Abstract
Studies of the distribution of ammonia oxidising archaea (AOA) and bacteria (AOB) suggest distinct ecological niches characterised by ammonia concentration and pH, arising through differences in substrate affinity and ammonia tolerance. AOA form five distinct phylogenetic clades, one of which, the ‘Nitrososphaera sister cluster’, has no cultivated isolate. A representative of this cluster, named ‘Candidatus Nitrosocosmicus franklandus’, was isolated from a pH 7.5 arable soil and we propose a new cluster name: ‘Nitrosocosmicus’. While phylogenetic analysis of amoA genes indicates its association with the Nitrososphaera sister cluster, analysis of 16S rRNA genes provided no support for a relative branching that is consistent with a ‘sister cluster’, indicating placement within a lineage of the order Nitrososphaerales. ‘Ca. N. franklandus’ is capable of ureolytic growth and its tolerances to nitrite and ammonia are higher than in other AOA and similar to those of typical soil AOB. Similarity of other growth characteristics of ‘Ca. N. franklandus’ with those of typical soil AOB isolates reduces support for niche differentiation between soil AOA and AOB and suggests that AOA have a wider physiological diversity than previously suspected. In particular, the high ammonia tolerance of ‘Ca. N. franklandus’ suggests potential contributions to nitrification in fertilised soils.
Keywords: Thaumarchaeota, Nitrososphaera sister cluster, soil, ammonia inhibition, Nitrosocosmicus, nitrification
This study describes ecophysiological characteristics of the first cultivated representative of a soil archaeal group performing an important biogeochemical process, ammonia oxidation, with significance for niche specialisation and archaeal phylogeny.
INTRODUCTION
Soil nitrification, the sequential oxidation of ammonia to nitrite and nitrate, is generally limited by the activity of ammonia oxidisers (AO), which perform the first step in this process. Traditionally, ammonia oxidation was thought to be dominated by ammonia oxidising bacteria (AOB), but the cultivation of autotrophic ammonia oxidising archaea (AOA) from marine (Könneke et al.2005) and subsequently terrestrial (Lehtovirta-Morley et al.2011; Tourna et al.2011) environments led to a reassessment of the microbial ecology of soil nitrification. Potential differences in archaeal and bacterial physiology presented the possibility of niche differentiation between AOB and AOA. This is exemplified by the high affinity for ammonia of the marine isolate Nitrosopumilus maritimus (Martens-Habbena et al.2009), which explains the dominance of AOA in oligotrophic sea water, and by acidophilic growth of the soil isolates, Nitrosotalea devanaterra and Nitrosotalea sp. Nd2, which provides an explanation for nitrification in acid soils (Lehtovirta-Morley et al.2011, 2014). There is limited evidence for greater sensitivity of AOA to ammonia, with inhibition of growth at micromolar concentrations of ammonia, while AOB typically grow at millimolar concentrations (Koper et al.2010; Prosser and Nicol 2012). In addition, differences in effects of organic compounds have been reported within and between AOA and AOB (Lehtovirta-Morley et al.2011, 2014; Sayavedra-Soto and Arp 2011; Tourna et al.2011) and there is evidence for greater nitrite sensitivity in N. maritimus (presented in terms of nitrous acid in Table 1). Cell mass and specific cell activity are approximately one order of magnitude lower in cultivated soil AOA than soil AOB but maximum specific growth rates are similar (Prosser and Nicol 2012; Table 1).
Table 1.
Characteristics of the ‘model’ AOA (N. maritimus) and AOB (N. europaea), six AOA soil isolates or enrichments and a range of soil AOB. Some data are not available for certain strains and cell yield and activity data for Nitrosotalea strains are not directly comparable when presented in terms of NH3. Cell yield and specific cell activity of C13 calculated using cell and amoA gene concentrations are labelled a and b, respectively.
| Cell yield | Specific cell activity | Inhibitory | Inhibitory | ||||
|---|---|---|---|---|---|---|---|
| Organism | Size (μm) | μ max (h−1) | (cells μM−1 NH3) | (fmol NO2− cell−1 h−1) | NH4+ conc. (mM) | HNO2 conc. (μM) | References |
| Nitrosopumilus maritimus SCM1 | 0.17–0.22 × 0.5–0.9 | 0.033 | 5 × 104 | 0.53 | 2 | 0.028 | Könneke et al. (2005) |
| Nitrososphaera viennensis EN76 | 0.6–0.8 diam | 0.024 | 20 | 0.79 | Stieglmeier et al. (2014a) | ||
| Nitrosoarchaeum koreensis MY1 | 0.3–0.5 × 0.6 – 1.0 | 1.1 × 105 | 2.5 | 20 | 0.50 | Jung et al. (2011) | |
| Nitrosotenuis chungbukensis MY2 | 0.2 × 0.7 | 3.4 × 105 | 20 | 0.50 | Jung et al. (2014) | ||
| Nitrosotalea devanaterra Nd1 | 0.33 × 0.89 | 0.011 | 4.5 × 105 | 0.072 | 50 | 0.91–3.5 | Lehtovirta-Morley et al. (2011) |
| Nitrosotalea sp. Nd2 | 0.2 × 0.7 | 0.025 | 4 × 105 | 0.065 | 1.61–5.7 | Lehtovirta-Morley et al. (2014) | |
| Nitrosocosmicus franklandus (C13) | 1.1 diam | 0.024 | a7.60 × 103/ b1.29 × 105 | a2.02 b0.58 | >100 mM | 1.58 | This study |
| Nitrosomonas europaea ATCC 19718 | 0.8–1.1 × 1.0–1.7 | 0.052–0.066 | 4.61–6.44 × 103 | 11 | >400 | 31.6 | Prosser (1989); Hunik, Meijer and Tramper (1992); Koops et al. (2006) |
| Soil AOB (data collated from approximately 25 strains) | 0.3–0.8 × 1.0–8.0 | 0.005–0.044 | 1.38–10.6 × 103 | 4–23 | 7–50 | Prosser (1989); Koops and Pommerening-Röser (2001); Koper et al. (2010); Prosser and Nicol (2012) |
Characterisation of soil AOA and comparison with AOB are limited by difficulties in isolation of pure cultures, due to low specific growth rates of AO and their susceptibility to contamination by faster growing heterotrophs. Pure cultures of AOB have been available since their first isolation from soil by Frankland and Frankland (1890), although few strains have been subjected to detailed physiological analysis. In contrast, only nine AOA have been isolated to date: N. maritimus strains SCM1 (from a salt water aquarium) (Könneke et al.2005), NAO2 and NAO6 (from marine surface water) (Elling et al.2015), N. devanaterra Nd1 and Nitrosotalea sp. Nd2 (from acidic soils) (Lehtovirta-Morley et al.2011, 2014), Nitrososphaera viennensis EN76 (from garden soil) (Tourna et al.2011), N. gargensis Ga9.2 (from a hot spring) (Palatinszky et al.2015) and Nitrosopumilus strains PS0 and HCA1 (closely related to N. maritimus SCM1, isolated from coastal waters) (Qin et al.2014). Only three of these isolates are from soil, but two soil enrichments have also been characterised, Nitrosoarchaeum koreensis MY1 (Jung et al. 2011) and Nitrosotenuis chungbukensis MY2 (Jung et al.2014).
AOA and AOB use the enzyme ammonia monooxygenase (AMO) in the first step of ammonia oxidation, whose three subunits, A, B, C, are encoded by amoA, amoB and amoC genes, respectively. The amoA gene has been used extensively as a marker in diversity analyses, and initial studies showed that AOA are widely distributed in most environments (soil, marine waters and sediments, freshwater, geothermal springs) (e.g. Francis et al.2005; Reigstad et al.2008). In the soil, AOA typically outnumber AOB (e.g. Leininger et al.2006) and the relative abundance of phylogenetic groups within the AOA varies in different soils (Gubry-Rangin et al.2011).
A phylogenetic study (Pester et al.2011) of publically available archaeal amoA gene sequences placed all within one of five major clusters. Four of these clusters have characterised, cultured representatives (the Nitrosopumilus, Nitrosotalea, Nitrosocaldus and Nitrososphaera clusters). The fifth has a distinct, but specific association with the Nitrososphaera cluster, it was termed the ‘Nitrososphaera sister’ cluster (Pester et al.2011) and its phylogenetic divergence occurred early in thaumarchaeotal diversification (Gubry-Rangin et al. 2015). Sequences within this group are widely distributed in soil and other environments, but typically do not comprise the major proportion of amoA gene sequences in high throughput sequencing studies of soil (e.g. Gubry-Rangin et al.2011). Pester et al. (2011) found amoA gene sequences representative of this group in seven of nine soils sampled from four different continents, comprising up to 13% of all archaeal amoA gene sequences. In addition, a genome sequence of an unpublished soil enrichment culture has been deposited in GenBank (accession number CP012850.1) and has provisionally been named ‘Candidatus Nitrosocosmicus oleophilus’, with the proposed genus name describing ‘a widely distributed AO’ (Rhee, pers. comm.).
The aim of this study was to characterise a novel AOA belonging to the Nitrososphaera sister cluster (Pester et al.2011), isolated from a near-neutral pH agricultural soil, and to consider the implications for niche differentiation of AOA and AOB in soil.
MATERIALS AND METHODS
Culture enrichment and isolation
Sandy loam soil of pH 7.5 was sampled from an agricultural plot on the Scottish Rural College's Craibstone estate, Aberdeen (grid reference NJ872104). AO enrichment cultures were obtained by inoculation of 0.5 g soil in 50 ml ‘fresh water medium’ (FWM), containing NaCl (1 g l−1), MgCl2·6H2O (0.4 g l−1), CaCl2·2H2O (0.1 g l−1), KH2PO4 (0.2 g l−1), KCl (0.5 g l−1), NaHCO3 (0.168 g l−1), NH4Cl (0.027 g l−1) and 1 ml l−1 additions of the following solutions: modified non-chelated trace elements (Könneke et al.2005), Fe-NaEDTA (7.5 mM) (Tourna et al.2011) and 50 mg l–1 streptomycin sulphate. Medium was adjusted to pH 7.5 at room temperature and filter-sterilised through a bottle-top filter (0.22 μm, Merck Millipore, Watford) into 100-ml Duran bottles (Sigma-Aldrich, Gillingham, UK). Soil suspensions were initially incubated statically at 37°C in the dark, monitored for nitrite accumulation and subcultured (2% inoculum) when nitrite concentration increased exponentially. Nitrite and ammonium concentrations were determined colorimetrically using Griess reagent and indophenol method, respectively, as previously described (Lehtovirta-Morley et al. 2014). Reactions and absorbance measurements were performed in clear polystyrene 96-well microplates (Greiner Bio-One, Stonehouse, UK) and measured using an Infinite F50 Microplate Reader and Magellan reader control and data analysis software (Tecan, Männedorf, Switzerland).
The presence of bacteria in enrichment cultures was determined by spread-inoculating 1 ml of culture onto Tryptone Soy, Nutrient and LB Agar plates (Sigma-Aldrich, Gillingham, UK) at 5%, 10% and 25% of standard concentrations, respectively, and 1.5% w/v agar. Agar plates were incubated in the dark at 30°C or 40°C. In addition, culture purity was assessed by PCR amplification of bacterial and fungal SSU rRNA genes. Cells were pelleted from liquid culture before extracting DNA using a standard bead-beating protocol with SDS extraction buffer and phenol:chloroform:isoamyl alcohol (Lehtovirta-Morley et al.2011). PCR was performed using BIOTAQ polymerase and dNTP mix (Bioline, London, UK) according to manufacturer's instruction (see Table 2 for details of primers and PCR cycling conditions).
Table 2.
Details of primers and PCR cycling conditions used in this study.
| Target gene | Primer | Sequence (5′-3′) | Reference | Cycling conditions |
|---|---|---|---|---|
| Bacterial 16S rRNA | 27f1492r | AGAGTTTGGATCMTGGCTCAGGYYACCTTGTTACGACTT | Lane (1991) Nicol et al. (2008) | 95°C 5 min; 10 cycles of 94°C 30 s, 55°C 30 s, 72°C 2 min; 25 cycles of 92°C 30 s, 55°C 30 s, 72°C 2 min; 72°C 5 min |
| Fungal 18S rRNA | Fung5fFF390r | GGGAACCAGGACTTTTACGAGGTCTCGTTCGTTATCG | Smit et al. (1999) Vainio and Hantula (2000) | 95°C 5 min; 10 cycles of 94°C 30 s, 55°C 30 s, 72°C 50 s; 25 cycles of 92°C 30 s, 55°C 30 s, 72°C 50 s; 72°C 5 min |
| Archaeal 16S rRNA | A109F 1492r | ACKGCTCAGTAACACGT GYYACCTTGTTACGACTT | Grosskopf, Janssen and Liesack (1998) Nicol et al. (2008) | 95°C 5 min; 10 cycles of 94°C 30 s, 55°C 30 s, 72°C 2 min; 25 cycles of 92°C 30 s, 55°C 30 s, 72°C 2 min; 72°C 5 min |
| Thaumarchaeal amoA | CrenamoA23fCrenamoA616r | ATGGTCTGGCTWAGACGGCCATCCATCTGTATGTCCA | Tourna et al. (2008) Tourna et al. (2008) | 95°C 5 min; 10 cycles of 94°C 30 s, 55°C 30 s, 72°C 1 min; 25 cycles of 92°C 30 s, 55°C 30 s, 72°C 1 min; 72°C 5 min |
| Nitrosotalea sp. Nd2 amoA gene cluster | Nd2F1Nd2R2 | GATACTTGCAGTGATACCTACCCCAGATATTCTTGTTTCAACAGAGG | This studyThis study | 95°C 5 min; 35 cycles of 94°C 30 s, 52°C 30 s, 72°C 4 min; 72°C 10 min |
To eliminate bacteria from enrichment cultures, their antibiotic sensitivity was determined by spread-inoculating 0.5 ml of an exponentially growing enrichment on FWM agar plates containing antibiotic discs and incubating at 40°C in the dark. Antibiotic discs were prepared by permeating sterile paper with 50 mg l−1 stock solutions (diluted in H2O) of ampicillin, carbenicillin, gentamycin, kanamycin, clindamycin, penicillin or rifampicin and dried in the dark at ambient temperature (Lehtovirta-Morley et al.2014). Antibiotics inhibiting bacterial contaminants were then added to FWM at a final concentration of 50 mg l−1, in an attempt to purify the enrichment culture.
Culture maintenance and preservation
The purified archaeal strain, initially termed C13, was routinely cultured in FWM as described above but with the addition of 1 ml l−1 of the following solutions: vitamin solution (excluding C5H14NO+) (Lehtovirta-Morley et al.2011), Se-We solution (Widdel and Bak 1992) and 10 ml l−1 of HEPES buffer (1 M HEPES, 0.6 M NaOH), with or without clindamycin (50 mg l−1) and ampicillin (100 mg l−1). All solutions were added prior to filter sterilisation.
C13 was cryopreserved and stored at –80°C using dimethylsulfoxide (DMSO) (VWR, Lutterworth, UK) as a cryoprotectant. DMSO was diluted to a concentration of 14% (v/v) using FWM and filter-sterilised (0.22-μm filter) before the addition of 0.5 ml DMSO solution to 0.5 ml of an exponentially growing culture in a sterile 1.5 ml microcentrifuge tube. Tubes were agitated for 5 min at 25°C in the dark to allow the DMSO to penetrate the cells before storage at –80°C. To resuscitate cells, samples were thawed at 25°C and immediately centrifuged at 4°C at 1200 × g for 10 min to minimise the cytotoxic effects of DMSO. Supernatant was then removed, pelleted cells were resuspended in 1 ml sterile FWM and centrifuged and washed three times, to remove residual DMSO, before final resuspension in 10 ml FWM. Growth of cells preserved for 3 weeks was detectable after resuscitation for 4 days.
Physiological characterisation
Unless otherwise stated, all physiological characteristics were determined during batch cultivation, with static incubation of triplicate cultures in FWM in the dark at 40°C as described above for enrichment cultures. The effects of ammonium concentration were determined in FWM supplemented with 0, 1, 2, 5, 10, 20, 50 or 100 mM NH4Cl and inhibition by nitrite concentration by supplementation with 0, 1, 2, 5, 10, 20 or 50 mM NaNO2. Ureolytic activity was investigated in ammonia-free FWM supplemented with 1 mM urea. Growth was also determined during incubation at temperatures ranging from 15°C to 60°C (5°C intervals). The effect of initial pH value in the range of 5–9 (0.5 pH intervals) was investigated in unbuffered medium containing a reduced concentration of 100 μm NH4Cl, to minimise the decrease in pH decline due to ammonia oxidation, and in medium buffered with 5 mM MES (2-(N-morpholino)ethanesulfonicacid) or 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonicacid). As MES and HEPES have pKa values of 5.91 and 7.31, respectively, at 40°C, neither possessed the buffering capacity to fulfil the full range of pH tested in this study, and MES buffer was used at pH 5.5–7 and HEPES buffer at pH 7–8.5.
Cell counts and microscopy
Cells were enumerated microscopically in 1 ml samples of liquid culture, fixed with 5% formaldehyde (final concentration; w/v) and stored at 4°C. Each sample was repeatedly passed through a syringe and needle (0.4 mm diameter) to disperse aggregated cells. Cells were then diluted and stained with 2 μg DAPI (4, 6-diamidino-2-phenylindole), incubated for 10–15 min at 4°C in the dark, filtered onto a black 0.2-μm polycarbonate membrane and washed with PBS. Dried filters were placed on a glass slide mounted with an antifadent (Citifluor AF2, Citifluor Ltd, Leicester, UK) and counted under oil immersion using an Olympus BH-2 microscope with a U-MWU2 fluorescence light source (Olympus, Southend-on-Sea, UK) at ×1000 magnification. For scanning electron microscopy, cells were fixed and gold-sputtered as previously described (Lehtovirta-Morley et al.2014) and images taken using an Evo MA 10 scanning electron microscope (Carl Zeiss, London, UK).
Abundance of amoA genes
Growth of strain C13 was also determined by quantification of amoA genes. DNA was extracted as described above, except that 20 μg glycogen was added as co-precipitant during DNA precipitation. qPCR amplification was performed in a BioRad MyIQ Single-Color Real-Time PCR Detection System (Hertfordshire, UK) using amplification-conditions and cycling-parameters described in Thion and Prosser (2014), except that 0.6 μm of each primer was used. A PCR product containing the amo gene cluster of Nitrosotalea sp. Nd2 (see Table 2 for details of primers and PCR cycling conditions) was used as a standard at dilutions giving 102–108amoA genes. The amplification efficiency was 97% and the r2 value 0.99. Positive amplification was confirmed by melting curve analysis and agarose gel electrophoresis.
DOPE-FISH analysis
Purity of strain C13 was also determined by DOPE-FISH (double labelling of oligonucleotide probes for fluorescence in situ hybridisation) using archaeal ARCH915 probe (Stahl and Amann 1991) labelled with Cy3 and bacterial EUB338 probe (Daims et al.1999) labelled with fluorescein. One ml of stationary phase culture was washed twice with PBS, concentrated in 100 μl of PBS and fixed with 96% ethanol (1:1 v/v). Before applying the cells to a teflon-covered microscope slide, cells in the fixed culture were dispersed by passing through a syringe and needle as described above. After dehydrating the cells on a microscope slide, 1 μl of each doubly labelled FISH probe (50 ng μl−1) was added to 10 μl of hybridisation buffer using 30% stringency (0.9 M NaCl, 20 mM Tris-HCl, 30% (v/v) formamide and 0.01% SDS) (Stoecker et al.2010). Cells were incubated at 46°C for 4 h in the dark, after which the microscope slide was transferred to the pre-warmed washing buffer (112 mM NaCl, 20 mM Tris-HCl and 5 mM EDTA) and incubated for 15 min at 48°C. Cells were then resuspended in 100 μl of pre-warmed washing buffer and incubated at 48°C for 20 min. Finally, the microscope slide was dried and counter- stained with DAPI in an antifadent oil (Citifluor AF1, Citifluor Ltd, Leicester, UK) and samples were visualised using a Zeiss Imager M2 fluorescent microscope (Carl Zeiss, London, UK).
Phylogenetic analysis
PCR was performed with primer sets targeting the archaeal amoA gene (CrenamoA23f/CrenamoA616r) and 16S rRNA gene (A109f/1492r) (Table 2). PCR products were purified using a Macherey–Nagel NucleoSpin PCR Clean-up Kit (Fisher Scientific, Loughborough, UK) and sequenced, with assembled 16S rRNA and amoA gene sequences deposited in GenBank with accession numbers KU290365 and KU290366, respectively. Sequences were aligned with those from all previously cultivated AOA and a selection of environmental clones using the ClustalW function within the BioEdit software (Hall 1999) before removing regions of ambiguous alignment. Phylogenetic analyses were performed using maximum-likelihood with General Time Reversible-correction (PhyML; Guindon and Gascuel 2003), Bayesian (MrBayes (Ronquist et al.2012), parsimony (MEGA5 (Tamura et al.2011)) and Tamura's 3-parameter pairwise distance analysis (MEGA5) for 16S rRNA gene analysis, and Jones-Taylor-Thornton (JTT)-corrected maximum-likelihood (PhyML), Bayesian (MrBayes), parsimony (MEGA5) and JTT-corrected pairwise distance (MEGA5) analyses of inferred translated amino acid sequences for amoA genes. Where appropriate, analyses used estimated variable sites only with gamma-distributed site variation and bootstrap support was calculated 1000 times for ML, parsimony, distance analyses, with Bayesian analysis performed with 100 000 iterations, sampling frequencies of 10, with an average standard deviation <0.01.
Statistical analysis
Maximum specific growth rate, μmax, was calculated as the gradient of semi-logarithmic plots of nitrite, cell or amoA gene concentration vs time during exponential growth. The effect of treatments on μmax was analysed by one-way ANOVA and linear regression analysis was performed on μmax using Excel. Cell activity was determined for each replicate culture by minimising the sum of squares of differences between nitrite concentrations determined experimentally and those predicted by assumption of constant specific cell activity during exponential growth using the Solver routine in Excel, with μmax and initial cell concentration or amoA gene concentration determined experimentally for each replicate culture.
RESULTS
Strain isolation
Enrichment cultures containing Thaumarchaeota, identified by amplification and sequencing of amoA genes, were obtained by inoculation of inorganic (FWM) medium containing 0.5 mM NH4+ with a sandy loam soil, pH 7.5. One culture containing a single amoA gene clonal population (designated C13) was selected for purification. Growth of heterotrophic bacteria was observed on Nutrient, Tryptone Soy and LB agar, but was eliminated after seven successive subcultures in FWM containing 500 mg l−1 kanamycin. Bacterial contamination was, however, detected by PCR amplification of 16S rRNA genes and by growth on solid FWM.
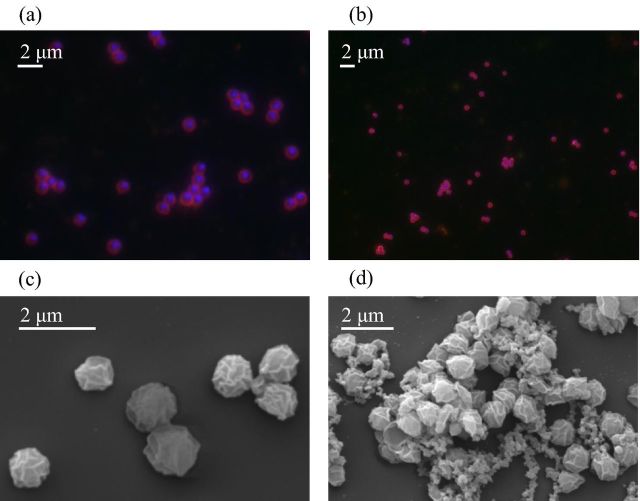
Heterotrophic contamination was reduced further by supplementation of medium with one of five antibiotics (ampicillin, carbenicillin, gentamycin, clindamycin and rifampicin) to which bacterial contaminants were sensitive. Nitrite was produced in medium containing each antibiotic except gentamycin, but was not detectable in subsequent subcultures, presumably because of a requirement for compounds produced by co-cultured bacteria. Growth was restored by the addition of spent (filter-sterilised) growth medium (1:10 ratio) and the addition of clindamycin and ampicillin. After a further eight subcultures with addition of spent medium and antibiotics, bacterial growth was not detected on FWM agar plates incubated for two weeks. Subsequent growth occurred in the absence of spent medium with no detectable amplification of bacterial 16S rRNA genes. The purity of strain C13 was also confirmed by DOPE-FISH which revealed the presence of archaeal cells, but never bacteria (Fig. 1a, b).
Figure 1.
Typical DOPE-FISH images of stationary-phase C13 culture (a, b) using archaeal ARCH915 probe (red), bacterial EUB338 probe (green) and counterstained with DNA DAPI stain (blue). Scanning electron micrographs of individual cells (c) and aggregates (d) of archaeal ammonia oxidiser strain C13.
Physiological characteristics
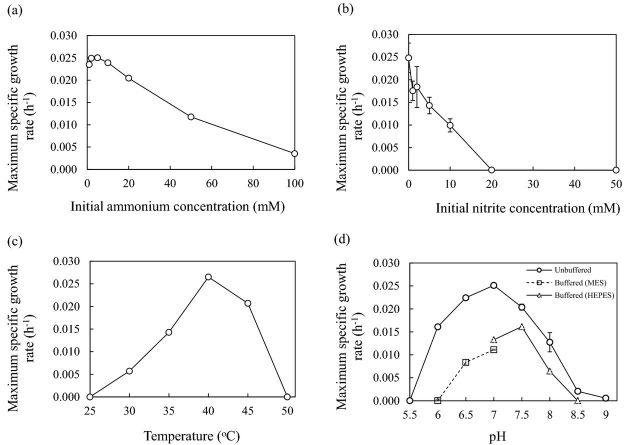
The influence of ammonia concentration on growth was determined by cultivation of strain C13 in medium containing 0–100 mM NH4+ (Fig. 2a). μmax was greatest (0.025 [s.e. 0.0002] h−1) at 5 mM, and lower at 1 mM. μmax decreased with increasing ammonium concentration, but growth was possible at the highest concentration tested, 100 mM, at 0.0035 [s.e. 0.0002] h−1. Nitrite was tolerated at initial concentrations of 0–10 mM NO2− (p = 0.13), but growth was completely inhibited by 20 mM and 50 mM NO2−, with no detectable increase in nitrite concentration after incubation for 21 days (Fig. 2b).
Figure 2.
The influence of (a) initial ammonium concentration, (b) initial nitrite concentration, (c) temperature and (d) pH on maximum specific growth rate of archaeal ammonia oxidiser isolate in batch culture. Growth in (d) was in unbuffered FWM or FWM buffered with MES or HEPES. Error bars are standard errors of means from triplicate cultures but are often smaller than the symbol size.
Strain C13 is mesophilic and grew at 30°C–45°C (Fig. 2c), with no detectable increase in cell abundance or nitrite concentration outside this temperature range after incubation for 21 days. Growth was optimal at 40°C, with a μmax of 0.027 [s.e. 0.00006] h−1. The isolate is neutrophilic, growing in the pH range 6–8.5 (Fig. 2d), and no detectable increase in cell abundance or nitrite concentration occurred outside this pH range during incubation for 21 days. Growth in unbuffered FWM was optimal at pH 7, with a maximum specific growth rate of 0.025 [s.e. 0.0002] h−1 (Fig. 2d), but pH in this medium varied during growth, increasing to 7.5 after incubation for 4 days, and subsequently decreasing to 7.3 when ammonium was completely utilised. Growth in FWM buffered by addition of MES or HEPES was optimal at pH 7.5 (Fig. 2d), but the μmax was lower than in unbuffered medium, at 0.016 [s.e. 0.00006] h−1, presumably due to inhibition by buffer.
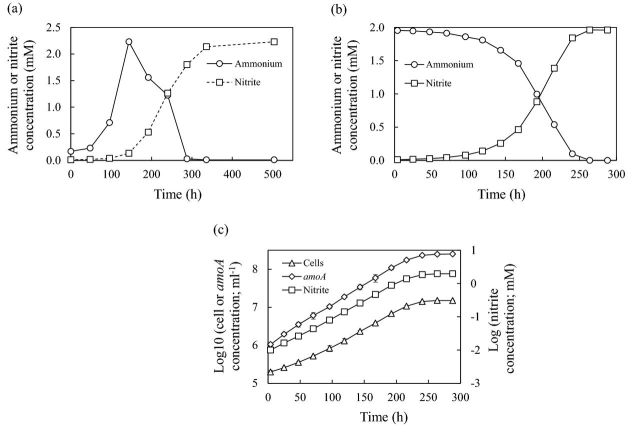
C13 is ureolytic and grew on medium containing 1 mM urea as the sole source of ammonia (Fig. 3a) with a μmax of 0.025 s.e. 0.0005 h−1. Ammonia concentration increased from 65 μm at day 0 (carried over in inoculum) to >2 mM during incubation for 6 days and was stoichiometrically converted to nitrite (Fig. 3b). μmax estimated as specific rates of increase in nitrite, cell and amoA concentration differed slightly (Fig. 3c, Table 3), although differences between μmax values are statistically significant (t-test, p > 0.05). amoA gene concentrations were approximately one order of magnitude greater than cell concentrations, and cell activities and cell yields calculated using both sets of data are presented in Table 3.
Figure 3.
(a) Changes in ammonium and nitrite concentration during growth of strain C13 during batch growth on FWM with urea as nitrogen source; (b) changes in ammonium and nitrite concentrations and (c) semilogarithmic plots of nitrite (□), cell (▵) and amoA (◊) concentrations during growth on FWM with 2 mM ammonium chloride. Error bars are standard errors of means from triplicate cultures but are often smaller than the symbol size.
Table 3.
Growth characteristics of strain C13 during batch growth at 40°C in HEPES-buffered FWM at pH 7.5 with initial ammonia concentration of 2 mM and determined on the basis of changes in concentrations of nitrite, cells and amoA genes. Means and standard errors are calculated from triplicate cultures.
| Basis for calculations | μ max (h−1) | Specific cell activity (fmol NO2− cell−1 h−1) | Cell yield (cells μM−1 NH3) |
|---|---|---|---|
| Nitrite concentration | 0.0237 (s.e. 0.0001) | ||
| Cell concentration | 0.0248 (s.e. 0.0011) | 2.02 (s.e. 0.19) | 7.60 × 103 (s.e. 0.19 × 103) |
| amoA concentration | 0.0239 (s.e. 0.0004) | 0.58 (s.e. 0.17) | 1.29 × 105 (s.e. 0.097 × 105) |
Morphology
Cells are irregularly shaped cocci with a diameter of 0.96 μm (standard error = 0.021 μm, range = 0.43–1.59 μm, n = 66) (Fig. 1c). Cells frequently occurred as chains of four cells or in irregular clusters, often with substantial extracellular material that was also observed detached from cells. These irregularly shaped aggregates varied in diameter from 7 to 15 μm (Fig. 1d).
Phylogenetic analyses
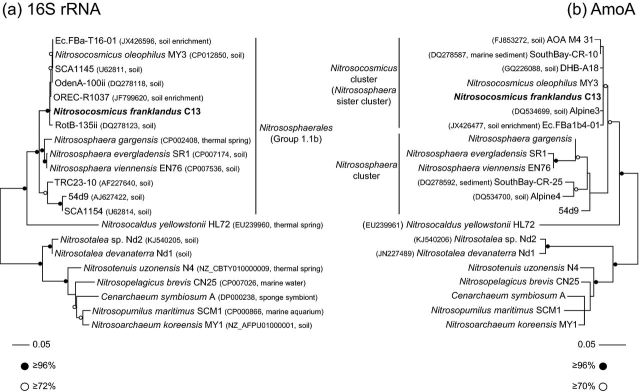
Phylogenetic analysis of 16S rRNA and amoA genes placed C13 within a well-supported, monophyletic lineage associated with organisms found in enrichment cultures and cloned sequences from various soil-based studies, with 16S rRNA and amoA genes sharing 99% and 92% identity, respectively, with those from the unpublished genome of ‘Ca. Nitrosocosmicus oleophilus’ and an enrichment culture obtained from arctic soil (Alves et al.2013). It was however, distinct from all cultured Nitrososphaera strains, with closest 16S rRNA similarity to N. viennensis at 95% identity (Fig. 4). While analysis of both genes shows a clear phylogenetic association with Nitrososphaera-like organisms, differences were observed when associating strain C13 (and closely related organisms) within a separate, sister cluster to the Nitrososphaera. Analysis of derived AmoA protein sequences demonstrated that C13 was placed in the clade designated by Pester et al. (2011) as the Nitrososphaera sister cluster (sharing 91%–99% identity with the 14 sequences analysed in that study), and distinct from those designated as belonging to the Nitrososphaera cluster. We thus propose the new clade delineation ‘Nitrosocosmicus’ cluster for these sequences. However, analysis of 16S rRNA gene sequences did not provide support for a separate lineage affiliation, with C13 being placed within a lineage that is associated with other members of the Nitrososphaerales (Stieglmeier et al.2014a).
Figure 4.
Maximum-likelihood phylogenetic analysis of (a) 16S rRNA genes and (b) derived AmoA protein sequences of isolated archaeon strain C13 with sequences from other cultivated organisms, metagenomes or cloned environmental sequences. Clade names are as described by Stieglmeier et al. (2014a) (16S rRNA) or Pester et al. (2011) (AmoA). Circles describing support represent the most conservative value from four methods used (bootstrap (ML, distance, parsimony) or posterior probabilities (Bayesian)) and the scale bar represents 0.05 changes per nucleotide or amino acid position. Accession number and environmental source are given in parentheses. If an accession number describes a genome or genomic fragment containing both a 16S rRNA and amoA gene, details are provided in the 16 rRNA tree only.
DISCUSSION
The thaumarchaeotal strain C13 is a mesophilic, neutrophilic autotrophic archaeal AO that grows within the temperature range 30°C–45°C and pH range 6–8.5. In these respects, it is similar to the soil AOA N. viennensis (28°C–45°C, pH 6–8.5) (Stieglmeier et al.2014b) and N. chungbukensis MY2 (25°C–40°C, pH 6.5–8) (Jung et al.2014). It also utilises urea as a source of ammonia, in common with several other members of the ‘Nitrososphaera’ lineage that possess genes encoding urease and urea transporters and N. viennensis and N. gargensis that can grow on urea (Tourna et al.2011; Spang et al.2012; Zhalnina et al.2014).
Cells of strain C13 are larger than those of N. viennensis (0.6–0.8 μm diam) (Stieglmeier et al.2014a) and other soil AOA isolates, with an estimated cell volume of 0.7 μm3. Cells were associated with extracellular polymeric material that led to aggregate formation and may facilitate biofilm formation, which has been shown to protect AOB from deleterious effects of low pH and nitrification inhibitors (de Boer et al.1991; Powell and Prosser 1991; Allison and Prosser 1993). μmax values were within the range found for other neutrophilic soil AOA (Table 1). Cell concentration was approximately one order of magnitude lower than amoA gene concentration, and similar differences have been reported in N. maritimus (Nakagawa and Stahl 2013). Cell concentration may be underestimated by incomplete dispersion of cells in aggregates, while amoA gene concentration may be overestimated by extracellular DNA following cell lysis. Cell yield and activity estimated using amoA gene concentrations are more consistent with those for other soil AOA and with N. koreensis MY1, respectively.
Strain C13 was more tolerant to ammonia than other soil AOA (Table 1), although the mechanisms for ammonia tolerance are unknown, and growth was possible at 100 mM NH4+, at which μmax was reduced to approximately 10% of that at optimal ammonium concentration. Strain C13 was also more tolerant to inhibition by nitrite than other neutrophilic soil AOA and grew at initial concentrations up to 20 mM NO2−. At pH 7.5, this is equivalent to 1.58 μm HNO2, which is believed to cause inhibition, and nitrous acid tolerance of strain C13 is therefore similar to that of N. devanaterra and Nitrosotalea sp. Nd2, greater than that of other cultivated soil AOA and one order of magnitude greater and lower than N. maritimus and Nitrosomonas europaea, respectively (Table 1).
Comparisons of the ‘model’ AOA and AOB, N. maritimus and N. europaea ATCC 19718, have led to the view that AOA are smaller, have lower growth rates and specific cell activity and lower tolerance to ammonia and nitrite than AOB; these differences have further been suggested to drive niche differentiation between AOA and AOB. While these distinctions are evident for the model AOA and AOB (Table 1), and exist across the breadth of AOA and AOB, they are less marked when considering only soil isolates (Table 1). Soil AOA and AOB have a similar range for μmax and are therefore likely to compete equally for ammonia when it is not growth-limiting. AOA are generally smaller and consequently have lower specific cell activity and greater cell yield than AOB, but this, in itself, provides no obvious competitive advantage. However, strain C13 extends the range of ammonia tolerance beyond that of characterised cultivated soil AOB (Prosser 1989; Koper et al.2010), reducing support for selective inhibition of AOA at high ammonia concentration, and suggesting a potential role for AOA in nitrification in fertilised soils. Care is therefore required when using information from a limited number of isolates to explain ecological phenomena and the differences between physiological characteristics of Nitrosotalea isolates (Lehtovirta-Morley et al.2014) highlight physiological diversity between strains that are very closely related in terms of 16S rRNA gene sequences. In addition, cultivation conditions will select strains that may not be representative of dominant and active members of natural communities and adaptation and selection will increase with continued cultivation. In this respect, C13 did not grow at 25°C and growth was optimal at 40°C, which is not typical of its environmental origin.
Phylogenetic analysis of predicted AmoA amino acid sequences places strain C13 in the Nitrososphaera sister cluster (Pester et al.2011), a lineage that is closely related to, but distinct from, the Nitrososphaera cluster. However, the same relative branching order was not resolved in 16S rRNA gene phylogenies, with strain C13 belonging to clade placed within, and not basal to, the order Nitrososphaerales, a lineage that is generally referred to as ‘Group 1.1b’ in environmental surveys (e.g. Nicol et al.2008). These contrasting topologies are consistent with a recent detailed comparison by Vico Oton et al. (2015) of thaumarchaeal 16S rRNA (clusters O and M) and amoA (C13_1 and C13_2) gene phylogenies. However, as strain C13 shares only 94%–95% 16S rRNA gene identity with other cultivated Nitrososphaera representatives, C13 probably represents a novel genus within the Nitrososphaerales.
This paper therefore reports the discovery of an AO that grows at neutral pH, but whose growth and activity are similar to those of soil AOB enrichments and isolates at neutral pH. Phylogenetic analysis of 16S rRNA and amoA genes places the organism within the Nitrososphaerales but distinct from previously isolated representatives of this order. We propose the following candidate status:
‘Nitrosocosmicus franklandus’ gen. et sp. nov.
Etymology. nitrosus (Latin masculine adjective): nitrous; cosmicus (Latin masculine adjective): cosmopolitan; Frankland (English noun): after Percy and Grace Faraday Frankland. The name reflects its ability to oxidise ammonia to nitrite, its widespread distribution in the environment, and provides a dedication to the authors of the first publication to describe isolation of organisms associated with ammonia oxidising activity (Frankland and Frankland 1890).
Source. Agricultural soil of pH 7.5, Craibstone, Aberdeen, Scotland, UK.
Description. A chemolithoautotrophic AO of the kingdom Thaumarchaeota, appearing as irregular cocci with a diameter 1.1 μm.
In accordance with other members of Nitrososphaerales, N. franklandus is an ammonia oxidising archaeon, isolated from a terrestrial environment, demonstrating the characteristic irregular coccoid cell morphology associated with this order (Stieglmeier et al.2014a). As with N. viennensis, it is able to grow on urea but it can tolerate higher ammonium concentrations, equivalent to those tolerated by oligotrophic bacterial AOs (Tourna et al.2011). Nitrosocosmicus franklandus represents an ecologically relevant strain of Thaumarchaeota with the potential to compete successfully with ammonium oxidising bacteria in fertilised soils where ammonium concentrations are high. The discovery of N. franklandus reduces support for niche differentiation between AOA and AOB in soil, with the possible exception in acid soils, and suggests that physiological diversity within each group may be more important in determining the composition of natural communities.
Acknowledgments
The authors would like to thank Mr Kevin Mackenzie and Mrs Gillian Milne (University of Aberdeen) for technical support with scanning electron microscopy, and Dr Robin Walker for access to the Woodlands Field experimental plots at the SRUC, Craibstone Estate, Aberdeen.
FUNDING
This work was financially supported by (standard grants and and fellowship ), , Grant Agreement No. , the AXA .
Conflict of interest. None declared.
REFERENCES
References
- Allison SM, Prosser JI. Ammonia oxidation at low pH by attached populations of nitrifying bacteria. Soil Biol Biochem. 1993;25:935–41. [Google Scholar]
- Alves RJ, Wanek W, Zappe A, et al. Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidising archaea. ISME J. 2013;7:1620–31. doi: 10.1038/ismej.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer W, Klein Gunnewiek PJA, Veenhuis M, et al. Nitrification at low pH by aggregated autotrophic bacteria. Appl Environ Microb. 1991;57:3600–4. doi: 10.1128/aem.57.12.3600-3604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Bruhl A, Amann R, et al. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–4. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Elling FJ, Könneke M, Mussmann M, et al. Influence of temperature, pH, and salinity on membrane lipid composition and TEX86 of marine planktonic thaumarchaeal isolates. Geochim Cosmochim Acta. 2015;151:238–55. [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, et al. Ubiquity and diversity of ammonia-oxidising archaea in water columns and sediments of the ocean. P Natl Acad Sci USA. 2005;102:14683–8. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland PF, Frankland GC. The nitrifying process and its specific ferment. Part I. Philos T Roy Soc B. 1890;181:107–28. [Google Scholar]
- Grosskopf R, Janssen PH, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microb. 1998;64:960–9. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubry-Rangin C, Hai B, Quince C, et al. Niche specialisation of terrestrial archaeal ammonia oxidisers. P Natl Acad Sci USA. 2011;108:21206–11. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubry-Rangin C, Kratsch C, Williams TA, et al. Coupling of diversification and pH adaptation during the evolution of terrestrial Thaumarchaeota. P Natl Acad Sci USA. 2015;112:9370–75. doi: 10.1073/pnas.1419329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. PHYML: a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid Symp Ser. 1999;41:95–8. [Google Scholar]
- Hunik JH, Meijer HJG, Tramper J. Kinetics of Nitrosomonaseuropaea at extreme substrate, product and salt concentrations. Appl Microbiol Biot. 1992;37:802–7. [Google Scholar]
- Jung M-Y, Park S-J, Kim S-J, et al. A mesophilic, autotrophic, ammonia-oxidising archaeon of thaumarchaeal group i.1a cultivated from a deep oligotrophic soil horizon. Appl Environ Microb. 2014;80:3645–55. doi: 10.1128/AEM.03730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M-Y, Park S-J, Min D, et al. Enrichment and characterisation of an autotrophic ammonia-oxidising archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl Environ Microb. 2011;77:8635–47. doi: 10.1128/AEM.05787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, et al. Isolation of an autotrophic ammonia-oxidising marine archaeon. Nature. 2005;437::543–46. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Koops H-P, Pommerening-Röser A. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol Ecol. 2001;72:1–9. [Google Scholar]
- Koops H-P, Purkhold U, Pommerening-Röser A, et al. Dworkin M, Falkow S, Rosenberg E, et al. The Prokaryotes. 3rd ed. vol 6. New York: Springer; 2006. The lithoautotrophic ammonia-oxidising bacteria; pp. 778–811. [Google Scholar]
- Koper TE, Stark JM, Habteselassie MY, et al. Nitrification exhibits Haldane kinetics in an agricultural soil treated with ammonium sulfate or dairy-waste compost. FEMS Microbiol Ecol. 2010;74:316–22. doi: 10.1111/j.1574-6941.2010.00960.x. [DOI] [PubMed] [Google Scholar]
- Lane DJ. Stackebrandt E, Goodfellow M. Nucleic Acid Techniques in Bacterial Systematics. New York: John Wiley and Sons; 1991. 16/23S rRNA sequencing; pp. 115–75. [Google Scholar]
- Lehtovirta-Morley LE, Ge C, Ross J, et al. Characterisation of terrestrial acidophilic archaeal ammonia oxidisers and their inhibition and stimulation by organic compounds. FEMS Microbiol Ecol. 2014;89:542–52. doi: 10.1111/1574-6941.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, et al. Cultivation of an obligate acidophilic ammonia oxidiser from a nitrifying acid soil. P Natl Acad Sci USA. 2011;108:15892–7. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, et al. Archaea predominate among ammonia-oxidising prokaryotes in soils. Nature. 2006;442:806–9. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, et al. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–9. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Stahl DA. Transcriptional response of the archaeal ammonia oxidiser Nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl Environ Microb. 2013;79:6911–6. doi: 10.1128/AEM.02028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol GW, Leininger S, Schleper C, et al. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia-oxidising archaea and bacteria. Environ Microbiol. 2008;10:2966–78. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Palatinszky M, Herbold, Jehmlich N, et al. Cyanate as an energy source for nitrifiers. Nature. 2015;524:105–8. doi: 10.1038/nature14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Rattei T, Flechl S, et al. amoA-based consensus phylogeny of ammonia-oxidising archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol. 2011;14:525–39. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SJ, Prosser JI. Protection of Nitrosomonas europaea colonising clay minerals from inhibition by nitrapyrin. J Gen Microbiol. 1991;137:1923–29. doi: 10.1099/00221287-137-8-1923. [DOI] [PubMed] [Google Scholar]
- Prosser JI. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1989;30:125–81. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 2012;20:523–31. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Qin W, Amin SA, Martens-Habbena W, et al. Marine ammonia-oxidising archaeal isolates display obligate mixotrophy and wide ecotypic variation. P Natl Acad Sci USA. 2014;111:12504–9. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reigstad LJ, Richter A, Daims H, et al. Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol Ecol. 2008;64:167–74. doi: 10.1111/j.1574-6941.2008.00466.x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayavedra-Soto LA, Arp DJ. Ward BB, Arp DJ, Klotz MG. Nitrification. Washington DC: ASM Press; 2011. Ammonia-oxidising bacteria: their biochemistry and molecular biology; pp. 11–37. [Google Scholar]
- Smit E, Leeflang P, Glandorf B, et al. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl Environ Microb. 1999;65:2614–21. doi: 10.1128/aem.65.6.2614-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Poehlein A, Offre P, et al. The genome of the ammonia oxidising candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14:3122–45. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- Stahl DA, Amann R. Stackebrandt E, Goodfellow M. Nucleic Acid Techniques in Bacterial Systematics. Chichester, UK: John Wiley & Sons Ltd; 1991. Development and application of nucleic acid probes; pp. 205–48. [Google Scholar]
- Stieglmeier M, Klingl A, Alves RJE, et al. Nitrososphaera viennensis sp. nov., an aerobic and mesophilic ammonia-oxidising archaeon from soil and member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol. 2014a;64:2738–52. doi: 10.1099/ijs.0.063172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglmeier M, Mooshammer M, Kitzler B, et al. Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidising archaea. ISME J. 2014b;8:1135–46. doi: 10.1038/ismej.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecker K, Dorninger C, Daims H, et al. Double labeling of oligonucleotide probes for fluorescence in situ hybridisation (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl Environ Microb. 2010;76:922–6. doi: 10.1128/AEM.02456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thion C, Prosser JI. Differential response of non-adapted ammonia‐oxidising archaea and bacteria to drying–rewetting stress. FEMS Microbiol Ecol. 2014;90:380–9. doi: 10.1111/1574-6941.12395. [DOI] [PubMed] [Google Scholar]
- Tourna M, Freitag TE, Nicol GW, et al. Growth, activity and temperature responses of ammonia oxidising archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–64. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, et al. Nitrososphaera viennensis, an ammonia oxidising archaeon from soil. P Natl Acad Sci USA. 2011;108:8420–5. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio EJ, Hantula J. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol Res. 2000;104:927–36. [Google Scholar]
- Vico Oton E, Quince C, Nicol GW, et al. Phylogenetic congruence and ecological coherence in terrestrial Thaumarchaeota. ISME J. 2015;10:85–96. doi: 10.1038/ismej.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F, Bak F. Ballows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H. The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Application. 2nd edn. New York: Springer; 1992. Gram-negative mesophilic sulfate-reducing bacteria; pp. 3352–78. [Google Scholar]
- Zhalnina K, de Quadros PD FAO Camargo et al. Drivers of archaeal ammonia-oxidising communities in soil. Front Microbiol. 2014;3:210. doi: 10.3389/fmicb.2012.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]