% for ordinary least squares). It is superior to robust methods using sandwich estimators because it adjusts both coefficient estimates and standard errors rather than just standard errors. The 25(OH)D explanatory variable was approximately normally distributed. All p-values and regression estimates reported are for 25(OH)D in nmol/l as a continuous variable, categorizing the 25(OH)D variable into quartiles made no difference. In the primary analysis, the models were adjusted for maternal pre-pregnancy body mass index (BMI, continuous), offspring sex, maternal education (4 categories), maternal smoking (yes/no), maternal folate plasma values (continuous), parity (3 categories), maternal age (continuous), birth season (four categories) and estimated cell type proportions (continuous matrix consisting of 6 (default) blood cell-types: CD4+ T cells, CD8+ T cells, NK-cells, B-cells, monocytes and granulocytes). The cell-type estimations were calculated with the minfi package [31], which is based on the method described by Houseman et al. [32]. In a sensitivity analysis, we ran additional models adjusting for various subsets of the covariates mentioned above:
Model 1. None
Model 2. Maternal Age + Folate + Offspring gender + Houseman cell counts
Model 3. Model 2 + Season
Model 4. Model 2 + Maternal education + Maternal smoking + Parity + Maternal BMI
Model 5. Model 4 + Season
We refer to the first as the ‘crude’ model and the last as the ‘full’ model.
We used the genomic control (λGC) [33] to assess model quality and all models tested were found to have λGC close to one. A λGC close to one suggests that the assumption of independent and identically distributed tests is fulfilled indicating that the less conservative FDR-based q-values [34] can be used to assess significance in the genome-wide models. Since none of the models produced any associations with FDR < 0.05 and concordant effect sizes, only the results from the crude analysis including no covariates are presented. The meta-analysis was performed using the Fisher method [35] on all the (2×) 473,731 p-values from both MoBa and ALSPAC crude models and FDR adjusted for multiple testing.
2.2. The Avon Longitudinal Study of Parents and Children study (ALSPAC)
Study population and sample acquisition. This study used DNA methylation data generated under the auspices of the Avon Longitudinal Study of Parents and Children (ALSPAC) [21], [36], [37]. DNA extracted from cord blood and peripheral blood samples along with a wide range of exposure and phenotypic data were used.
25-Hydroxyvitamin D levels. Approximately a quarter of the 25(OH)D samples were collected in each of the first two trimesters and half in the third trimester of pregnancy. Because 25(OH)D levels are known to fluctuate during the year (season) and perhaps by gestational week, 25(OH)D were pre-adjusted for season and gestational age at blood sample collection as previously described [38].
DNA methylation profile generation. Cord blood DNA methylation was assayed using the Illumina HumanMethylation450 platform and data pre-processed using procedures identical to those used for the MoBa dataset. Bisulfite conversion was performed using the EZ DNA Methylation kit (Zymo Research Corporation, Irvine, CA) according to manufacturer instructions. All steps were performed at the University of Bristol as part of the Accessible Resource for Integrated Epigenomic Studies (ARIES) project (http://www.ariesepigenomics.org.uk). During the data generation process a wide range of batch variables were recorded in a purpose-built laboratory information management system (LIMS). The LIMS also reported QC metrics from the standard control probes on the 450 K BeadChip. Samples failing quality (samples with >20% probes with p-value > = 0.01) were repeated. Samples from all three time points in ARIES were randomized across arrays to minimize the potential for batch effects. As an additional QC step, genotype probes on the 450 K BeadChip were compared between samples from the same individual and against SNP-chip data to identify and remove any sample mismatches. The ALSPAC samples were not analyzed in a single day as were the MoBa samples. Consequently, the dataset was normalized using an alternative approach optimized to minimize the effects of resulting technical artefacts. Specifically, data normalization included background correction and subset quantile normalization using the pipeline described by Touleimat and Tost [39] and implemented in the watermelon R package [29].
2.3. Statistical analyses
Associations between DNA methylation and 25(OH)D were tested using procedures and covariate subsets identical to those used for the MoBa study. The only exception was that maternal plasma folate was omitted as it has not been measured in ALSPAC. Due to potentially lingering batch effects present following normalization, additional analyses were performed that included covariates generated using independent surrogate variable analysis (ISVA) [40]. Two versions were considered called ‘isva0’ and ‘isva1’. In ‘isva0’, ISVA was applied to the 25(OH)D levels and DNA methylation data. In ‘isva1’, ISVA was applied as in ‘isva0’ but additionally all covariates, a batch variable (sample plate), and all ‘isva0’ surrogate variables were included as input for generating surrogate variables. The results ‘isva0’/’isva1’ were meta-analyzed with results from the crude/full MoBa models.
3. Results
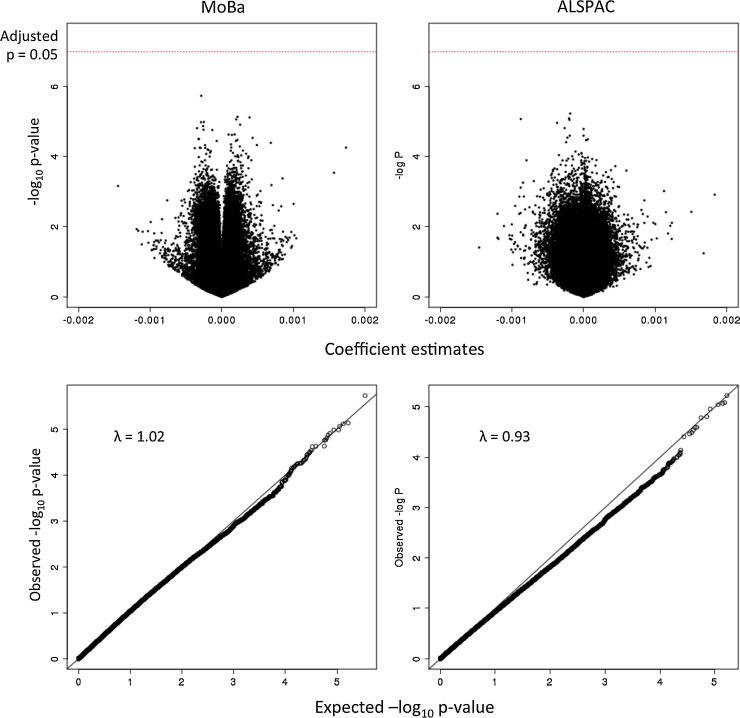
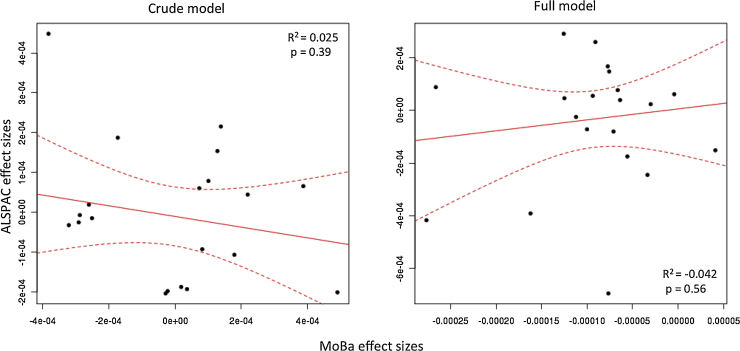
There was no association between mid-pregnancy 25(OH)D and cord blood DNA methylation at any single site on the Illumina HumanMethylation450 BeadChip among the 819 mother and child-pairs in MoBa and the 597 mother and child-pairs in ALSPAC (at FDR < 0.05; 473,731 tests). Adjustment for potential confounding variables (See Table 1, as well as the Materials and Methods section) and cell-type estimations as well as a meta-analysis, based on Fisher's method, comprising results from both MoBa and ALSPAC cohorts did not result in any association between maternal 25(OH)D levels and DNA methylation in offspring (at FDR < 0.05; 473,731 tests). The 1000 strongest associations are provided in Supplementary Information file 1. Regression estimates tended to be very small, and all p-values were greater than 0.05 after correction for multiple tests (FDR > 0.2, Bonferroni adjusted p > 0.2, 473731 tests). Fig. 1 shows QQ- and volcano plots for both MoBa and ALSPAC cohorts based on coefficient estimates and p-values for 473731CpG probes. Fig. 2 shows the lack of agreement in effect sizes in MoBa and ALSPAC for the top 20 meta-analyzed associations. Information regarding maternal circulating 25(OH)D levels and other covariates used throughout the study can be found in Table 1.
Table 1.
Population characteristics.
| MoBa | ALSPAC | |
|---|---|---|
| Number of participants used | 819 | 597 |
| Maternal age (years) | ||
| Mean ± SD | 29.9 (4.33) | 30.1 (4.38) |
| Maternal mid-pregnancy 25-hydroxy vitamin D (ng/mL) | ||
| Mean ± SD | 73.5 (23.42) | 68.3 (32.3) |
| Maternal folate levels (nmol/L) | ||
| Mean +/− SD | 12.0 (7.99) | N/A |
| Maternal pre-pregnancy BMI (kg/m2) | ||
| Median ± SD | 23.2 (4.15) | 22.9 (3.71) |
| Maternal education | ||
| Low | 61 | 52 (CSE) |
| High school | 258 | 52 (Vocational) |
| College | 367 | 198 (O level) |
| University | 129 | 172 (A level) |
| 114 (Degree) | ||
| Maternal smoking during pregnancy | ||
| Yes | 110 | 88 |
| No | 702 | 463 |
| Maternal parity | ||
| First child | 346 | 275 |
| Second child | 333 | 215 |
| Third or more | 140 | 89 |
| Offspring sex | ||
| Male | 449 | 288 |
| Female | 370 | 309 |
| Offspring asthma at 3 yrs | ||
| Yes | 328 | 81 (wheezing) |
| No | 491 | 516 |
| Birth season | ||
| Feb–Apr | 267 | 121 |
| May–Jul | 172 | 161 |
| Aug–Oct | 136 | 181 |
| Nov–Jan | 244 | 134 |
Fig. 1.
Association statistics consistent with null distributions. Volcano- and qq-plots for regression models having offspring methylation betas as the response variable and maternal vitamin D levels as the explanatory variable for both MoBa (left) and ALSPAC (rights) cohorts.
Fig. 2.
Study effect sizes not correlated between MoBa and ALSPAC. Each scatterplot shows the effect sizes of the top 20 meta-analyzed associations. The left plot shows the effect sizes for the crude model (no covariates), and the right plot shows those for the full model. The dashed lines mark the 95% confidence interval for the regression line.
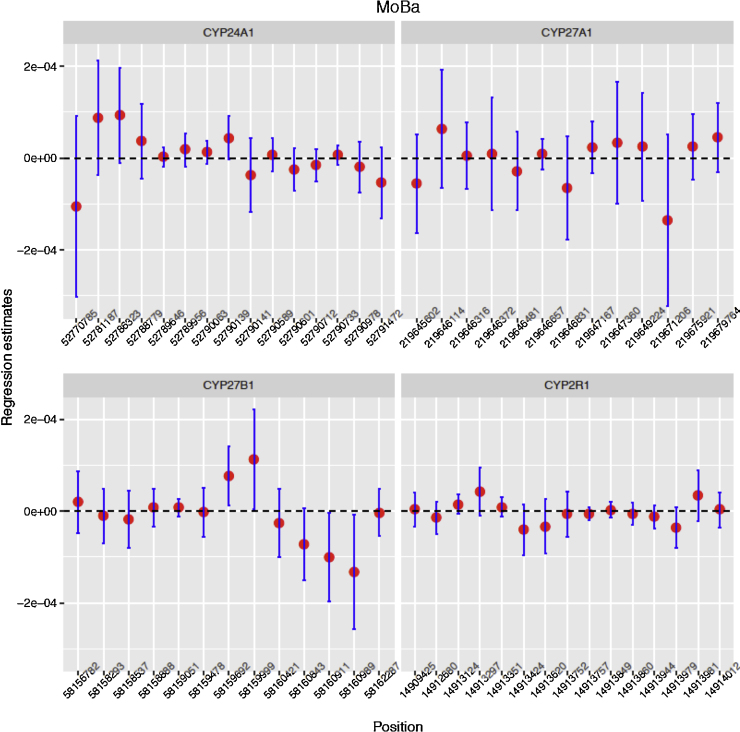
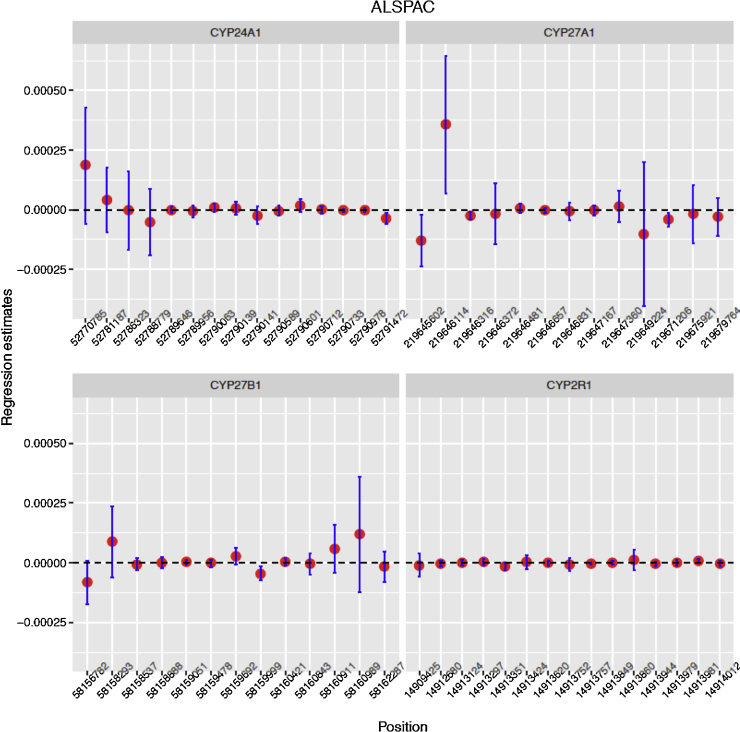
Furthermore, detailed analysis of CpG’s linked to the four a priori defined candidate genes (CYP24A1, CYP27B1, CYP27A1 and CYP2R1) yielded weak associations with very small regression coefficient estimates (Fig. 3; FDR > 0.6 for 56 tests; see Supplementary information File 2). Repeating the analysis within the ALSPAC cohort, there were similarly only weak associations between maternal 25(OH)D and cord blood DNA methylation among 597 mother and child pairs both at the genome wide and at the four candidate loci (Fig. 4).
Fig. 3.
MoBa regression coefficient estimate confidence intervals typically contain zero. Estimated slope between DNA methylation levels of offspring CpG sites and maternal vitamin D levels from the crude model for 4 candidate genes in the MoBa study. The horizontal axis represents the genomic positions of the CpGs, while the vertical axes represent slope estimates. The vertical lines represent ±1.96 × estimated standard error for the regression coefficient.
Fig. 4.
ALSPAC regression coefficient estimate confidence intervals typically contain zero. Estimated slope between methylation levels of offspring CpG sites and maternal vitamin D levels from the crude model for 4 candidate genes in the ALSPAC study. The horizontal axis represents the genomic positions of the CpGs, while the vertical axes represent slope estimates. The vertical lines represent ±1.96 × estimated standard error for the regression coefficient.
4. Discussion
We explored the potential influence of maternal mid-pregnancy 25(OH)D on fetal DNA methylation using the Illumina 450 K BeadChip. Despite the existing hypothesis that maternal vitamin D status may influence offspring health [41], we found no evidence for any DNA methylation based effect in cord blood, either genome-wide or in four candidate genes.
The strength of our study is the large sample size from two well-characterized cohorts with 25(OH)D status and genome-wide Illumina HumanMethylation450k data available for a total of 1416 mother and child-pairs. Despite the fact that the Illumina HumanMethylation450 BeadChip only covers 485512CpG sites out of a possible ∼28 million, most of the targeted sites are found in the promoter region [42] which is the predominant region reported to influence gene expression with regards to circulating 25(OH)D levels [19], [43], [44]. Epigenetic effects of maternal vitamin D levels on offspring methylomes can nevertheless not be excluded; the CpG’s in the neighborhood of the candidate genes, as mapped by the Illumina HumanMethylation450 BeadChip, may not be the same as the ones reported from other studies. Although the sample size is relatively large compared to previous DNA methylation studies, it may possibly be too small to detect weak epigenetic effects.
Our analysis in both cohorts was limited to cord blood DNA methylation. It is possible that DNA methylation levels are strongly affected by maternal 25(OH)D levels in some other tissue. Analysis was complicated by the fact that cord blood is composed of several different fluctuating cell types, each with their own distinct DNA methylation profiles. We attempted to control for this by including estimates of cell type proportions in regression models [32]. Although this approach is not ideal, it is currently the only feasible solution [45]. We did not observe strong associations with 25(OH)D using regression models that included nor excluded cell count estimates.
We are not aware of any previously published studies of maternal 25(OH)D and cord blood DNA methylation. However, a few other studies have explored relationships between vitamin D supplementation or circulating 25(OH)D and methylation at a few CpG sites in two to four candidate genes in adults [18], [46], and one genome-wide DNA methylation study explored association with vitamin D deficiency in African children [17].
Given that vitamin D status fluctuates throughout pregnancy, it is possible that the fetus is more sensitive to vitamin D levels at certain gestational periods [6]. In the MoBa study, measurements were taken around 18 weeks gestation, and in the ALSPAC study they were measured throughout pregnancy but normalized to 28 weeks gestation. It is therefore possible that measurements at other time points might have provided stronger associations with cord blood DNA methylation. Hopefully future studies will be designed to systematically investigate timing.
5. Conclusions
We found no strong associations between DNA methylation in neonatal genomes and maternal plasma 25(OH)D concentration. Further scrutiny of a set of specific candidate genes did not indicate any association. Our results suggest that similarly powered studies of maternal 25(OH)D in relation to cord blood DNA methylation with the Illumina HumanMethylation450 BeadChip will be unlikely to identify true associations, if they exist. Any future study should utilize DNA methylation profiles of alternative cell types, expanded genomic coverage, larger sample sizes, or measurements of 25(OH)D at different gestational time points.
Acknowledgements
For MoBa this research was supported [in part] by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES-49019). The Norwegian Mother and Child Cohort Study is supported by the Norwegian Ministry of Health and the Ministry of Education and Research, NIH/NIEHS (contract no. N01-ES-75558), NIH/NINDS (grant no. 1 UO1 NS 047537-01) and the Norwegian Research Council/FUGE (grant no. 151918/S10), and the present study by the Norwegian Research Council/Human Biobanks and Health (grant number 221097). We are grateful to all families participating in the Norwegian Mother and Child Cohort Study.
ARIES was funded by the BBSRC (BBI025751/1 and BB/I025263/1). Core programme support for ALSPAC is provided by the Medical Research Council (MRC) and the Wellcome Trust (Grant ref: 102215/2/13/1) and the University of Bristol. ARIES is maintained under the auspices of the MRC Integrative Epidemiology Unit at the University of Bristol (MC_UU_12013/2 and MC_UU_12013/8). We are grateful to all the families who took part in the ALSPAC study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsbmb.2016.03.005.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Berry D., Hyppønen E. Determinants of vitamin D status: focus on genetic variations. Curr. Opin. Nephrol. Hypertens. 2011;20(4):331–336. doi: 10.1097/MNH.0b013e328346d6ba. [DOI] [PubMed] [Google Scholar]
- 2.Jones G., Strugnell S.A., DeLuca H.F. Current understanding of the molecular actions of vitamin D. Physiol. Rev. 1998;78(4):1193–1231. doi: 10.1152/physrev.1998.78.4.1193. [DOI] [PubMed] [Google Scholar]
- 3.Pludowski P., Holick M.F., Pilz S., Wagner C.L., Hollis B.W., Grant W.B., Shoenfeld Y., Lerchbaum E., Llewellyn D.J., Kienreich K. Vitamin D effects on musculoskeletal health, immunity, autoimmunity, cardiovascular disease, cancer, fertility, pregnancy, dementia and mortality—a review of recent evidence. Autoimmun. Rev. 2013;12(10):976–989. doi: 10.1016/j.autrev.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Theodoratou E., Tzoulaki I., Zgaga L., Ioannidis J.P. Vitamin D and multiple health outcomes: umbrella review of systematic reviews and meta-analyses of observational studies and randomised trials. BMJ. 2014;348:g2035. doi: 10.1136/bmj.g2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouillon R., Van Schoor N.M., Gielen E., Boonen S., Mathieu C., Vanderschueren D., Lips P. Optimal vitamin D status: a critical analysis on the basis of evidence-based medicine. J. Clin. Endocrinol. Metab. 2013;98(8):E1283–1304. doi: 10.1210/jc.2013-1195. [DOI] [PubMed] [Google Scholar]
- 6.Brannon P.M., Picciano M.F. Vitamin D in pregnancy and lactation in humans. Annu. Rev. Nutr. 2011;31:89–115. doi: 10.1146/annurev.nutr.012809.104807. [DOI] [PubMed] [Google Scholar]
- 7.Javaid M.K., Crozier S.R., Harvey N.C., Gale C.R., Dennison E.M., Boucher B.J., Arden N.K., Godfrey K.M., Cooper C. Maternal vitamin D status during pregnancy and childhood bone mass at age 9 years: a longitudinal study. Lancet. 2006;367(9504):36–43. doi: 10.1016/S0140-6736(06)67922-1. [DOI] [PubMed] [Google Scholar]
- 8.Beckhaus A.A., Garcia-Marcos L., Forno E., Pacheco-Gonzalez R.M., Celedon J.C., Castro-Rodriguez J.A. Maternal nutrition during pregnancy and risk of asthma, wheeze, and atopic diseases during childhood: a systematic review and meta-analysis. Allergy. 2015;70(12):1588–1604. doi: 10.1111/all.12729. [DOI] [PubMed] [Google Scholar]
- 9.Sørensen I.M., Joner G., Jenum P.A., Eskild A., Torjesen P.A., Stene L.C. Maternal serum levels of 25-hydroxy-vitamin D during pregnancy and risk of type 1 diabetes in the offspring. Diabetes. 2012;61(1):175–178. doi: 10.2337/db11-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christesen H.T., Elvander C., Lamont R.F., Jørgensen J.S. The impact of vitamin D in pregnancy on extraskeletal health in children: a systematic review. Acta Obstet. Gynecol. Scand. 2012;91(12):1368–1380. doi: 10.1111/aogs.12006. [DOI] [PubMed] [Google Scholar]
- 11.Harvey N.C., Holroyd C., Ntani G., Javaid K., Cooper P., Moon R., Cole Z., Tinati T., Godfrey K., Dennison E. Vitamin D supplementation in pregnancy: a systematic review. Health Technol. Assess. 2014;18(45):1–190. doi: 10.3310/hta18450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chawes B.L., Bonnelykke K., Stokholm J., Vissing N.H., Bjarnadottir E., Schoos A.M., Wolsk H.M., Pedersen T.M., Vinding R.K., Thorsteinsdottir S. Effect of vitamin D3 supplementation during pregnancy on risk of persistent wheeze in the offspring: a randomized clinical trial. JAMA. 2016;315(4):353–361. doi: 10.1001/jama.2015.18318. [DOI] [PubMed] [Google Scholar]
- 13.Litonjua A.A., Carey V.J., Laranjo N., Harshfield B.J., McElrath T.F., O’Connor G.T., Sandel M., Iverson R.E., Jr., Lee-Paritz A., Strunk R.C. Effect of prenatal supplementation with vitamin D on asthma or recurrent wheezing in offspring by age 3 years: the VDAART randomized clinical trial. JAMA. 2016;315(4):362–370. doi: 10.1001/jama.2015.18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanley B., Dijane J., Fewtrell M., Grynberg A., Hummel S., Junien C., Koletzko B., Lewis S., Renz H., Symonds M. Metabolic imprinting, programming and epigenetics—a review of present priorities and future opportunities. Br. J. Nutr. 2010;104(1):S1–S25. doi: 10.1017/S0007114510003338. [DOI] [PubMed] [Google Scholar]
- 15.Carlberg C., Seuter S., Heikkinen S. The first genome-wide view of vitamin D receptor locations and their mechanistic implications. Anticancer Res. 2012;32(1):271–282. [PubMed] [Google Scholar]
- 16.Fetahu I.S., Hobaus J., Kallay E. Vitamin D and the epigenome. Front. Physiol. 2014;5:164. doi: 10.3389/fphys.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H., Wang X., Shi H., Su S., Harshfield G.A., Gutin B., Snieder H., Dong Y. A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. J. Pediatr. 2013;1009:e1001. doi: 10.1016/j.jpeds.2012.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wjst M., Heimbeck I., Kutschke D., Pukelsheim K. Epigenetic regulation of vitamin D converting enzymes. J. Steroid Biochem. Mol. Biol. 2010;121(1–2):80–83. doi: 10.1016/j.jsbmb.2010.03.056. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y., Zhao L.J., Xu X., Ye A., Travers-Gustafson D., Zhou B., Wang H.W., Zhang W., Lee Hamm L., Deng et al H.W. DNA methylation levels of CYP2R1 and CYP24A1 predict vitamin D response variation. J. Steroid Biochem. Mol. Biol. 2014;144(Pt A):207–214. doi: 10.1016/j.jsbmb.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnus P., Irgens L.M., Haug K., Nystad W., Skjaerven R., Stoltenberg C., MoBa Study G. Cohort profile: the Norwegian Mother and Child Cohort Study (MoBa) Int. J. Epidemiol. 2006;35(5):1146–1150. doi: 10.1093/ije/dyl170. [DOI] [PubMed] [Google Scholar]
- 21.Fraser A., Macdonald-Wallis C., Tilling K., Boyd A., Golding J., Davey Smith G., Henderson J., Macleod J., Molloy L., Ness A. Cohort profile: the avon longitudinal study of parents and children: aLSPAC mothers cohort. Int. J. Epidemiol. 2013;42(1):97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsen R.M., Vollset S.E., Gjessing H.K., Skjaerven R., Melve K.K., Schreuder P., Alsaker E.R., Haug K., Daltveit A.K., Magnus P. Self-selection and bias in a large prospective pregnancy cohort in Norway. Paediatr. Perinat. Epidemiol. 2009;23(6):597–608. doi: 10.1111/j.1365-3016.2009.01062.x. [DOI] [PubMed] [Google Scholar]
- 23.Schreuder P., Alsaker E. The Norwegian Mother and Child Cohort Study (MoBa)—MoBa recruitment and logistics. Nor. Epidemiol. 2014;24:1–2. [Google Scholar]
- 24.Rønningen K.S., Paltiel L., Meltzer H.M., Nordhagen R., Lie K.K., Hovengen R., Haugen M., Nystad W., Magnus P., Hoppin J.A. The biobank of the Norwegian Mother and Child Cohort Study: a resource for the next 100 years. Eur. J. Epidemiol. 2006;21(8):619–625. doi: 10.1007/s10654-006-9041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Midttun Ø., Ueland P.M. Determination of vitamins A, D and E in a small volume of human plasma by a high-throughput method based on liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2011;25(14):1942–1948. doi: 10.1002/rcm.5073. [DOI] [PubMed] [Google Scholar]
- 26.Joubert B.R., Haberg S.E., Nilsen R.M., Wang X., Vollset S.E., Murphy S.K., Huang Z., Hoyo C., Midttun O., Cupul-Uicab L.A. 450 K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 2012;120(10):1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M.C., Joubert B.R., Kuan P.F., Haberg S.E., Nystad W., Peddada S.D., London S.J. A systematic assessment of normalization approaches for the Infinium 450 K methylation platform. Epigenetics. 2014;9(2):318–329. doi: 10.4161/epi.27119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pidsley R., Wong C.C.Y., Volta M., Lunnon K., Mill J., Schalkwyk L.C. A data-driven approach to preprocessing Illumina 450 K methylation array data. BMC Genom. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yohai V., Stahel W., Zamar R. vol. 34. Springer; New York: 1991. pp. 365–374. (A Procedure for Robust Estimation and Inference in Linear Regression). [Google Scholar]
- 31.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics. 2016:2014. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J., Weedon M.N., Purcell S., Lettre G., Estrada K., Willer C.J., Smith A.V., Ingelsson E., O’Connell J.R., Mangino M. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 2011;19(7):807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher R.A. Genesis Publishing Pvt Ltd.; 1925. Statistical Methods for Research Workers. [Google Scholar]
- 36.Boyd A., Golding J., Macleod J., Lawlor D.A., Fraser A., Henderson J., Molloy L., Ness A., Ring S., Davey Smith G. Cohort profile: the ‘children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol. 2013;42(1):111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Relton C.L., Gaunt T., McArdle W., Ho K., Duggirala A., Shihab H., Woodward G., Lyttleton O., Evans D.M., Reik W. Data resource profile: accessible resource for integrated epigenomic studies (ARIES) Int. J. Epidemiol. 2015;44(4):1181–1190. doi: 10.1093/ije/dyv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawlor D.A., Wills A.K., Fraser A., Sayers A., Fraser W.D., Tobias J.H. Association of maternal vitamin D status during pregnancy with bone-mineral content in offspring: a prospective cohort study. Lancet. 2013;381(9884):2176–2183. doi: 10.1016/S0140-6736(12)62203-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Touleimat N., Tost J. Complete pipeline for Infinium(®) Human Methylation 450 K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4(3):325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 40.Teschendorff A.E., Zhuang J., Widschwendter M. Independent surrogate variable analysis to deconvolve confounding factors in large-scale microarray profiling studies. Bioinformatics. 2011;27(11):1496–1505. doi: 10.1093/bioinformatics/btr171. [DOI] [PubMed] [Google Scholar]
- 41.Ponsonby A.L., Lucas R.M., Lewis S., Halliday J. Vitamin D status during pregnancy and aspects of offspring health. Nutrients. 2010;2(3):389–407. doi: 10.3390/nu2030389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulahannan N., Greally J. Genome-wide assays that identify and quantify modified cytosines in human disease studies. Epigenet. Chromatin. 2015;8(1):5. doi: 10.1186/1756-8935-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Novakovic B., Galati J.C., Chen A., Morley R., Craig J.M., Saffery R. Maternal vitamin D predominates over genetic factors in determining neonatal circulating vitamin D concentrations. Am. J. Clin. Nutr. 2012;96(1):188–195. doi: 10.3945/ajcn.112.035683. [DOI] [PubMed] [Google Scholar]
- 44.Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M.A., Bibikova M., Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 45.Jaffe A.E., Irizarry R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15(2):R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou J., Lippert C., Heckerman D., Aryee M., Listgarten J. Epigenome-wide association studies without the need for cell-type composition. Nat. Methods. 2014;11(3):309–311. doi: 10.1038/nmeth.2815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.