Abstract
Background
The plant-specific TCP transcription factor family, which is involved in the regulation of cell growth and proliferation, performs diverse functions in multiple aspects of plant growth and development. However, no comprehensive analysis of the TCP family in watermelon (Citrullus lanatus) has been undertaken previously.
Results
A total of 27 watermelon TCP encoding genes distributed on nine chromosomes were identified. Phylogenetic analysis clustered the genes into 11 distinct subgroups. Furthermore, phylogenetic and structural analyses distinguished two homology classes within the ClTCP family, designated Class I and Class II. The Class II genes were differentiated into two subclasses, the CIN subclass and the CYC/TB1 subclass. The expression patterns of all members were determined by semi-quantitative PCR. The functions of two ClTCP genes, ClTCP14a and ClTCP15, in regulating plant height were confirmed by ectopic expression in Arabidopsis wild-type and ortholog mutants.
Conclusions
This study represents the first genome-wide analysis of the watermelon TCP gene family, which provides valuable information for understanding the classification and functions of the TCP genes in watermelon.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0765-9) contains supplementary material, which is available to authorized users.
Keywords: TCP, Transcription factors, Watermelon, Internode elongation
Background
The TCP gene family, a small group of transcription factors (TF) exclusive to higher plants, was first described in 1999 [1]. The family plays important roles in regulating diverse physiological and biological processes, including phytohormone biosynthesis and signal transduction, leaf morphogenesis and senescence, branching, flower development, pollen development and the circadian clock [2–15]. TCP proteins are characterized by a 59-amino-acid non-canonical basic-Helix-Loop-Helix (bHLH) motif that is responsible for DNA binding, nuclear targeting and pair-wise protein–protein interaction [1, 16]. This domain was first identified from four proteins with critical roles in the evolution and developmental control of plant morphology: TEOSINTE BRANCHED 1 (TB1) of maize (Zea mays), CYCLOIDEA (CYC) of snapdragon (Antirrhinum majus) and the PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) of rice (Oryza sativa) [16–18]. Thus the name of the TCP TF family is derived from the acronym for these proteins. TCP genes can be divided into two subfamilies based on the homology of the TCP domains: class I (or TCP-P) and class II (or TCP-C) [19]. TCP class I, also known as the PCF subfamily, contains rice OsPCF1 and OsPCF2, whereas TCP class II is further subdivided into the CIN and CYC/TB1 subclades [7]. The most obvious difference between the two classes is a four-amino-acid deletion in the basic region of the TCP domain of class I compared with that of class II proteins. Moreover, the DNA binding sequence for the two classes differs slightly but partly overlaps (GGNCCCAC for class I and GTGGNCCC for class II) [20, 21].
Accumulating evidence confirms that class I TCP proteins mainly play a role in cell growth and proliferation [13, 20], whereas the CIN proteins may be involved in lateral organ development and the CYC/TB1 clade is mainly involved in the development of axillary meristems giving rise to either flowers or lateral shoots [5, 7, 9, 22–27]. Generally, the two classes of TCP genes are considered to act antagonistically on specific biological processes. Class I genes are usually assumed to promote plant growth, mainly based on the finding that OsPCF1/OsPCF2 and AtTCP20 act as transcriptional activators of PCNA and CYCB1;1 genes [7, 20, 28]. In practice, most class I single mutants do not show conspicuous phenotypic variation, which might be because of functional redundancies. For example, increasing evidence demonstrates that AtTCP14 and AtTCP15 function redundantly to regulate biological processes and influence plant structure. The two genes also mediate responses of leaves and flowers to cytokinin and promotion of seed germination by gibberellin (GA) [29–31]. More recently, AtTCP14 and AtTCP15 were shown to repress endoreduplication by directly regulating the expression of cell-cycle genes to influence cell and organ growth [32]. Notable plant morphological changes are observed in the tcp14 tcp15 double mutant, such as shortened internode length as well as varied leaf and sepal morphology, whereas single mutants show mild phenotypic defects [29, 33]. Moreover, AtTCP9 and AtTCP19 play a positive role in a redundant manner with AtTCP20 in the control of leaf senescence, as tcp9 tcp20 and tcp19 tcp20 double mutants exhibit earlier onset of senescence in comparison with the wild type, whereas none of the single mutants exhibit accelerated senescence [13, 15].
By contrast, many phenotypic observations on mutants suggest that the class II TCP proteins usually have preventative roles in cell growth and proliferation. CIN-type genes limit cell proliferation at the margins of the developing leaf primordium. In snapdragon, Arabidopsis and tomato cin-type mutants, leaf cells exhibit the ability to continue to divide for a longer period compared with the wild type, thus generating larger leaves of altered shape and/or with a crinkled surface [2, 21, 25, 34, 35]. In addition, many TB1-type TCP genes act as axillary bud-specific regulators, such as TB1 of maize [18, 22], AtBRC1 and AtBRC2 of Arabidopsis [4, 36], PsBRC1 of pea (Pisum sativum) [37] and OsFC1/OsTB1 of rice [38, 39]. Defects in these genes result in excessive shoot branching, which are indicative of a negative function of these TCP genes on bud activity [4, 36–39]. In some instances, class II TCP genes may also play positive roles in plant growth and development. AtTCP1, a CYC/TB1 subclade member, is implicated in the control of floral symmetry [40]. Over-expression of a dominant-negative form of TCP1, TCP1-SRDX, results in a dwarfed phenotype as well as defects in the longitudinal elongation of cotyledonary petioles, rosette leaves and inflorescence stems in Arabidopsis [9, 40].
To date, only a small number of TCP TFs have been identified and functionally characterized in model plants such as Arabidopsis and rice. Watermelon (Citrullus lanatus L.), an important cucurbit crop, is widely cultivated throughout the world. However, little information is available on the watermelon TCP family. In this study, a global analysis of the TCP gene family in watermelon was carried out for the first time. Twenty-seven ClTCP genes were identified in the watermelon genome and a systematic analysis, including determination of chromosomal location, phylogenetic relationships, gene duplication, conserved motifs and expression pattern was performed. Plant height is an important agronomic trait of watermelon. Normally, watermelon genotypes of reduced plant height are more suitable for intensive culture and early maturation in a greenhouse. ClTCP genes involved in the regulation of plant height in watermelon were identified in this research.
Results and discussion
Identification of TCP genes in Citrullus lanatus
To identify TCP protein-coding genes in watermelon, Arabidopsis and rice TCP proteins sequences were employed as the query for a BLAST search against the Cucurbit Genomics Database (http://www.icugi.org/cgi-bin/ICuGI/index.cgi). Twenty-seven putative TCP TFs, which contained the conserved TCP domain, were identified (Table 1). The results of a search for watermelon TCP family members in the Plant Transcription Factor Datebase (PlantTFDB; http://planttfdb.cbi.pku.edu.cn) were in agreement with the former search. Due to the lack of standard annotations designated to the 27 TCP genes in watermelon, we named the genes ClTCP1a to ClTCP21 consistent with the Arabidopsis TCP proteins that showed the highest sequence similarity and following the gene nomenclature system applied to Arabidopsis. The length of the 27 newly identified ClTCP TFs ranged from 182 to 517 amino acids with an average of 332.8 amino acids. Other characteristics of the ClTCP TFs, including molecular weight (Mw), isoelectric point (pI), type and chromosome location, are listed in Table 1. The ClTCP TFs can be classified into the two TCP classes based on the differences within their TCP domains: 12 of the TFs belong to Class I because of the presence of a four-amino-acid deletion in the basic domain relative to the other TFs; the 15 Class II ClTCP TFs can be further clustered into the CIN subclass and the CYC/TB1 subclass (Additional file 1: Figure S1). The genomic location of each ClTCP in watermelon is shown in Fig. 1. The 27 ClTCP genes were mapped to nine chromosomes. Moreover, based on a neighbor-joining (NJ) phylogenetic tree constructed from the full-length amino acid sequences, a putative orthologous relationship between the 27 ClTCP TFs and 24 AtTCP TFs was established (Fig. 1 and Additional file 2: Table S1). The number of TCP genes in watermelon is similar to that in Arabidopsis, which is in strong agreement with the fact that the number of protein-coding genes in the watermelon genome (23,440 genes) [41] approximates that in Arabidopsis (25,498 genes) [42]. A number of Arabidopsis TCP genes have more than one counterpart in the watermelon genome, which might be a result of differential gene expansion in watermelon and Arabidopsis after their divergence from a common ancestor.
Table 1.
TCP gene family in Citrullus lanatus
| Name | Identifier | Length | MW | PI | Type | Chr. Location |
|---|---|---|---|---|---|---|
| (aa) | (Da) | |||||
| ClTCP1a | Cla016452 | 372 | 41900.2 | 9.2502 | CYC/TB1 | Chr11:21507938-21509176 |
| ClTCP1b | Cla007113 | 391 | 43810.3 | 9.3686 | CYC/TB1 | Chr05:15872127-15873302 |
| ClTCP2a | Cla009785 | 453 | 49073 | 8.9768 | CIN | Chr01:32896256-32897617 |
| ClTCP2b | Cla019567 | 473 | 51428.6 | 7.4337 | CIN | Chr03:7077952-7079373 |
| ClTCP3 | Cla023342 | 438 | 47583.7 | 6.2952 | CIN | Chr11:19833922-19835238 |
| ClTCP4 | Cla002428 | 432 | 46917.3 | 6.9235 | CIN | Chr07:23567484-23568782 |
| ClTCP5 | Cla019050 | 361 | 39578.9 | 8.5739 | CIN | Chr06:24664979-24666064 |
| ClTCP7 | Cla009096 | 264 | 27651.8 | 10.4014 | PCF | Chr01:22327174-22327968 |
| ClTCP8 | Cla022939 | 517 | 53272.4 | 7.5207 | PCF | Chr11:15780378-15781931 |
| ClTCP9 | Cla006939 | 389 | 41942.8 | 9.9295 | PCF | Chr06:180401-181570 |
| ClTCP10a | Cla013523 | 319 | 34941.8 | 8.0631 | CIN | Chr02:28431330-28432289 |
| ClTCP10b | Cla016274 | 286 | 31374 | 9.3142 | CIN | Chr09:10452307-10453167 |
| ClTCP11 | Cla018312 | 206 | 22074.9 | 8.0236 | PCF | Chr04:20715863-20716483 |
| ClTCP12a | Cla020363 | 359 | 41067.2 | 9.3598 | CYC/TB1 | Chr05:30498979-30500058 |
| ClTCP12b | Cla002323 | 241 | 28002.1 | 10.0495 | CYC/TB1 | Chr01:4771786-4772872 |
| ClTCP13 | Cla010762 | 300 | 33416.1 | 8.7256 | CIN | Chr07:29613042-29613944 |
| ClTCP14a | Cla019117 | 398 | 42508.6 | 7.7033 | PCF | Chr06:25224480-25225676 |
| ClTCP14b | Cla018622 | 283 | 30621.1 | 9.3136 | PCF | Chr04:23795732-23797562 |
| ClTCP15 | Cla010176 | 353 | 37417.4 | 9.7333 | PCF | Chr05:31441525-31442586 |
| ClTCP16 | Cla008721 | 182 | 20185.6 | 5.3317 | PCF | Chr02:31590819-31591367 |
| ClTCP17 | Cla018555 | 209 | 23699.4 | 9.5856 | CIN | Chr04:23273928-23274557 |
| ClTCP18a | Cla018516 | 235 | 26945.5 | 10.6516 | CYC/TB1 | Chr04:22871510-22872505 |
| ClTCP18b | Cla018993 | 326 | 37626.9 | 8.6485 | CYC/TB1 | Chr06:24195608-24196588 |
| ClTCP19 | Cla006219 | 345 | 36656.2 | 8.4939 | PCF | Chr05:7945454-7946491 |
| ClTCP20a | Cla002099 | 312 | 33447.1 | 6.5662 | PCF | Chr05:18535808-18536746 |
| ClTCP20b | Cla001506 | 285 | 30795.3 | 9.1123 | PCF | Chr06:2011967-2012824 |
| ClTCP21 | Cla020050 | 257 | 26673.7 | 10.5046 | PCF | Chr02:24455585-24456358 |
aa amino acid, MW molecular weight, PI isoelectric point, Chr chromosome
Fig. 1.
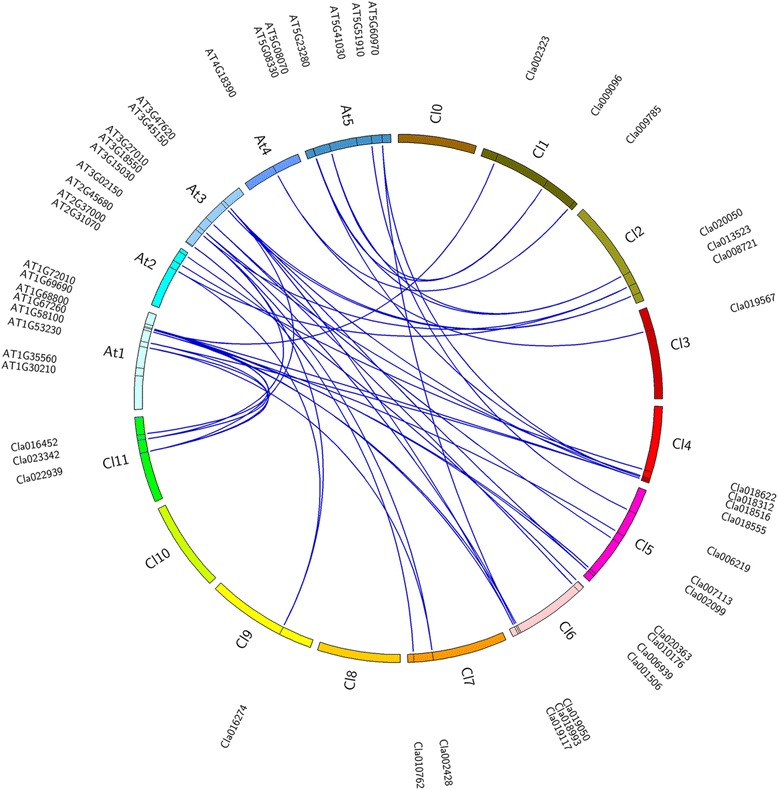
Visualization of the TCP Maps linkage groups. A Circos diagram illustrates the relative positions of TCP genes. The genes are plotted against their linked counterpart chromosomes. Chromosomal locations were determined according to chromosomal location information gathered from TAIR (https://www.arabidopsis.org) and Cucurbit Genomics Database (http://www.icugi.org). The map was obtained using Circos software
Phylogenetic analysis and conserved motifs
To evaluate the phylogenetic relationships among the TCP proteins in watermelon, Arabidopsis and rice, an unrooted phylogenetic tree was constructed using the NJ method from a multiple sequence alignment of 27 watermelon, 24 Arabidopsis and 21 rice TCP proteins (Fig. 2). The TCPs were divided into 11 subgroups, designated Group A to Group K, according to their sequence features within and outside the TCP domain. The TCPs in Groups A, B and C belonged to the Class II subfamily CIN-type, Group D belonged to the Class II CYC/TB1-type, whereas the remainder of the TCPs belonged to the Class I subfamily (Fig. 2). The TCP genes from the three species were distributed in almost all clades, which indicated that the TCP family diversified before the divergence of these plants. Notably, rice TCP was absent in Group E and a similar result was observed for Sorghum bicolor (Fig. 2 and Additional file 3: Figure S2). This finding implies that this clade may have been lost in rice and sorghum, or was acquired in an ancestor of Arabidopsis and Citrullus after the divergence of monocots and dicots.
Fig. 2.
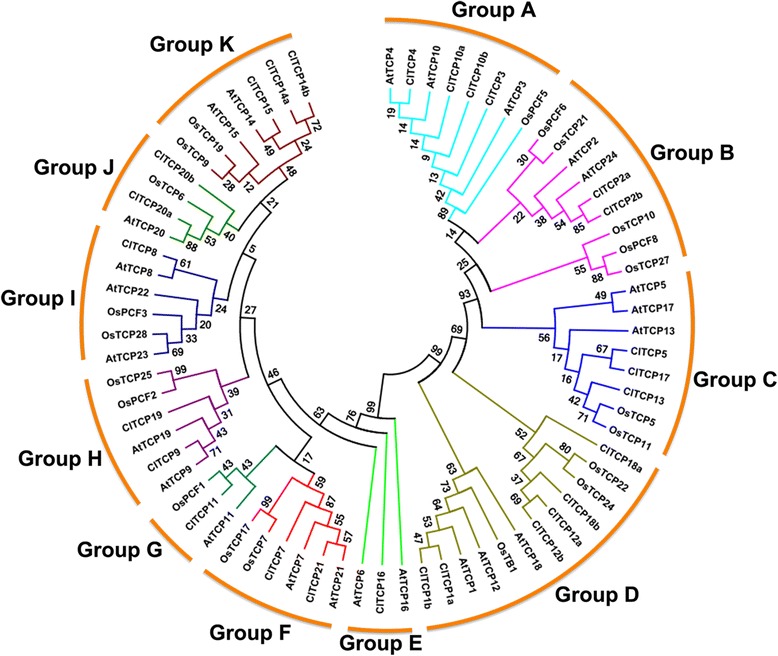
Phylogenetic relationships of TCP transcription factors from watermelon, Arabidopsis and rice. The unrooted phylogenetic tree was constructed using MEGA 5.0 with the neighbor-joining method. Support for the topology was assessed by means of a bootstrap analysis with 1000 replicates
Analysis of the conserved motif structure was performed to confirm the validity of the phylogenetic tree. The R domain, an 18–20 residue arginine-rich motif, is absent in all Class I proteins and is mainly present in CYC/TB1 proteins. The miR319 site is only present in a subset of the CIN-like genes (Fig. 3). In Arabidopsis, miR319 modulates jasmonate biosynthesis, negatively regulates leaf growth, positively regulates leaf senescence and affects petal development. These functions are dependent on post-transcriptional regulation of the miR319-targeted TCP genes (AtTCP2, AtTCP3, AtTCP4, AtTCP10 and AtTCP24) [2, 5, 27, 43]. In the present study, five CIN-type ClTCP genes (ClTCP2a, 2b, 3, 4 and 10a) contained the putative miR319 target site and shared the highest sequence similarity with the Arabidopsis miR319-targeted TCP genes (Fig. 3). These findings indicated that regulation of hormone response and leaf development by miRNA-targeted homologous TCP TFs may be conserved in watermelon and Arabidopsis. In addition, exon/intron structure analysis showed that most of the ClTCP genes lacked an intron, with the exception of ClTCP1a and ClTCP12b, which contained one intron, and ClTCP18a contained two introns (data not shown). Interestingly, these three ClTCP genes belong to the CYC/TB1-type subclade.
Fig. 3.
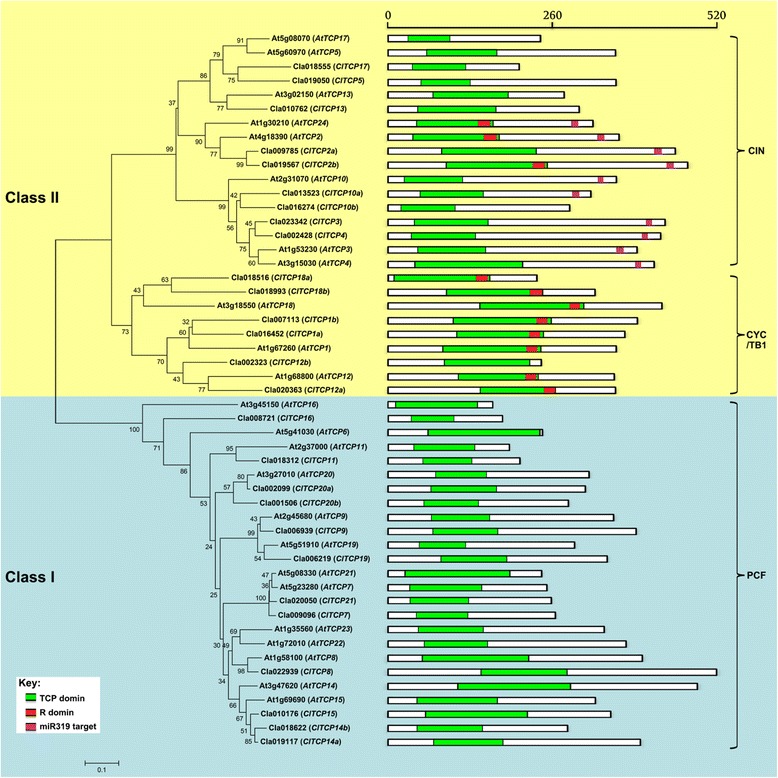
Phylogenetic analysis and conserved motifs of TCP family members in Arabidopsis thaliana and Citrullus lanatus. An unrooted phylogenetic tree, showing relationships between all TCP transcription factors in A. thaliana (At) and C. lanatus (Cl), was constructed using MEGA 5.0 with the neighbor-joining method. Support for the topology was assessed by means of a bootstrap analysis with 1000 replicates. Class I is highlighted in blue, and Class II is highlighted in yellow. On the right is the protein structure constructed using DOG 2.0 indicating conserved motifs: TCP domain (green) (http://pfam.xfam.org), R domain (red) (PlntTFDB database). The position of the microRNA miR319 recognition sequence in the mRNA is indicated in light purple (not drawn to scale). The scale bar represents amino acid length
Expression profiles of TCP genes in Citrullus lanatus
To predict possible functions of TCP genes in watermelon, we performed semi-quantitative PCR (semi-qPCR) analysis of transcripts in different organs, including the seed, cotyledon, leaf, root, internode, shoot apical meristem (SAM), male and female flower buds, and fruit. Interestingly,expression analysis showed that every class/clade showed a characteristic expression profile. As indicated in Fig. 4, most CIN-type ClTCP genes were not expressed or only weakly expressed in the root, flower or fruit, and were more highly expressed in the seed, cotyledon and leaf, which suggested that these genes may perform important roles in the shoot. Most CYC/TB1-type ClTCP genes were relatively weakly expressed in the seed, leaf of early stage and root, but were relatively highly expressed in specific tissues. For example, ClTCP18b and ClTCP12b were relatively highly expressed in the internode and SAM beyond the six-leaf stage, and in the flower and fruit, whereas ClTCP1a, ClTCP1b and ClTCP12a were only expressed in the internode and SAM beyond the six-leaf stage. These results indicated that CYC/TB1-type ClTCP genes might play important roles in the development of internodes and flowers. Generally, Class II TCP genes, which function in a similar manner mainly by suppressing cell division and plant growth, exhibit tissue-specific expression pattern. CYC/TB1 subclade genes have been long considered to be key players in the development of axillary meristems giving rise to either flowers or lateral shoots. AtTCP1, the gene most closely related to CYC, is involved in the longitudinal elongation of leaves. The Arabidopsis gain-of-function tcp1-1D mutant shows an elongated-leaf phenotype, whereas expression of a TCP1-SRDX chimeric repressor gene in the wild type results in the opposite phenotype to the tcp1-1D mutant [9, 40]. Moreover, mutation of the HaCYC2c gene, a TCP1/CYC homolog in sunflower, promoted the developmental switch from sterile to hermaphrodite flowers [44]. Expression of AtTCP1 is strong in the petiole, lower portion of the inflorescence stem, and the midrib and distal region of expanding rosette leaves. Two ClTCP1 genes, which are closely related to AtTCP1, were strongly expressed in the internode and SAM of watermelon (Fig. 4). This result is partly consistent with the expression pattern of AtTCP1 in Arabidopsis and implies that ClTCP1 genes may play roles in internode and inflorescence development in watermelon. AtTCP18, which is also known as BRANCHED1 (BRC1) and TEOSINTE BRANCHED1-LIKE1 (TBL1), acts downstream of auxin and strigolactone to coordinate axillary bud outgrowth [4, 36]. AtTCP18 also represses the floral transition of the axillary meristems by interacting with FLOWERING LOCUS T (FT) [45]. AtTCP12, also known as BRC2, exhibits a weaker or no mutant phenotype compared with AtTCP18 [4, 36]. Furthermore, no interactions between the BRC2 and FT proteins have been detected in yeast two-hybrid experiments [45]. In watermelon, the expression level of ClTCP12b and ClTCP18b was significantly higher in the internode, SAM, flower and fruit. Expression of ClTCP12a was detected only in the internode and SAM (Fig. 4). These observations suggested that these genes are likely to perform similar roles in branch and/or inflorescence development in watermelon to those of the Arabidopsis homologs. In contrast, CIN-type TCP genes are considered to have originated prior to CYC/TB1-type TCPs and are important for generation of the flat surface and smooth margin of the leaf. Thus, cin-type mutants usually exhibit crinkly and/or serrated leaves [2, 23, 27, 35]. In watermelon, expression of all CIN-type TCP genes was detected in the cotyledon, leaf and SAM (Fig. 4). This result is consistent with their predicted roles in leaf and lateral-organ development.
Fig. 4.
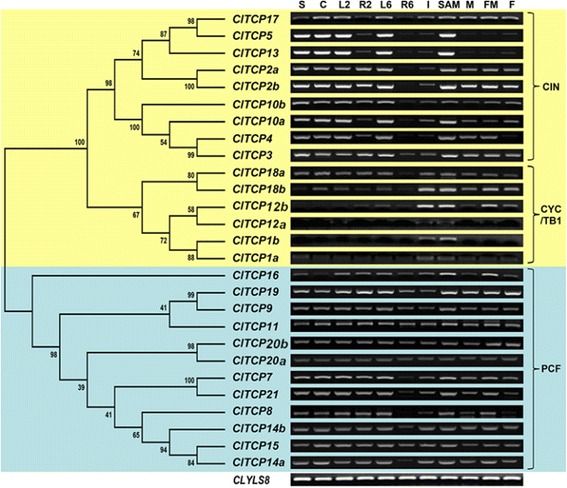
Expression patterns of Citrullus lanatus TCP genes in different tissues. The expression profile of ClTCP genes in the seed, leaf, internode, shoot tip, root, flowers and fruit was obtained through semi-quantitative PCR analysis. Expression of the CLYLS8 gene was monitored as an internal control. The phylogenetic tree of all TCP transcription factors in Citrullus lanatus was constructed using MEGA 5.0 with the neighbor-joining method. Support for the topology was assessed by means of a bootstrap analysis with 1000 replicates. S: Seeds at germination; C: cotyledons; L2: leaves at two-true-leaf stage; R2: roots at two-true-leaf stage; L6: leaves at six-true-leaf stage; R6: roots at six-true-leaf stage; I: internodes; SAM: shoot apical meristem; M: male flower; FM: female flower; F: immature fruit 3 hours after pollination
In contrast, most Class I genes, which usually promote plant growth and cell proliferation, showed more widespread and less tissue-specific expression patterns, such as in leaf, flower, and at an early stage of fruit development (Fig. 4). This finding implied that these genes may play diverse regulatory roles at multiple development stages. In Arabidopsis, several important functions of Class I TCP TFs have been discovered even though few phenotypic variations are observed in the single mutants. For example: AtTCP8 is proposed to be involved in mitochondrial biogenesis [46]. AtTCP14 and AtTCP15 are reported to modulate cell proliferation during seed, leaf, floral and internode development [31, 33, 47]. AtTCP15 may also be important for endoreduplication [48]. AtTCP16 plays a role in early pollen development [3]. AtTCP20, which acts upstream of AtTCP9, controls leaf development via the jasmonate signaling pathway [13, 15, 28]. All of these AtTCP genes have at least one counterpart in watermelon, implying that Class I TCP in watermelon may perform similar functions. Taken together, the above-mentioned findings from model plants highlight that the TCP family performs diverse functions in multiple biological processes. ClTCP genes are likely to share conserved functions with Arabidopsis homologs, as they show not only high sequence similarity but also similar expression patterns.
Role of ClTCP14a and ClTCP15 in plant height
ClTCP14a and ClTCP15 are members of the Class I subfamily of TCP TFs in watermelon (Fig. 3). These two ClTCP genes are closely related to Arabidopsis AtTCP14 and AtTCP15 as well as Antirrhinum TCP TF TIC [33, 49]. Given the unavailability of a TCP-related mutant in watermelon, we examined the function of these two ClTCP genes in four independent transgenic lines (p35S:ClTCP14a-WT, p35S:ClTCP15-WT, p35S:ClTCP14a-tcp14 tcp15 and p35S:ClTCP15-tcp14 tcp15), which over-expressed ClTCP14a or ClTCP15 in both Arabidopsis Col-0 and tcp14 tcp15 double-mutant backgrounds. After growth under long-day conditions for 42 days, the double-mutant seedlings showed a significant reduction in inflorescence height than that of the wild type (Fig. 5). No visible phenotype was identified in any single mutant, similar to the observations of Kieffer et al. [33]. Ectopic expression of either ClTCP14a or ClTCP15 was sufficient to restore the inflorescence height and stem internodes length of tcp14 tcp15 double mutant to that of the wild type. The p35S:ClTCP14a-WT and p35S:ClTCP15-WT lines exhibited an increase in inflorescence height compared with that of the wild type (Fig. 5). These results suggested that ClTCP14a and ClTCP15 function redundantly to control Arabidopsis plant height and may play positive roles in stem internode elongation in watermelon. Scanning electron microscopy revealed that the double mutant bore excessively branched trichomes compared with those of the wild type, and that overexpression of ClTCP14a or ClTCP15 inhibited trichome branching in both backgrounds (Additional file 4: Figure S3). Furthermore, ectopic expression of each watermelon TCP gene in both backgrounds increased the relative chlorophyll content in mature leaves (Additional file 5: Figure S4). These findings suggested that ClTCP14a and ClTCP15 may also be involved in leaf development.
Fig. 5.
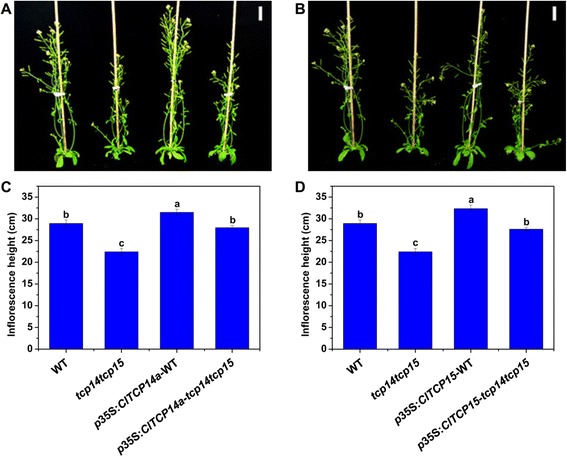
Morphological effects of constitutive expression of ClTCP14a and ClTCP15 in transgenic Arabidopsis. A Seedlings of the wild type (WT; Col-0), double mutant (tcp14 tcp15) and p35S:ClTCP14a in WT and double-mutant backgrounds were grown under long-day conditions for 42 days. B Seedlings of the WT, double mutant and p35S:ClTCP15 in WT and double-mutant backgrounds were grown under long-day conditions for 42 days. C Inflorescence height of seedlings as shown in (A). D Inflorescence height of seedlings as shown in (B). Scale bars = 3 cm. Different lower-case letters denote a significant difference in inflorescence height among genotypes (P < 0.05, one-way ANOVA and then Tukey’s test for multiple comparisons). Values are means ± SD (n = 20)
Given that GA is a regulator of plant height, we investigated whether overexpression of ClTCP14a and ClTCP15 affected GA biosynthesis and metabolism. AtKO1 and AtGA2ox3, which are involved in GA biosynthesis and degradation, were more weakly and more highly expressed, respectively, in the tcp14 tcp15 double mutant compared with those of the wild type. Ectopic expression of each watermelon TCP in the tcp14 tcp15 background revealed positive and negative impacts on the expression of AtKO1 and AtGA2ox3, respectively (Fig. 6A and B). However, these effects were not observed in the wild-type background. The GA receptor, AtGID1a, was slightly but significantly up-regulated in ClTCP14a- and ClTCP15- overexpressing Arabidopsis (Fig. 6C). These results suggested that overexpression of ClTCP14a and ClTCP15 may enhance GA accumulation and signaling in tcp14 tcp15 and the effects of these TCP genes on plant height may be associated with the GA pathway. Interestingly, the expression of all GA-related genes differed significantly between ClTCP14a- and ClTCP15-transgenic Arabidopsis, which might reflect the higher expression level of ClTCP14a compared with that of ClTCP15 in each transgenic line (Additional file 6: Figure S5).
Fig. 6.
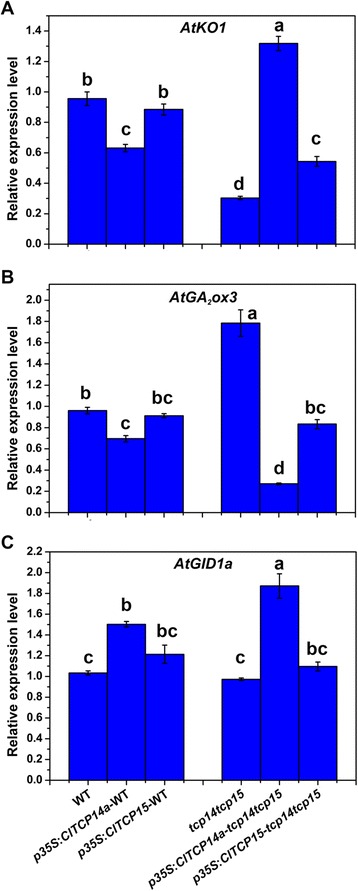
Expression of gibberellic acid (GA)-related genes in transgenic Arabidopsis. The relative expression level of A the GA biosynthesis gene AtKO1, B the GA degradation gene AtGA 2 ox3 and C the GA receptor gene AtGID1a in seedlings of the wild type (WT), double-mutant (tcp14 tcp15), p35S:ClTCP14a and p35S:ClTCP15 in both WT and double-mutant backgrounds was determined by quantitative RT-PCR. Expression of the CLYLS8 gene was monitored as an internal control. Different lower-case letters denote a significant difference in relative expression level (P < 0.05, one-way ANOVA and then Tukey’s test for multiple comparisons). Values are means ± SD (n = 3)
In addition, the effects of GA and chlormequat chloride (CCC, a GA biosynthesis inhibitor) on plant height as well as ClTCP14a and ClTCP15 expression were examined in watermelon. The results revealed that GA and CCC were a functional enhancer and inhibitor, respectively, of watermelon plant height (Fig. 7a and b). Both regulators likely function by affecting internode length rather than internode number, as no differences in internode numbers were observed. Expressions of both ClTCP14a and ClTCP15 was significantly up- and down-regulated by GA and CCC treatment, respectively (Fig. 7c and d). These results confirmed that ClTCP14a and ClTCP15 might positively regulate watermelon plant height and internode length via a GA-related pathway.
Fig. 7.
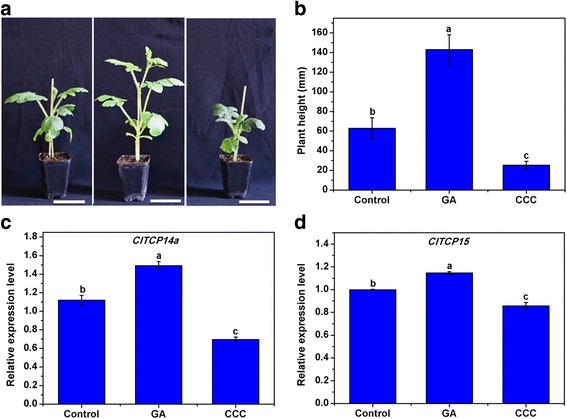
Effects of gibberellic acid (GA) and chlormequat chloride (CCC) on plant height and expression of ClTCP genes in watermelon. Watermelon seedlings were treated with water (Control), GA and CCC at the two-true-leaf stage. a and b Plant height of watermelon seedlings at the six-true-leaf stage. Scale bars = 7 cm. Different lower-case letters denote a significant difference in plant height between treatments (P < 0.05, one-way ANOVA and then Tukey’s test for multiple comparisons). Values are means (n = 5) ± SD. c Relative expression level of ClTCP14a in watermelon seedlings 1 day after treatment. d Relative expression level of ClTCP15 in watermelon seedlings 1 day after different treatments. Different lower-case letters denote a significant difference in relative expression level (P < 0.05, one-way ANOVA and then Tukey’s test for multiple comparisons). Values are means ± SD (n = 3)
Plant height is an important agronomic trait in watermelon, which dramatically affects planting density and fruiting position in the field. TCP TFs, as well known cell proliferation regulators, are undoubtedly important participants in internode and plant elongation. The present results revealed that ClTCP14a and ClTCP15 redundantly regulated internode length and plant height via a GA-related pathway in transgenic Arabidopsis (Figs. 5, 6 and 7). In Arabidopsis, AtTCP14 and AtTCP15 are reported to regulate internode development by promoting cell proliferation, based mainly on the phenotypes observed in double-mutant and TCP14:SRDX lines [33]. The present results provide direct evidence for this genotype–phenotype correlation. Moreover, AtTCP14 and AtTCP15 are expressed in internodes of young inflorescence stems, young flower pedicels, cotyledons and leaf primordia [33]. These results are generally consistent with the present expression analysis of ClTCP genes in watermelon (Fig. 4). Moreover, it was reported recently that AtTCP14 and AtTCP15 mediate GA-dependent activation of the cell cycle during seed germination [31]. Thus, we hypothesize that ClTCP14a and ClTCP15 may also act downstream of GA and promote cell proliferation during internode formation in a similar manner. Interestingly, our findings suggest that ClTCP14a and ClTCP15 might also affect GA biosynthesis and signaling (Fig. 6), which might result from a feedback regulatory mechanism.
Conclusions
In this study, 27 TCP genes were identified in the watermelon genome, which were distributed on nine chromosomes with different densities. These TCP genes were classifiable into two classes based on the similarity in TCP domain. Expression analysis showed that members of each class/clade show a similar expression pattern. Moreover, many ClTCP genes showed a similar expression pattern to that of their Arabidopsis homologs, which suggests that the TCP family shows conserved functions in the two species. In addition, the function of two ClTCP genes, ClTCP14a and ClTCP15, in the regulation of internode elongation was confirmed. Ultimately, these findings will lead to potential applications for the improvement of watermelon cultivars via genetic engineering.
Methods
Plant materials and growth conditions
Watermelon (Citrullus lanatus L. cv. IVSM9, an inbred line developed by the Laboratory of Germplasm Innovation and Molecular Breeding, Zhejiang University) was used as the main plant material. Plants were grown under a photoperiod of 16 h at 27 °C (day) and 8 h at 24 °C (night) in a phytotron with a photosynthetic photon flux density of 600 μmol m−2 s−1 and relative humidity of 70–80 %.
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the wild type. All Arabidopsis materials, including tcp14-4, tcp15-3, tcp14-4 tcp15-3 and their background were obtained from the University of Leeds, UK, and were genotyped by PCR as described by Kieffer et al. [33]. Plants were grown in Sanyo growth chambers (Sanyo, http://www.sanyobiomedical. co.uk) at 20 °C under long-day conditions with a photoperiod of 16 h/8 h (day/night), photosynthetic photon flux density of 200 μmol m−2 s−1 and 60 % relative humidity.
Chromosomal analysis
Information on the chromosomal locations of all AtTCP genes was obtained from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org), and that for all ClTCP genes was obtained through BLASTN searches against the Cucurbit Genomics Database (http://www.icugi.org). All TCP genomic data were visualized in a circos map using CIRCOS software (http://circos.ca).
Sequence alignment and phylogenetic analysis
The sequences of 24 TCP family members in the genome of Arabidopsis were retrieved from TAIR (http://www.arabidopsis.org) or PlantTFDB (http://planttfdb.cbi.pku.edu.cn/). Twenty-seven ClTCP genes were identified from a BLAST analysis of the Cucurbit Genomics Database (http://www.icugi.org). A multiple sequence alignments of the amino acid sequences of the TCP proteins of Citrullus lanatus and Arabidopsis was generated with ClustalX 2.0 software with the default settings as described by Thompson et al. [50]. An unrooted phylogenetic tree based on the sequence alignments was constructed using MEGA 5.0 software (http://www.megasoftware.net/) [51] and the neighbor-joining method with the following parameters: pairwise alignment, 1000 bootstrap replicates, Poisson correction model, uniform substitution rates and complete deletion. In addition, a separate phylogenetic tree was constructed for all of the TCP protein sequences from Citrullus lanatus for further analysis.
Identification of conserved motifs
AtTCP and ClTCP protein sequences were submitted to online searches with the Pfam (http://pfam.xfam.org) and SMART (http://smart.embl-heidelberg.de) tools to identify conserved TCP domains. The R domain was obtained from PlantTFDB (http://planttfdb.cbi.pku.edu.cn/). The method of identifying miR319-targeting TCP genes was described previously [2]. To visualize protein domain structures, IBS 1.0 software (http://www.mybiosoftware.com/ibs-illustrator-of-biological-sequences.html) was used.
RNA isolation and RT-PCR analysis
Total RNA was isolated from tissues using the RNAprep Pure Plant Kit and treated with DNase I (Tiangen, http://www.tiangen.com). RNA concentration and quality were assessed using a Thermo 2000 Bioanalyzer with a RNA NanoDrop (Thermo Scientific, http://www.thermo.com). Reverse transcription was performed with 1 μg total RNA in a 20-μl volume, using the ReverTra Ace® qPCR RT Master Mix with gDNA Remover kit (Toyobo) and diluted to 200 μl with water. For semi-qPCR and PCR, 1 μL reverse-transcription product was used as the PCR template in a 20-μl volume reaction. Different PCR annealing temperatures were applied to optimize results and the PCR was terminated after 35 cycles. PCR products were separated by 1.5 % agarose gel electrophoresis and visualized under an ultraviolet scanner. For RT-qPCR analysis, a 20-μl qPCR mixture was employed, which contained 2.5 μl first-strand cDNAs, 10 μl 2× FastStart Universal SYBR Green Master (Roche) and 0.25 μM of the forward and reverse primers for each gene. Relative expression levels of each gene were normalized to mRNA levels of yellow-leaf-specific protein 8 (CLYLS8) as a loading control. Three biological replicates were analyzed in each case. CT values were obtained with the Real-Time PCR System StepOne version 2.1 software (Applied Biosystems). Relative fold expression changes were calculated by the comparative CT method: fold change was calculated as 2−∆∆CT. The ∆CT values were calculated as the difference between the CT value and the CT value of CLYLS8. ∆∆CT was the difference between the ∆CT value of TCP genes and the ∆CT value of the reference gene. The gene-specific primers for semi-quantitative PCR and RT-qPCR procedures are listed in Additional file 7: Table S2.
Vector constructs
To study the function of ClTCP14a and ClTCP15, two constructs were developed using the CaMV35S promoter. Full-length ClTCP14a cDNA (1197 bp) and ClTCP15 cDNA (1062 bp) were amplified by two-round PCR: the first round with the gene-specific primers ClTCP14a-Fl-F and ClTCP14a-Fl-R, and ClTCP15-Fl-F and ClTCP15-Fl-R, respectively; the second round with the common primers attB1-F and attB2-R. Finally, both sequences were cloned into the Gateway™ vector pMDC83 (Invitrogen) via the BP and LR reactions as described by Curtis and Grossniklaus [52]. The primers used for vector construction are listed in Additional file 8: Table S3.
Transformation of Arabidopsis
The two constructs were transformed into Agrobacterium tumefaciens strain GV3101 (pMP90) [53]. Transformation of both wild-type and tcp14 tcp15 plants was conducted by means of the floral dip method [54]. After transformation, plants were kept in a growth chamber until seed set. Transformant selection was done on germination medium containing 50 μg ml−1 Hygromycin-B (Roche, http://www.roche.com) for 10 days, after which germinated T1 seedlings were transferred to soil and grown until seed set. During selection of T1 plants, a plant line negative for hygromycin resistance was selected and maintained as a negative control. In addition, overexpression analysis of TCP14a or TCP15 in candidate transgenic Arabidopsis was employed to confirm the successful transformation (Additional file 6: Figure S5). Homozygous T3 plants were used in this study. Sixty primary transformants were identified and analyzed in most experiments.
Internode measurement
Inflorescence height and internode length of 42-day-old Arabidopsis plants were measured. All measurements were obtained from three independent experiments, and at least 10 replicate seedlings were measured in each experiment.
Microscopy
Samples dissected and prepared for scanning electron microscopic analysis were analyzed as previously described [33].
Phytohormone treatment
Gibberellic acid (150 mg L−1) (Biotech Grade biosharp, http://www.biosharp.cn) and 150 mg L−1 chlormequat chloride (Shanghai BioRc Co., Ltd.) were sprayed onto the leaf surface of watermelon seedlings at the two-true-leaf stage. Water was applied as the control. Three days later, the treatment was repeated. The plant height was measured at the six-true-leaf stage.
Availability of supporting data
The datasets supporting the results of this article are available at http://dx.doi.org/10.5061/dryad.9pp6q
Acknowledgements
This work was supported by the earmarked fund for Modern Agro-Industry Technology Research System of China (CARS-26-17), National Natural Science Foundation of China (31501782), Zhejiang Provincial Natural Science Foundation of China (LQ16C150002), Education Department Research Program of Zhejiang province (Y201329960) and Fundamental Research Funds for the Central Universities (2013QNA6013). We thank Dr. Martin Kieffer for donation of the mutants.
Additional files
Alignment of the predicted amino acid sequences for members of the watermelon TCP family. (PDF 3445 kb)
Orthologous relationship of TCPs between watermelon and Arabidopsis. (XLSX 39 kb)
Phylogenetic relationships of TCP transcription factors from watermelon, Arabidopsis, rice and sorghum. (PDF 2881 kb)
Effect of ClTCP14a and ClTCP15 on trichome branching on Arabidopsis leaf. (TIF 3851 kb)
Relative chlorophyll content in Arabidopsis leaves. (PDF 371 kb)
ClTCP14a and ClTCP15 overexpression level in transgenic Arabidopsis. (PDF 893 kb)
Gene-specific primers used for semi-qPCR and RT-qPCR. (XLSX 47 kb)
Primers used for vector construction. (XLSX 30 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZH, JY and MZ conceived and designed the study; PS, WW, BF and KG performed the experiments and analyzed the data; ZH and PS wrote the paper; all authors have read and approved the final version.
Contributor Information
Pibiao Shi, Email: 1032175660@qq.com.
Kateta Malangisha Guy, Email: gabrielkateta@gmail.com.
Weifang Wu, Email: 13735846154@163.com.
Bingsheng Fang, Email: 21216072@zju.edu.cn.
Jinghua Yang, Email: yangjinghua@zju.edu.cn.
Mingfang Zhang, Email: mfzhang@zju.edu.cn.
Zhongyuan Hu, Email: huzhongyuan@zju.edu.cn.
References
- 1.Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999;18:215–222. doi: 10.1046/j.1365-313X.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 2.Palatnik J, Allen E, Wu X, Schommer C, Schwab R, Carrington J, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 3.Takeda T, Amano K, Ohto M, Nakamura K, Sato S, Kato T, Tabata S, Ueguchi C. RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol Biol. 2006;61:165–177. doi: 10.1007/s11103-006-6265-9. [DOI] [PubMed] [Google Scholar]
- 4.Aguilar-Martínez J, Poza-Carrion C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nag A, King S, Jack T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:22534–22539. doi: 10.1073/pnas.0908718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pruneda-Paz J, Breton G, Para A, Kay S. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martín-Trillo M, Cubas P. TCP genes: a family snapshot ten years later. Trends Plant Sci. 2010;15:31–39. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Van Aken O, Millar AH, Murcha M, Whelan J. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell. 2010;22:3921–3934. doi: 10.1105/tpc.110.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Z, Fujioka S, Blancaflor E, Miao S, Gou X, Li J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell. 2010;22:1161–1173. doi: 10.1105/tpc.109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarvepalli K, Nath U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011;67:595–607. doi: 10.1111/j.1365-313X.2011.04616.x. [DOI] [PubMed] [Google Scholar]
- 11.Viola I, Uberti Manassero N, Ripoll R, Gonzalez D. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem J. 2011;435:143–155. doi: 10.1042/BJ20101019. [DOI] [PubMed] [Google Scholar]
- 12.Yanai O, Shani E, Russ D, Ori N. Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 2011;68:571–82. doi: 10.1111/j.1365-313X.2011.04716.x. [DOI] [PubMed] [Google Scholar]
- 13.Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk A, Muino J, Cutri L, Dornelas M, Angenent G, Immink R. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012;159:1511–1523. doi: 10.1104/pp.112.200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balsemão-Pires E, Andrade L, Sachetto-Martins G. Functional study of TCP23 in Arabidopsis thaliana during plant development. Plant Physiol Biochem. 2013;67:120–125. doi: 10.1016/j.plaphy.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Danisman S, van Dijk A, Bimbo A, van der Wal F, Hennig L, de Folter S, Angenent G, Immink R. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J Exp Bot. 2013;64:5673–5685. doi: 10.1093/jxb/ert337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- 18.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 19.Navaud O, Dabos P, Carnus E, Tremousayque D, Hervé C. TCP transcription factors predate the emergence of land plants. J Mol Evol. 2007;65:23–33. doi: 10.1007/s00239-006-0174-z. [DOI] [PubMed] [Google Scholar]
- 20.Kosugi S, Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002;30:337–348. doi: 10.1046/j.1365-313X.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- 21.Masuda H, Cabral L, De Veylder L, Tanurdzic M, de Almeida Engler J, Geelen D, Inzé D, Martienssen R, Ferreira P, Hemerly A. ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J. 2008;27:2746–2756. doi: 10.1038/emboj.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doebley J, Stec A, Gustus C. Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nath U, Crawford B, Carpenter R, Coen E. Genetic control of surface curvature. Science. 2003;299:1404–1407. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- 24.Cubas P. Floral zygomorphy, the recurring evolution of a successful trait. Bioessays. 2004;26:1175–1184. doi: 10.1002/bies.20119. [DOI] [PubMed] [Google Scholar]
- 25.Crawford B, Nath U, Carpenter R, Coen E. CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 2004;135:244–253. doi: 10.1104/pp.103.036368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koyama T, Furutani M, Tasaka M, Ohme-Takaqi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell. 2007;19:473–484. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schommer C, Palatnik J, Aggarwal P, Chételat A, Cubas P, Farmer E, Nath U, Weigel D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6:1991–2001. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Potuschak T, Colón-Carmona A, Gutiérrez R, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci U S A. 2005;102:12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steiner E, Efroni L, Gopalraj M, Saathoff K, Tseng T, Kieffer M, Eshed Y, Olszewski N, Weiss D. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell. 2012;24:96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steiner E, Yanai O, Efroni I, Ori N, Eshed Y, Weiss D. Class I TCPs modulate cytokinin-induced branching and meristematic activity in tomato. Plant Signal Behav. 2012;7:807–810. doi: 10.4161/psb.20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resentini F, Felipo-Benavent A, Colombo L, Blázquez M, Alabadí D, Masiero S. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol Plant. 2015;8:482–485. doi: 10.1016/j.molp.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Peng Y, Chen L, Lu Y, Wu Y, Dumenil J, Zhu Z, Bevan M, Li Y. The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell. 2015;27:649–662. doi: 10.1105/tpc.114.132274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieffer M, Master V, Waites R, Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011;68:147–158. doi: 10.1111/j.1365-313X.2011.04674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ori N, Cohen A, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, Alvarez J, Blum E, Zamir D, Eshed Y. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet. 2007;39:787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- 35.Efroni I, Blum E, Goldshmidt A, Eshed Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell. 2008;20:2293–2306. doi: 10.1105/tpc.107.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finlayson S. Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol. 2007;48:667–677. doi: 10.1093/pcp/pcm044. [DOI] [PubMed] [Google Scholar]
- 37.Braun N, de Saint Germain A, Pillot J, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, Luo D, Bendahmane A, Turnbull C, Rameau C. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 2012;158:225–238. doi: 10.1104/pp.111.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003;33:513–520. doi: 10.1046/j.1365-313X.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- 39.Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S, Kyozuka J. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 2010;51:1127–1135. doi: 10.1093/pcp/pcq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koyama T, Sato F, Ohme-Takagi M. A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci Biotechnol Biochem. 2010;74:2145–2147. doi: 10.1271/bbb.100442. [DOI] [PubMed] [Google Scholar]
- 41.Guo S, Zhang J, Sun H, Salse J, Lucas W, Zhang H, Zheng Y, Mao L, Ren Y, Wang Z, Min J, Guo X, Murat F, Ham B, Zhang Z, Gao S, Huang M, Xu Y, Zhong S, Bombarely A, Mueller L, Zhao H, He H, Zhang Y, Zhang Z, Huang S, Tan T, Pang E, Lin K, Hu Q, Kuang H, Ni P, Wang B, Liu J, Kou Q, Hou W, Zou X, Jiang J, Gong G, Klee K, Schoof H, Huang Y, Hu X, Dong S, Liang D, Wang J, Wu K, Xia Y, Zhao X, Zheng Z, Xing M, Liang X, Huang B, Lv T, Wang J, Yin Y, Yi H, Li R, Wu M, Levi A, Zhang X, Giovannoni J, Wang J, Li Y, Fei Z, Xu Y. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet. 2013;45:51–58. doi: 10.1038/ng.2470. [DOI] [PubMed] [Google Scholar]
- 42.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 43.Palatnik J, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, Carrington J, Weigel D. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Fambrini M, Salvini M, Pugliesi C. A transposon-mediate inactivation of a CYCLOIDEA-like gene originates polysymmetric and androgynous ray flowers in Helianthus annuus. Genetica. 2011;139:11–12. doi: 10.1007/s10709-012-9652-y. [DOI] [PubMed] [Google Scholar]
- 45.Niwa M, Daimon Y, Kurotani K, Higo A, Pruneda-Paz JL, Breton G, Mitsuda N, Kay SA, Ohme-Takagi M, Endo M, Araki T. BRANCHED1 Interacts with FLOWERING LOCUS T to Repress the Floral Transition of the Axillary Meristems in Arabidopsis. Plant Cell. 2013;25:1228–1242. doi: 10.1105/tpc.112.109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammani K, Gobert A, Hleibieh K, Choulier L, Small I, Giegé P. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell. 2011;23:730–40. doi: 10.1105/tpc.110.081638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313X.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Li B, Dong A. The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol Plant. 2012;5:270–280. doi: 10.1093/mp/ssr086. [DOI] [PubMed] [Google Scholar]
- 49.Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B. CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development. 2004;131:915–922. doi: 10.1242/dev.00993. [DOI] [PubMed] [Google Scholar]
- 50.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koncz Cand Schell J. The promoter of TL -DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. doi: 10.1007/BF00331014. [DOI] [Google Scholar]
- 54.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated trans- formation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]


