Abstract
Chromatin biology and epigenetics are scientific fields in rapid expansion due to their fundamental role in understanding cell development, heritable characters and progression of diseases. Histone post-translational modifications (PTMs) are major regulators of the epigenetic machinery, due to their ability to modulate gene expression, DNA repair and chromosome condensation. Large- scale strategies based on mass spectrometry have been impressively improved in the last decade, so that global changes of histone PTM abundances are quantifiable with nearly routine proteomics analyses and it is now possible to determine combinatorial patterns of modifications. Presented here is an overview of the most utilized and newly developed proteomics strategies for histone PTM characterization and a number of case studies where epigenetic mechanisms have been comprehensively characterized. Moreover, a number of current epigenetics therapies are illustrated, with an emphasis on cancer
Keywords: mass spectrometry, histone, histone variants, cancer, epigenetics, post-translational modifications, proteomics, chromatin, translational medicine
1. Introduction/Epigenetic overview
Epigenetics is the scientific field that studies heritable but reversible changes in gene expression that is not modulated by modification in DNA sequence. Cell differentiation is a typical example; multipotent cells such as embryonic stem cells diverge and get specialized into different tissues, despite having the same genome (Figure 1). The specific gene expression patterns of a cell that reflect its developmental state can be tightly and dynamically regulated by the activity of the underlying chromatin (chromatin structure modulation, epigenetic reprogramming). This epigenetic machinery includes DNA methylation [1,2], non-coding RNA-mediated changes of gene expression [3], use of histone variants, and histone posttranslational modifications (PTMs) mainly on histone tails [4]. Histones are small basic proteins (11 to 22 kDa) and highly conserved throughout eukaryotes. Histones are assembled into nucleosomes, which are the basic repeating units of chromatin, composed of eight histones wrapped by approximately 147 base pairs of DNA. Each nucleosome contains two copies of the four core histones (H3, H4, H2A, H2B) [5], tied together by linker histone H1. In addition to the canonical histones, several other non-allelic histone isotypes have evolved to distinct histone variants, replacing the canonical isoforms under specific requirements of the particular chromosomal processes. For instance, H2A.X has a C-terminal sequence which is more easily phosphorylated in case of DNA damage compared to canonical H2A [6], and it is also involved in sex chromosome inactivation [7] and meiotic silencing of unpaired chromatin [8]. H3.3 is a histone H3 isotype more enriched near promoters [9], and CENP-A substitutes canonical histone H3 in centromeres [10]. In overall, these five classes of histone proteins (H3, H4, H2A, H2B and H1) constitute the major protein components of the chromatin and provide a tight packing of the DNA to accomplish the first step in chromatin assembly, while still dynamically allowing regulatory proteins access to the DNA to fine tune gene expression, chromosome segregation and DNA repair. All histones undergo various combinations of diverse post-translational modifications (PTMs) such as acetylation and phosphorylation, which can be responsible for activation or repression of gene expression via opening and closing of chromatin structure [11] in a heritable manner. For instance, ε-N-acetylation neutralizes the positive charge of lysine residues of histones (i.e. H4K16ac), thereby weakening DNA-histone interaction; thus chromatin structure becomes loose and hence more accessible [12].
Figure 1. The genome and the phenotype during development and role of epigenetics.
Representation of the massive change of a human phenotype during development, while maintains a nearly identical DNA sequence through the entire lifecycle. Epigenetics is a major effector in this process, as gene expression is modified and then modifications inherited during embryonic development of specialized tissues.
In this review, we present a summary of the state-of-the-art proteomics techniques adopting mass spectrometry (MS) used for the characterization of histones and their combinatorial PTMs. We describe how these techniques are being used to better understand basic biology and human diseases with an emphasis on cancer.
2. Epigenetics in Health and Disease: Translational and Clinical Implications
Epigenetic processes are fundamental to normal development (Figure 1) and have been linked to a variety of human diseases [13]. Aberrations in epigenetic processes, such as modifications on DNA and histones, can significantly disrupt gene regulation, and may lead to developmental disorders and disease development such as cancer. Cancer is a disease long known to have genetic causes. However, it has been recently demonstrated to also have epigenetic alterations, like changes in DNA methylation and histone PTMs. The earliest evidence of the link between cancer and epigenetics was obtained from DNA methylation studies [14]. However, over the past decade it has become increasingly apparent that covalent modifications of histone proteins and alteration in the activity or expression of histone-modifying enzymes play a critical role in the development and progression of cancer. For example, Lee et al. shown that global histone acetylation levels and hence the regulation of the epigenome are altered in cancer cells related to metabolic reprogramming mediated by oncogenic activation of a signal transduction pathway [15]. A comprehensive understanding of epigenetic mechanisms, their interactions and alterations in health and disease has significant implications for the prevention, diagnosis and treatment of major human diseases and for ageing. These epigenetic alterations can potentially be used in clinical practice as biomarkers for early detection or progression, chemosensitivity, and prognosis of cancer. The development of these biomarkers to clinical practice will require a deep knowledge of the epigenetic state of a cell at certain times or conditions. Yet, despite the importance and impact of epigenetics in basic biology and medicine, it remains poorly understood. Therefore, there is a large effort in developing new technologies to unravel the mechanisms underlying disease development and progression, including finding the genes and proteins with PTMs that are altered in function or abundance, affecting the epigenetic state. Greater understanding of the epigenetic landscape of specific cancer tissues or cell types may lead to identification of potential cancer biomarkers for disease detection, monitoring and molecular characterization. This may also facilitate personalized cancer treatment.
A cell's chromatin acquires developmentally important epigenetic marks during differentiation that regulate the expression of specific genes. These regulatory control marks, such as histone PTMs, survive cell division and could potentially be transmissible across multiple cell generations. In this regard, Alabert and Groth reviewed the mechanisms, which lead to the recycling of chromatin histones in the newly replicated DNA during cell replication [16]. Although epigenetic processes are heritable, they are responsive to developmental, behavioral and environmental cues. Hence, unlike genetic alterations, epigenetic abnormalities are potentially reversible, which makes them ideal targets for drug treatment. Steps have been made in this regard. The United States Food and Drug Administration (FDA) has approved drugs targeting two epigenetic targets for cancer treatment: the DNA methyltransferases (DNMTs) and histone deacetylases (HDACs). A number of other drugs are in different stages of pharmaceutical development, so that Dawson and Kouzarides recently stated that “we have now entered an era of epigenetic cancer therapies” [17].
DNA methylation – Among epigenetic marks, DNA methylation abnormalities, including hypomethylation and hypermethylation, are the most studied in cancer to date. DNA methylation naturally occurs on cytosine bases at CpG islands, leading to gene inactivation [2]. DNA hypomethylation at CpG dinucleotides was the first epigenetic abnormality to be identified in cancer cells [14]. Feinberg and Vogelstein made the first link between hypomethylation and cancer, showing that a substantial proportion of CpGs that were methylated in normal tissues were unmethylated in cancer cells [14]. DNA hypomethylation in tumor cells is primarily caused by loss of methylation from repetitive regions of the genome causing genomic instability and changes in gene imprinting. In contrast, Baylin's group subsequently demonstrated a relationship between hypermethylation and gene silencing [18] in which abnormal methylation is linked to cancer via silencing of tumor-suppressor genes by DNA hypermethylation [19]. In some cases, DNA hypermethylation can be prevented using DNMT inhibitors (DNMTi's) such as 5-azacytidine (see `Epigenetic therapies' below for details), leading to DNA hypomethylation and cytotoxicity (at higher doses) in abnormal hematopoietic cells in the bone marrow. Numerous studies have demonstrated the role of DNA methylation in different cancer types in the recent past [20]. It is now commonly accepted that inactivation of tumor-suppressor genes occurs as a consequence of DNA hypermethylation within the promoter regions and many researchers have demonstrated a broad range of genes silenced by DNA methylation in different cancer types.
Histone PTMs – The N-terminal sequences (tails) of the core histones protrude from the tightly packed globular region, making them accessible to histone modifying enzymes [5]. This region of the sequence is heavily modified by dynamic post-translational modifications (PTMs) [11]. These covalent modifications include acetylation of lysine residues, methylation of arginine (mono- or di-) and lysine (mono-, di-, or tri-) residues, phosphorylation of serine, threonine and tyrosine residues, and other yet poorly characterized PTMs such as ubiquitination [21], SUMOylation [22], crotonylation, formylation and propionylation/butyrylation [23–25] of lysine, ADP ribosylation [26], citrullination and O-linked N-acetylglucosamine (O-GlcNAc) [27,28]. Many combinations of these covalent modifications are possible simultaneously on the core histones, so that in the early 2000s the presence of a `histone code' comparable to the genetic code was proposed [29]. More recently, the term histone code has been replaced by `histone PTM cross talk' [30], as the difficulties in assigning to the myriad of combinatorial PTMs a specific function became rapidly evident, and opened a debate whether a proper code really exists. It is now appreciated that histones and their accompanying PTMs are essential components of the epigenetic machinery responsible for cell development, and that aberrations in this process can be associated with initiation and progression of various diseases including cancer and neurodegenerative diseases. Hence, the characterization of histone PTMs and their combinatorial patterns gained greater importance and has become a priority in biomedical research.
The characterization of the role of histone modifications in cancer is highly complex, due to the large variety of PTM combinatorial patterns and the incomplete knowledge of how histone PTMs modify gene expression. A number of histone PTMs and mutations are known to be directly linked to aberrations and diseases. For example, loss of acetylation at lysine 16 (H4K16Ac) and loss of trimethylation at lysine 20 (H4K20me3) of histone H4 were the common histone marks reported to be deregulated in many different type of cancer cells of humans and mouse [31]. However, there is still much more to be learned about the link between histones and the occurrence and progression of diseases such as cancer.
Histone readers – Even though histone PTMs regulate the chromatin structure by directly altering the chemical environment of the surrounding chromatin, e.g. lysine acetylation neutralizing the positive charge of this residue [12], their major roles at molecular level is recruiting other proteins (known as readers) by providing ligands for their specific domains, e.g. proteins containing bromodomain [32], chromodomain and Tudor domains [4]. It is anticipated that any alterations on these epigenetic regulators may potentially drive tumorigenesis. Indeed, the PHD finger of JARID1A (JARID1APHD3) or PHF23 (PHF23PHD), which binds histone H3 lysine4 trimethylation (H3K4me3), is implicated in acute myeloid leukemia (AML) [33,34]. A genetic fusion protein containing nucleoporin protein 98 (NUP98) with the PHD finger of JARID1A [33] or PHF23 [35] expression was reported in AML patients. Later studies reported that a PHD finger that specifically recognizes H3K4me3/2 marks is essential for leukemogenesis and mutations in PHD fingers that abrogated H3K4me3-binding also abolished leukemic transformation [34]. Therefore, it is concluded that the oncogenic properties of the NUP98-PHD finger fusion proteins are directly potentiated by the ability of the PHD finger to bind chromatin.
Histone writers and erasers – The deposition and removal of histone PTMs is performed by histone modifying enzymes such as histone acetyltransferases (HATs), histone methyltransferases (HMTs) and kinases (known as the “writers”) or histone deacetylases (HDACs), histone demethylases (HDMs) and phosphatases (known as the “erasers”) [36]. Mutations or altered expression of these enzymes have been implicated in progression of a number of diseases including cancer, and inhibition of aberrant enzyme activities is expected to attenuate pathogenesis resulting from misregulated gene transcription. For example, it has been reported that the HMT Smyd3 (SET and MYND domain containing protein 3) which catalyzes histone H4 lysine 5 methylation (H4K5me) is highly overexpressed in several cancers including liver, breast and rectal carcinomas [37]. Another HMT EZH2 (Enhancer of zeste homolog 2) or related EZH1 as a catalytic subunit of the polycomb repressive complex 2 (PRC2) has been shown to help maintain epigenetic gene silencing and hematopoietic lineage specification at various developmental stages through enzymatic di- and trimethylation of K27 on histone H3 (H3K27me2/3) under normal conditions [38–40]. Elevated levels of EZH2 are correlated with aggressiveness and poor prognosis in solid tumors including cutaneous melanoma, pediatric glioblastoma, prostate, breast, bladder and endometrial cancers [41–44]. In agreement with these observations, mutation of lysine to methionine in position 27 of histone H3 (H3K27M) observed in pediatric brain tumors inhibits the enzymatic H3K27 tri- and di-methyltransferase activity of the PRC2 by blocking free PRC2 in the cells [44]. Taken together, EZH2 has received much attention as a promising epigenetic target, with dedicated efforts towards identifying effective, selective inhibitors (see `Epigenetic therapies' below).
Histone variants – Another mechanism of changing epigenetic properties of active genes is the replacement of a canonical histone by a non-canonical variant. In most organisms, the canonical histones H1, H2A, H2B, H3, and H4 are encoded by multiple gene copies, which are highly similar in sequence. All these canonical histones are almost exclusively expressed during the DNA replication/S phase of the cell cycle to supply histone proteins to package the newly replicated genome [45,46]. The histone variants, however, are typically present as single-copy genes and in contrast to canonical histones, variants are not restricted in their expression to the S phase but expressed throughout the cell cycle [47,48]. For that reason, they are also known as “replacement histones”. Interestingly, the core histones differ with respect to their propensity to diversify into multiple variants in a range from very few amino acid differences to extremely divergent changes. For instance, most eukaryotes including humans have a single histone H4, coded by 14 genes, but several H2A paralogs that exhibit the highest variety of histone variants, mostly due to divergences in length and sequence in their C-terminal tails. Replacement of the canonical histones with the histone variants alters the biochemical and biophysical nature of the nucleosome by affecting PTMs, protein interactions or higher-order chromatin structure, thereby these may play a role affecting distinct cellular processes. For example, one of the most extensively studied histone variant H2A.Z, which is highly conserved and essential for many organisms, has been linked to a wide variety of different nuclear functions, including DNA replication and cell-cycle control [49], DNA damage repair [50], transcriptional activation and repression [51–53], maintenance of heterochromatin [54], anti-silencing [55], chromosome segregation [56], and genome integrity [57,58]. The diverse roles of histone variants in many major epigenetic processes lead to the expectation that dysregulation can result in pathological disorders, and indeed altered expression of variants (i.e. many H2A variants) has been implicated in cancer. In support of this hypothesis, H2A.Z is highly expressed in embryonic stem cells and in several cancers including colorectal tumors [59], prostate [60], bladder [61], and breast cancer [62]. Overexpression of H2A.Z in breast cancer, where H2A.Z role has been best characterized, leads to increased metastasis risk and decreased patient survival. Another histone H2A variant macroH2A has been shown to suppress melanoma progression by regulating CDK8 [63]. Although many aspects are not fully understood as yet, growing evidence indicates that the combinatorial histone PTMs and variants along with histone modifier enzymes are of clinical importance. Understanding the function and regulation of these PTMs and variants in epigenetic processes remains a major challenge for the future.
3. Strategies to analyze histone PTMs and variants by mass spectrometry-based proteomics
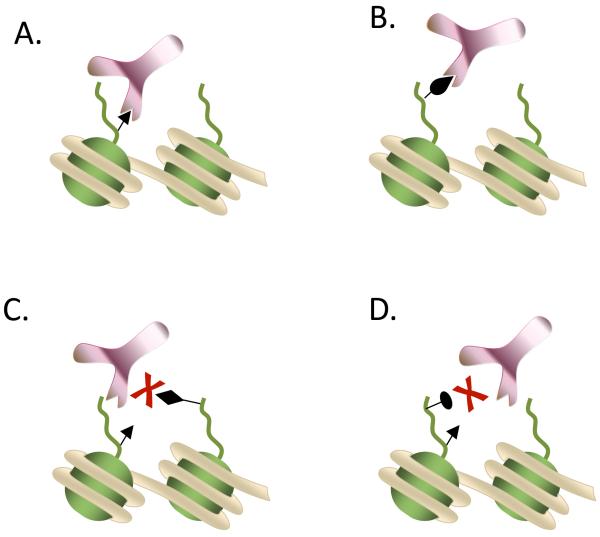
Identifying histone modifications and how they differ across diverse biological conditions and cell types is one of the current challenges in chromatin biology. Originally, this challenge was addressed by using Edman degradation for protein sequence determination (microsequencing) [64,65]. More recently, this method was replaced by antibody-based methods (Figure 2A) such as western blotting, immunofluorescence analysis and chromatin immunoprecipitation (ChIP) which are still widely used for low throughput studies. Microsequencing was used in early histone PTM studies, but it is time consuming and requires a large amount of highly purified sample, making it not ideal. Although antibody-based strategies are highly sensitive, employment of this methodology to identify the wide array of histone modifications presents a number of hurdles that prevent their practical use (Table 1, Figure 2). First, antibody-based assays require a priori knowledge of the type and position of the modification of interest. Besides, modification-specific antibodies are not always straightforward to generate. For example histone antibodies are not always specific for their intended PTM targets, and often recognize `off-target' proteins and PTMs (Figure 2B), reducing the efficacy of many approaches. The high sequence homology between variants as well as the high diversity and multivalency of the PTMs in histones, further hinders the ability to produce high-specificity antibodies for individual histones and the variants. In addition, antibodies can suffer from cross-reactivity with similar modifications embedded within an identical amino acid context on the same or a different histone protein as well as non-histone proteins carrying similar modifications and sequences [66]. Moreover, modifications that exist on adjacent or closely located residues within the same histone can potentially generate an epitope-masking effect (epitope occlusion) [67]. As a result, modifications may escape detection by antibodies that are not specifically designed to recognize both modifications on the same epitope (Figure 2C and 2D). Finally, traditional antibody-based methods lack high throughput capabilities, and typically analyze one PTM at a time by targeting specific isoforms. This makes it virtually impossible to measure combinatorial PTMs occurring within the same histone molecule.
Figure 2. Antibody steric hindrance during histone post-translational modification recognition.
Schematic representation of an antibody that is (A.) targeting its epitope without any interference, (B.) recognizing `off-target' epitope, (C.) not recognizing the epitope due to a nearby secondary histone modification and (D.) not recognizing the epitope due to a secondary histone modification distantly located from the target.
Table I.
Comparison between antibody and mass spectrometry based characterization of histone modifications
| Advantages | Limitations | |
|---|---|---|
|
| ||
| Antibody-based assays | •High sensitivity of detection for low abundant proteins if specific antibodies are available | •Only known modifications can be studied |
| •Do not require expensive instrumentation | •Limited by the availability of antibodies, requires generation of custom antibodies for each target | |
| •Rapid sample preparation and interpretation of the results | •Specificity and performance of the antibodies is sometimes not sufficient (antibody cross reactivity, no detection) | |
| •Allows to characterize DNA-protein interactions (i.e. ChIP-Seq) | •Epitope occlusion might occur because of neighboring marks | |
| •Very rarely can be applied to multiply modified peptides/proteins | ||
| •Lack high-throughput capabilities | ||
| •High cost for large-scale studies | ||
|
| ||
| MS-based assays | •Capable of simultaneously monitoring multiple PTMs in single experiments | •Sample preparation often requires more steps, before the sample is suitable for LC-MS |
| •Accurate relative quantification of global changes of histone PTMs | •Limited in high sensitivity as compared to antibody-based techniques | |
| •Allow discovery of previously unknown modification patterns | •Intact protein analysis (top-down) is still technically challenging, implying that MS analysis is still peptide centric, with all of the limitations related to a partial view of the protein sequence (e.g. discrimination of protein variants) | |
| •Can be adopted to investigate combinatorial codes | •High expertise is required for data analysis, in particular for middle-down and top-down datasets | |
| •Can be used in a high-throughput manner | •Validation of the MS results with antibody-based techniques is still recommended, as it is more widely accepted strategy | |
Mass spectrometry (MS)-based proteomics has emerged as a high throughput strategy to characterize proteins and protein PTMs in general, introducing an effective tool for histone analysis. The high speed (scan rate >10 Hz), high resolution (>60,000) and the possibility to combine MS acquisition with online separative techniques such as nano liquid chromatography (nLC), which led to highly sensitive analyses (<fmol), has made MS the technique of choice for discovery and quantification of histone PTMs. MS based methodology have several advantages over traditional histone analysis methods (Table 1). MS has the ability to perform unbiased profiling that accounts for diverse modifications simultaneously as opposed to restricting detection to priori-selected particular modifications as in antibody based methods. Moreover, as the MS signal is related to the concentration of the analyte, it can also be used in a quantitative manner for comparative analysis of protein expression levels as well as differential expression analysis of protein modifications. Furthermore, MS-based proteomics integrates bioinformatic tools that allow discovery of previously unknown modification patterns.
Mass spectrometry based histone analysis technology is well suited to study of a system-level view of histone modifications, however it is currently unable to map the PTM patterns to defined domains of the genome or to specific gene and promoter regions. Chromatin immunoprecipiation techniques have been used for this type of analysis. Recently, different comprehensive combinatorial approaches have been developed to capture the histone marks at functionally distinct chromatin (i.e. chromatin affinity purification with mass spectrometry (ChAP-MS) and Chromatin Proteomics (ChroP)/ChIP-MS) [68,69]. Thereby, the parallel analysis of histone marks and their binding proteins at functionally distinct chromatin regions can potentially be facilitated [70]. We anticipate that this new methodologies will allow harnessing of the power of MS to study regional histone modifications.
The MS-based proteomics strategies – MS strategies have been conventionally divided into three groups, defined as bottom-up, middle-down and top-down, depending on the portion of histone analyzed (Figure 3 and Figure 4A). The traditional approach is `bottom-up' proteomics, where the target protein is proteolytically digested into short (5–20 aa) peptides prior to MS analysis. On the other hand, the `top-down' approach is the analysis of the intact protein. The top-down approach is the ultimate goal to define the histone proteoforms present in a sample, where the proteoform is defined as a protein with a unique sequence and PTM combination [71]. However, performing the top-down analysis in a high throughput manner is still challenging, as smaller molecules such as peptides are easier to separate by LC and analyze by MS. Moreover, the larger the portion of protein analyzed, the higher is the probability to have isobaric proteoforms, which are species with same mass and nearly identical physico-chemical properties (e.g. H3K27me1K36me2 and H3K27me2K36me1) (Figure 4B). Therefore, a third strategy named `middle-down' has been developed to takes advantage of the fact that histone N-terminal tails can be cleaved off by specific proteases, generating a polypeptide with accessible size for separation and detection (50–70 aa). These technologies differ in their capacity and compatibility for shotgun or large-scale discovery and requirement for MS instrumentation (Figure 3). In the following section, we discuss such techniques used to characterize histones.
Figure 3.
Summary of bottom-up, top-down and middle down approaches in histone post-translational modification analysis
Figure 4. The mass spectrometry-based proteomics strategies for histone analysis.
(A.) Histones are digested into short peptides when adopting the bottom-up strategy (top left). The use of short peptides prevents the acquisition of combinatorial post-translational modification (PTM) data, as the peptides detected cannot be used to verify whether two modifications (e.g. red and yellow circles) were present simultaneously on the same histone protein. The middle-down strategy partially overcomes to this issue, since it allows for the analysis of the entire histone tail (top right), but it cannot provide data of histone PTMs localized on the nucleosome core. Top-down allows for the analysis of the intact histone (bottom left). The ultimate goal would be the analysis of the entire nucleosome (most probably cross-linked), but mass spectrometry of histone complexes, especially if modified, is still highly challenging (bottom right). (B.) Example of the increasing complexity in going from bottom-up to top-down analysis of histone H3 (green colored and highlighted by a pink glow). For example considering only methylation, a histone H3 peptide has on average 2–3 modifiable residues, i.e. arginine and lysine residues. An average bottom-up peptide with two lysine and one arginine residue could exist in 64 different combinations (K can be unmodified, me1, me2 and me3; R can be unmodified, me1, me2). Of these, 54 have at least one isobaric form, defined as alternative PTM combination with the same intact mass (e.g. H3K27me2K36me1 vs H3K27me2K36me2). In the middle-down and the top-down strategy this issue grows exponentially, as the only species with no isobaric forms are the completely unmodified and the completely methylated sequence. Even though it is less likely that all the possible combinations are exist in nature, this issue is challenging as it leads to highly complex MS/MS spectra, due to the myriad of co-fragmenting proteoforms.
3.1. Bottom-up analysis of histones
The main challenge in characterization of histone proteoforms is the high degree of complexity of the histone proteins due to the allelic variants and the diversity and multivalency of the PTMs in histones. The `bottom-up' approach is the most commonly used method at present when dealing with high complexity samples for large-scale analysis. The bottom-up approach is also well suited for analyzing chemical modification of peptides along with the peptide and protein semi-quantification. A typical strategy for analysis for histone modifications by bottom-up MS approach involves enzymatic digestion of histone proteins into small (<3 kDa) peptides and further separation of the digested peptides often by reversed-phase (RP) nLC prior to MS analysis. Burlingame and colleagues were the first to use bottom-up proteomics technology in analysis of histone proteins. They reported the first comprehensive analysis of human H4 modification [72] and chicken H3 modifications [73]. In this study, enzymatically digested purified histones were analyzed using RP-LC followed by MS. By using this technique, they were not only able to confirm known methylation and acetylation sites, but also identified H3K79 as novel methylation site on chicken histone H3.
Although the confident assignment and quantification of redundant peptides with modification can be a key issue for the peptide centric approach, analysis of short peptides has the advantage of a better front-end separation of peptides as compared to proteins (Figure 5A), higher sensitivity and mass accuracy and simpler tandem mass spectra (MS/MS) that results in predictable fragmentation products and straightforward sequence database searching for peptide identification and PTM assignments. However, the most widely used proteases in bottom-up proteomics, such as trypsin that cleaves the peptide bonds at the carboxyl end of lysine and arginine residues, are problematic for histone analysis as histone proteins are highly enriched in basic residues. Trypsin digestion of histones tends to yield many small (1–3 aa) and hydrophilic peptides that are difficult to retain on RP-HPLC columns and analyze by MS as they are below the mass dynamic range of MS detectors. While reducing trypsin incubation time and the enzyme to substrate ratio is one way to alleviate this difficulty, it is still challenging to generate reproducible homogenous peptides between samples due to limited digestion. Redundant missed-cleavage events can cause a non-homogenous pool of peptides in which the same PTM site exists on several different peptides, making quantitation of these marks extremely difficult. Thus, chemical derivatization has been proposed as a means to increase peptide hydrophobicity and to modify cleavable residues such as amine groups in histones (N-terminal amines, and unmodified and monomethylated lysine ε-amino groups) before trypsin digestion [74]. In a recent study, different methods to generate histone proteolytic peptides were systematically evaluated to maximize both sequence coverage and sensitivity for a comprehensive histone PTMs analysis [25]. Lysine propionylation of core histones generating highest number of complementary peptide sequences were noted, which boosted the sequence coverage of peptide mapping by MS, and hence has led to the identification of many novel PTM sites, including 28 lysine crotonylation (Kcr) sites, 18 lysine monomethylation (Kme) sites, 1 lysine dimethylation (Kme2) site, 4 lysine formylation (Kfo) sites, 2 lysine acetylation (Kac) sites, 8 arginine monomethylation (Rme) sites, and 6 tyrosine hydroxylation (Yoh) sites [25]. Treatment of histones with propionic anhydride converts any free amine groups including the N-termini of peptides and ε-amino groups of unmodified and monomethylated lysine residues to propionyl amides, thereby blocking them from trypsin cleavage. This leads to protease cleavage exclusively at arginine residues, resulting in generation of peptides of a length that is suitable for LC-MS analysis. Propionylation also increases the hydrophobicity of the peptide, which leads to enhanced chromatographic resolution on RP-HPLC. An alternative chemical derivatization strategy is the use of deuterated acetic acid/anhydride to acetylate all lysines, blocking them from tryptic digestion [75]. Even though deuterated acetic acid/anhydrate derivatization is less commonly in use, a number of groups employed the technique due to its advantages that enables MS to measure protein abundances quantitatively, thereby allows for the differentiation of endogenously acetylated residues from chemically acetylated residues [76]. Overall, chemical modification of histones prior to trypsin digestion addresses some of the limitations initially associated with trypsin cleavage.
Figure 5. Ion map of a typical bottom-up and middle-down LC-MS run.
(A.) Bottom-up LC-MS ion map of histone extract. A typical bottom-up analysis is performed with a relatively short gradient (< 1 hour) using C18 chromatography, and histone peptides are eluted in sharp peaks (<1 min) with a precise but disperse order. Different peptide sequences and with multiple modifications are spread all over the chromatogram. However, some trends are conserved, and can be used to confidently identify histone peaks. For instance, a peptide carrying only one modification and derivatized with propionic anhydride is eluted with the order di- and trimethylated, acetylated, unmodified (propionylated) and monomethylated (propionylated). (B.) Middle-down LC-MS ion map of histone H3 N-terminal tails. A typical middle-down analysis is performed using a long gradient (2–3 hours) with WCX-HILIC chromatography. Histone variants are usually fractionated in different tubes, in order to achieve a more in depth characterization of a given variant. Histone tails elution has a specific trend, where more modified species are eluted first. Acetylation (ac) has a larger influence in the retention time as compared to methylation (me).
Most bottom-up MS experiments use high resolution and high mass accuracy MS for detection of intact peptide masses. Fragmentation (MS/MS) is typically accomplished and detected in the linear ion trap by collision induced dissociation (CID) because of its speed and sensitivity. This type of analysis was used, for instance, to investigate global alterations of histone PTMs in colon cancer samples with their normal counterparts [77]. In this study, the bottom-up MS was not only able to identify 41 distinct PTM sites of histone peptides, but also revealed a differentially regulated mark, histone H3 lysine 27 acetylation (H3K27Ac), that had not been previously shown to be altered in colon cancer. The bottom-up MS approach has been used in combination with diverse platforms and quantification strategies such as stable isotope labeling by amino acids in cell culture (SILAC) [78,79] or label-free approaches [77,80–82] to compare the histone PTM profiles within diverse biological backgrounds. For instance, using the SILAC approach, Jaffe et al. monitored 42 distinct combinations of histone modifications on H3 across 115 Cancer Cell Line Encyclopedia (CCLE) lines [79]. In another study, SILAC [83] has been successfully used to quantify H3 and H4 PTMs of four cancer cell lines in comparison with normal epithelial breast cells to determine potential breast cancer histone PTM signatures [78]. Results demonstrated significant changes in relative abundance of several marks in the N-terminal tails of histones H3 and H4 between cancer cells and normal cells, which can potentially be considered as hallmarks of human tumors. SILAC can also be used pulse-chase style by labeling proteins [84] or by introducing the isotopic labels into modifications [85] at different time points to investigate the kinetics of histone synthesis, degradation and turnover rate of histone PTMs. Data can be obtain via these approaches can be particularly important to better understand modification kinetics of tumor cells as epigenetic instability often observed in cancer cells. Label-free quantitative MS analysis has been used more often in many studies. For example, a recent study used this methodology to characterize quantitative histone modification signature of 20 different cancer cell lines from a diverse variety of tissue origins, including cervix, prostate, lung and breast tissues [86]. Ion intensity-based label-free method has been successfully used to quantify all core histones PTMs from small cell lung cancer cells upon treatment with an HDAC inhibitor [80]. These and many other successful histone analyses studies [87,88] were accomplished using CID fragmentation [86]. However, as previously mentioned the bottom-up strategy is limited in protein sequence coverage, which also affects PTM assignment to the proper histone variant. For instance, this method has limited ability to distinguish the histone H3 variant as several peptides are common between the variants H3.1, H3.2 and H3.3, while H3.3 has different roles in the chromatin as compared to the others [9]. A similar limitation exists in the case of PTMs localized on distant region of the histone sequences such as H3K9me, H3K27me and H3K36me as they are known to cross talk with each other, and the analysis of short peptide sequences prevents the quantification of their co-existence frequency. Overall, bottom-up is the most sensitive and high throughput strategy for histone analysis, but it provides incomplete information about the different proteoforms.
3.2. Top-down analysis of histones
In order to characterize PTM cross talk along the entire protein sequence, which is crucial for in-depth understanding of the role of histone marks in chromatin regulation, PTMs must be detected on the same analyzed molecule. Top-down analysis of intact histone proteins by MS [89] has the advantages of characterizing combinatorial modifications across the entire protein, but currently it is technically challenging. The difficulty to obtain quality MS/MS spectra for intact proteins as compared to short tryptic peptides is one of the major factors responsible for the low analytical sensitivity of top-down MS. The complexity of the data generated is also a limiting obstacle that needs to be addressed, as well as the efficiency of separation of the different proteoforms. The impossibility of separating species with the same type and number of modifications, but scrambled into many different combinatorial patterns, leads to the generation of mixed MS/MS spectra containing a large number of proteoforms (Figure 4B). Despite the advances in data analysis software such complex spectra are still far from having their information completely deconvoluted.
Traditionally, FT-ICR-MS has been most commonly used in top-down studies [89], as high resolution MS and MS/MS is mandatory for the characterization of intact histones. Due to the improvements in high resolution mass analyzers this method is currently performed also using the orbitrap and time-of-flight (TOF) analyzers. Early top-down MS experiments utilized low-energy CID methods to induce protein fragmentation in which the peptide bond is cleaved upon collision with an inert gas, generating b- and y- ions. For example, one of the early top-down MS/MS analyses of a small intact protein (ribonuclease A) has been done on a triple quadrupole mass spectrometer using CID fragmentation [90]. Similarly, a number of different intact proteins have been successfully analyzed using CID-based MS/MS following the in-source fragmentation in a quadrupole TOF [91]. However, this approach is not suitable for histone analysis, due to the incomplete fragmentation generated by CID, which prevents accurate mapping of PTMs. This issue is enhanced in histones, as their most abundant modifications (acetylation on lysine and methylations on lysine and arginine) can be catalyzed on amino acid residues often localized next to each other in the protein sequence. Moreover, histones with near isobaric coexisting PTMs such as acetylation (42.011 Da) and trimethylation (42.047 Da) are very difficult to distinguish. CID is also not ideal for proteins/peptides containing labile PTMs such as phosphorylation on serine (Ser) and threonine (Thr) residues, since the phosphate group competes with the peptide backbone as the preferred site of cleavage. Upon collision activation, phosphoric acid might be displaced from the peptide, and result in the peptide backbone bonds remaining intact, preventing efficient MS/MS fragmentation. Electron capture dissociation (ECD) [92] and electron transfer dissociation (ETD) [93] are more suitable for long and heavily charged protein sequences such as intact histones. Moreover, ECD and ETD are both softer fragmentation techniques in which low energy electrons react with the positively charged analyte, generating a rearrangement of bonds with consecutive cleavage of the peptide backbone. While ECD is a slow reaction requiring FT-ICR mass analyzers to trap both analyte and electrons, ETD is a more modern technology that can be performed on low resolution analyzers (e.g. ion trap) [93] allowing higher speed [94] and cost effective instrumentation. This makes it compatible with LC-MS/MS timescales [95]. Top-down has been applied to histones since the early 2000s, where Banks et al. characterized using C4 chromatography and a Q-TOF the differential dephosphorylation of histone H1 isotypes under the effect of the hormone dexamethasone [96]. This technique was largely improved by Kelleher and his co-workers, introducing MS/MS fragmentation, direct infusion of purified histone variants and ECD fragmentation. This led to the accurate characterization of all histone canonical variants, including H3 [97], H2A [98], H2B [99] and H4 [100].
Top-down proteomics is often coupled to some type of up-front protein separation and purification prior to MS in order to reduce the sample complexity. The separation is mostly performed off-line from MS analysis. Achievement of more effective separation will allow identifying and distinguishing distinct proteoforms with improved sensitivity by reducing the masking effect of highly abundant proteins. As previously mentioned, the efficiency of protein separations presents a major analytical challenge [101]. Currently there is no method that is capable of separating the components of a complex sample in a single-step action. Therefore most of the time, multidimensional fractionation is necessary for deep exploration of complex proteomes and low abundant proteins. Earlier, traditional 2D-PAGE with isoelectric focusing and SDS-PAGE followed by different visualization techniques was the closest achieved top-down proteomics method providing a snapshot view of proteins in a sample in one or more biological states. This technique has been applied in cancer research, such as esophageal cancer and ovarian, breast, colon, kidney, lung and stomach tumors [102,103]. However, it is not straightforward for top-down MS due to the difficulty in eluting proteins out of the gel for intact protein MS analysis. Regardless, multiple research groups continue to use and develop various gel-based methods for top-down proteomics analysis. For example, capillary zone electrophoresis (CE) coupled to HPLC as front-end separation for proteins prior to MS has been widely studied with success. Recently, Sarg et al. reported the first comprehensive capillary electrophoresis coupled to electrospray-MS analysis application of histone PTMs [104]. In this study, using core histones from hyperacetylation-induced mouse erythroleukemia cancer cell lines, intact histone subtypes and their multiply modified forms were successful separated and the extent of their PTMs were detected in a fast and reproducible way. Another approach to fractionate intact proteins for top-down MS analysis has been developed using acid-labile surfactant (ALS) with gel based fractionation followed by gel-free RP-LC [105].
A more widely adopted strategy is LC that can work well to separate intact proteins. Traditional RP-LC using C18/C4 columns is frequently performed for separation of extracts of intact histones. In fact, the first modern MS analyses of histone modifications employed RP-HPLC to separate histone isotypes for top–down MS analysis [96]. An alternative method for LC separation of histone isotypes is hydrophilic interaction liquid chromatography (HILIC), which will be discussed in the next section.
3.3. Middle-down analysis of histones
Both bottom-up and top-down MS approaches have a unique set of limitations as well as a complementary set of advantages as discussed above. An intermediate approach to these two methods is middle-down MS [106], which has emerged as an alternative high throughput technology to analyze proteins and co-occurring PTMs. The middle–down approach is still a peptide-based technique employing proteolytic digestion, but it differs from bottom–up in terms of the size of peptides generated. This can be accomplished by enzymes other than trypsin such as Asp-N and Glu-C, which cleave less frequently-occurring amino acid residues in proteins [107,108] or by microwave-accelerated acid digestion [109], to yield larger peptides around 4–7 kDa. This characteristic of middle-down is particularly appealing for the characterization of histones and their PTMs, as it is possible to obtain intact N-terminal tails where the majority of the PTMs reside (Figure 4A and 6). For example, endoproteinase AspN is commonly used to generate the H4 1–24 peptide, which contains all known H4 PTM sites, and endoproteinase GluC is often used to generate the H3 1–50 peptide, which contains most of the known H3 PTM sites. These larger peptides can be further purified and analyzed by MS and MS/MS using CID and ETD as discussed earlier. Thus, middle-down MS combines most benefits of the top-down and bottom-up approaches by exploiting the ability to detect protein isoforms, variants, and combinations of multiple PTMs on an intact histone tail, while reducing technical challenges of whole histone protein analysis.
Figure 6. Deconvoluted MS/MS spectrum of a modified histone N-terminal tail.
MS/MS spectra generated using the middle-down strategy are usually rich in heavily charged product ions, that are deconvoluted into single charges to assist database searching and identification. The present spectrum was obtained using ETD fragmentation, which generates c and z ions, and split into three rows for simpler visualization.
Until recently, separation of intact proteins prior to MS analysis has not been as readily utilized for middle-down and top-down proteomics. RP-LC is often in use to separate histone polypeptides to some extent. An alternative method adapted for LC separation of histone tail peptides and intact histone proteins is weak cation exchange (WCX)-HILIC [110,111]. The resin is a mix of normal phase and ion exchange chromatography separation [111], where separation can be performed by using a gradient of water/organic solvent and salt concentration off-line from MS [112]. However, more recent improvements replaced salt with gradient of pH, which is more compatible for online LC-MS [113–115]. In this technique the order of elution is opposite that of RP-HPLC, with hydrophilic analytes being retained longer than hydrophobic ones (Figure 5B).
Although the technology is improving at a fast rate for protein separation technologies as well as MS instrumentation and MS/MS fragmentation techniques for proteomics applications, there is still room for further improvement. Encouraging progress has been accomplished in recent years indicating that top-down and middle-down approaches may become a truly high-throughput compliment to bottom-up MS in near future. For instance, Tran et al. reported an early glimpse of high-resolution LC-ESI-MS/MS approach for high-throughput top-down proteomics, demonstrating proof of-principle evidence of the efficacy of this methodology [116]. More recently, Gault et al demonstrated a case study of the method in characterizing the PTMs of bacterial protein proteoforms where multiple modifications of the same mass are present [117]. Sidoli et al. reported an optimized middle-down setup, where a hybrid LC setup and bioinformatics tools were introduced to increase the automation of LC-MS and data analysis [114]. One major issue of top- and middle-down proteomics remains their lower sensitivity as compared to bottom-up as larger peptides can occupy a larger distribution of charge states, resulting in a dilution of signal for any given charge state compared to smaller peptides. Moreover, currently available bioinformatics tools for middle-down and top-down are still limited.
As aberrations in the relative abundance of histone PTMs have been linked to several disease states including cancer, it is of fundamental importance that MS technology become high-throughput and highly reproducible for use with a potentially large number of clinical samples. The development of lower cost mass spectrometers that are able to analyze intact proteins will put the technology into more hands in the near future, which will speed up innovation and development in many fields such as clinical and translational proteomic applications. Until we reach the bottom-up like sensitivity and throughput for top-down MS, it is sensible to take the advantage of combining the different approaches.
4. Epigenetic Biomarkers for early detection, diagnostic and prognostic use
Cancer mortality could be reduced significantly with earlier detection of the disease, as survival is markedly better for early-stage patients for most types of cancer. Thus, much effort is being put into the development of accurate and timely detection strategies. The earliest studies on development of epigenetic biomarkers for cancer detection were focused on DNA methylation. The addition of a methyl group to the cytosine residues is catalyzed by the DNA methyltransferase (DNMT) family enzymes, including DNMT1, DNMT3A and DNMT3B [118]. The initial discovery of global hypomethylation of DNA in human tumors was followed by the identification of hypermethylated tumor-suppressor genes that are targeted for development of epigenetic biomarkers. For example commonly observed changes of hypermethylated cancer genes such as SEPT9, CDH13, MYOD1, MGMT, p16INK4b, and RASSF1A genes were detected in DNA purified from resected tumors, body fluids and plasma [119–121]. However, biomarkers developed based on DNA-methylation are still emerging and the utility of this information for the diagnosis, prognosis and treatment of disease has been limited to date. As dysregulation of histone modifications is often associated with the initiation of cancer formation and progression, it has been thought that histone modification patterns may serve as global biomarkers for screening or surveillance of cancer [122]. Relative to DNA methylation, far less is known about the significance of histone PTMs for clinical applications. Moreover, the extensive diversity in such modifications introduces a remarkable complexity that we are just beginning to elucidate. Although, histone PTMs have not been introduced into clinical use yet, numerous research studies have demonstrated that histone modification patterns are indicative for prognosis of various cancers. For example, enrichment in acetylation of histone tails leads to loosening of histone-DNA interactions by neutralizing a positive charge of lysine residues, thereby promoting transcriptional activation of genes. In addition, acetylation is also implicated in DNA repair, replication and chromatin condensation. Global loss of histone H4K16 acetylation has been shown to be a very common alteration present in several cancer cells [31], and is considered as a biomarker for cancer. Disruption of both HAT and HDAC activity is associated with the development of various cancer types. There are four classes of HDACs with 18 members (HDACs 1 to 11 and Sirtuins 1 to 7). For example, it was reported that H4K16 is deacetylated by SIRT1 (Sirtuins 1 - a class III histone deacetylase). SIRT1 is significantly elevated in several cancers such as leukemia, prostate, primary colon, and all non-melanoma skin cancers and therefore may render an attractive target for prognostic interventions [123]. Although the exact molecular mechanism is poorly understood, another sirtuin family member SIRT7, a highly selective H3K18Ac deacetylase, has been studied using quantitative mass spectrometry and ChIP-Seq along with the other biochemical tools and reported to stabilize tumorigenicity of human cancer cells in vivo [124]. As depletion of H3K18Ac has been associated with aggressive cancer phenotypes and poor patient prognosis [125], increased activity of SIRT7 may serve as a biomarker for screening for disease progression. Another study reported that in acute promyelocytic leukemia (PML) the PML-RAR gene chromosomal translocation recruits HDACs that change the chromatin structure from active to silenced, and contribute to leukemic transformation [126].
Genes that encode HAT enzymes are deregulated in various neoplasms including hepatocellular carcinomas, leukemia, lymphoma, colorectal, gastric primary tumors, and other epithelial cancers [127]. HATs are often part of multisubunit protein complexes. Chromodomains, bromodomains, PHD finger, tudor, WD40 are some of the major “reader” domains found in HATs that confer specificity. Among several HAT families, four of them (the GNAT family; the p300/CBP family; the MYST family and the Rtt109 family) have been studied extensively. For example, the MYST family HATs have been shown to be closely linked to cancer. The MYST family members KAT6A and KAT6B form stable multisubunit complexes, and are responsible for acetylation of a substantial portion of histone H3. It was reported that KAT6A/B fusions are associated with poor prognosis and decreased survival rate in AML, suggesting that inhibitors of these enzyme complexes might have therapeutic value [128].
Histone methylation patterns also provide prognostic and diagnostic information in cancer. Histone methylation occurs on both lysine and arginine residues. Lysine residues can be mono-, di- or tri- methylated whereas arginines can be mono- or symmetrically or asymmetrically di-methylated. Methylation of histones is involved to a lesser extent in modifying the histone charge state as compared to acetylation, but methylation patterns are highly involved in generating docking sites for histone readers. The degree of methylation on each site as well as the position of methylated residues often leads to different biological outcomes, i.e. transcriptional activation or repression of genes. For instance, methylation of H3K4 (H3K4me2/3) is usually enriched around transcriptionally active gene promoters, while methylation of H3K9 (H3K9me2/3) and H3K27 (H3K27me2/3) are typical marks of condensed heterochromatin [129]. Although the complete molecular mechanism is poorly understood, in both lung and kidney cancers lower levels of H3K4me2 along with H3K18ac correlated with poor survival rates with significantly increased risk of tumor recurrence [125]. Thus, it has been proposed that cellular levels of histone modifications may be used as general predictor of clinical outcome in adenocarcinomas.
Three families of HMTs have been identified to date that catalyze the addition of methyl groups donated from S-adenosylmethionine to histones. Among them, the SET-domain-containing proteins family and DOT1-like proteins family have been shown to methylate lysines (lysine methyltransferases), whereas the protein arginine N-methyltransferase (arginine methyltransferases-PRMT) family proteins have been shown to methylate arginine residues. For example, histone methyltransferases such as MLL1, DOT1L, EZH2, and SETD2 are recurrently deregulated in leukemia, either directly by gene mutations, or indirectly as a result of alterations of the other components in these complexes [130]. The removal of methyl groups is mediated by histone demethylases (HDMs). Only very recently histone methylation was found to be a reversible PTM. The first protein showing KDM activity was LSD1/KDM1A (lysine-specific demethylase 1), which specifically mediates the demethylation of mono- or dimethylated lysine 4 at histone H3 (H3K4me1 and H3K4me2) but not trimethylated lysine from H3K4me3 [131]. LSD1 has been found to be deregulated in several types of tumors. For example, expression of LSD1 is upregulated in bladder, small cell lung, and colorectal clinical cancer tissues [132]. Overexpressed LSD1 found at an early stage (i.e. in bladder tumors) suggested that LSD1 expression could be an early detection marker. As another example, the newly identified histone H3K4me1/2 demethylase (LSD2/AOF1/KDM1B) has been implicated in regulation of DNA methylation and gene silencing and highly expressed in breast cancer [133] suggesting that LDS2 may be a marker and potential target for a novel epigenetic therapy of breast cancer.
In addition, histone variants are also correlated with cancer [134]. Histone H3.3, which is a highly conserved variant differing from canonical H3.1 and H3.2 in five amino acids, has been found to be over expressed in various human tumors. Overexpression of H2A.Z has been reported in colorectal, breast, lung, and bladder cancer. For example, the H2A.Z overexpression in breast tumors correlated with increased probabilities of cancer metastasis and decreased patient survival. These results and many other studies suggest that this histone variant might be a biomarker of tumor progression.
Many possible combinations of modifications that can occur on a variety of sites on histones with their huge number of regulatory proteins provide an enriched pool for potential epigenetic biomarker discovery. These examples give an outlook on the potential of histone modification profiling as an important tool for diagnosis and prognosis of cancer, although their widespread use in clinical practice needs to be implemented. In combination with currently used markers (i.e. DNA methylation and cytogenetic and genetic markers) histone modifications might highly improve the prognostic accuracy in the future.
5. Epigenetic therapies
Azacytidine is a DNA hypomethylating agent and was the first epigenetic drug approved in 2004 by the US Food and Drug Administration (US-FDA). It is used for treatment of myeloid diseases including myeloid leukemia and myelodysplastic syndromes. We now have a growing number of research studies reporting the discovery of new drugs with epigenetic targets. However, to date only four epigenetic drugs are approved by FDA, which include two main classes; (i) DNA methyltransferase (DNMT) inhibitors, and (ii) histone deacetylase (HDAC) inhibitors. As briefly mentioned, the first epigenetic drug azacitidine (5-azacytidine, Vidaza, Celgene) is a DNA methylation inhibitor (DNMTi) and blocks DNA methylation in the abnormal (myelodysplastic) cells, thereby allowing the activation of tumor suppressor genes that had been turned off in abnormal cells as a result of the methylation changes. Another DNA methylation inhibitor decitabine (Dacogen; Eisai) was approved in 2006 for the treatment of the same disease (high risk myelodysplastic syndrome), which is often referred to as a type of pre-leukemia. However, these nucleoside analogs show poor activity against solid tumors and are associated with severe toxic side effects at higher doses, thus mostly low doses are used either alone or in combinational therapy. Moreover, these drugs are not targeted specifically to tumor cells and therefore modify the methylation patterns in tumor cells as well as in normal cells. Therefore, there is a need for more specific and less toxic new therapeutics. In fact, several new inhibitors are currently under development at various phase of drug development [135].
Another class of agents is the HDAC inhibitors (HDACi). HDACs are the category of histone modifier enzymes with the highest number of dedicated inhibitors (HDACi) for disease treatment [136]. This is also due to the early discovery of the first HDACi in 1971. In that year, dimethyl sulfoxide (DMSO) was found to induce erythroid differentiation [137]. From these observations, Marks and co-workers generated suberoylanilide hydroxamic acid (SAHA) as specific HDACi for class I and II HDACs [138]. Since October 2006 this molecule is known as vorinostat and it is the first FDA approved epigenetic drug targeting histone modifying enzymes. Two HDACis vorinostat (suberoyl- anilide hydroxamic acid, SAHA, Zolinza, Merck) and romidepsin (F-228, Istodax, Celgene) were approved by US-FDA (vorinostat in 2006; romidepsin in 2010) originally for the treatment of rare cutaneous T cell lymphoma (CTCL). However, HDACis also have been shown to suppress the growth of ovarian, endometrial, pancreatic and breast cancers and acute promyelocytic leukemia (APL) in vitro and in vivo. Clinical trials show that HDAC inhibitors are well tolerated. HDACis are now extensively studied epigenetic drugs there are at least 20 structurally different HDAC inhibitors that are in clinical trial for human cancers [139]. Moreover, other potentially therapeutic novel agents are currently under investigation for the treatment of various cancers. For example, selective inhibitors for EZH2 have been developed and shown to be effective in killing lymphoma cells with EZH2 mutation [140–142]. The first selective dual inhibitor of both EZH2 and EZH1 enzymes has been very recently developed as a promising therapeutics for tumors that rely on both wild-type EZH2 and EZH1 as in the case of mixed lineage leukemia (MLL) [143]. Although further research is needed for more specific and better targeted epigenetic therapeutics, such overall progress and on-going studies hold promise in management of cancer and other diseases, indicating a great potential for effective therapies. Of note, however, monitoring of the efficacy of such therapies, may require implementation of clinical monitoring of histone PTM's to assess pharmacologic activity, highlighting another potential use of clinical proteomics.
6. Expert Commentary
It is now clear that epigenetic changes play significant roles in development and progression of cancer, thus constitute important targets for therapy. As discussed, epigenetic therapeutics are already in use demonstrating that these changes can be modified therapeutically. Due to its increasing importance in basic and translational research, the number of studies on epigenetics has massively increased in the last decade. Its exponential growth has led to a variety of accomplishments, both in technology and biological discoveries, which open new challenges for discovery. The existence of an epigenetic code is constantly under debate, as the complexity of the epigenetic network is currently beyond a full mechanistic understanding. Even not considering DNA methylation and RNA interference, many questions remain about the histone code. This is in part due to the complexity of MS data for middle-down and top-down analyses. A major breakthrough required is advances in separation techniques, as likely the number of co-fragmented isobaric peptides or proteins will not be resolved when this exceeds a reasonable number (two-six). However, the impressive achievements in technologies and medicine have generated an optimistic environment among the epigenetic community, so that epigenetics is now one of the most funded fields in science. Taken together, it is now a very exciting period to work on epigenetics, and the large variety of possible studies, including inheritance, PTM cross talk, drug development, signaling pathway and new mechanisms, offer ample space to new scientific groups for novel discoveries.
7. Five-year view
Very likely, in the coming years more therapies will be approved, as many are currently in advanced clinical trials. Several new dynamic histone PTMs have been recently discovered [144–146], which are likely going to provide an even more complicated network of cross talk involving intra and inter histone variants. Even though this is already an impressive achievement, much needs to be done to understand their role and define whether any of these low abundant PTMs have a critical role in disease pathogenesis, or are part of steady state regulation of the cell and DNA structure. Importantly, several advances have been made in the MS platforms for histone characterization. For instance, data-independent acquisition methods are gaining popularity in proteomics [147]. SWATH™-MS is a data independent workflow that uses a first isolation window to step across a mass range, collecting full scan composite MS/MS spectra. The use of high-resolution instrumentation will lead to a map of high mass accuracy fragment ions from all detectable precursor masses. This type of data can be mined as a virtual selected reaction monitoring (SRM), even if at the time of the acquisition there is no knowledge about the analyte. This approach is particularly suitable for histone analysis and for their quantification, as they contain several isobaric peptides/proteoforms. While this has been proved successful for bottom-up type of analysis [148] their applicability for middle-down and top-down still has to be demonstrated. Collectively, epigenetics is a science in continuous expansion. In the coming years these technological improvements will likely assist to deeper understandings of the dynamic patterns of the histone modifications and their corresponding changes in cancer, thereby will enable the design of better treatment strategies, and eventually will allow personalized therapy.
Key Issues.
Histone post-translational modifications (PTMs) have a fundamental function in chromatin biology, and aberrations of histone PTM levels have been linked to development of diseases such as cancer.
Mass spectrometry (MS) has made many significant contributions to various biological and translational researches, mostly in the area of protein PTMs and especially in the histone biology.
Currently, the initial characterization of histones and mapping of their PTMs as well as quantification of global changes of their abundances can be accomplished using highly established bottom-up approach as nearly routine proteomics analyses.
Top down MS technique is ideal for analysis of the combinatorial histone PTMs that are simultaneously occurring on the same proteoform and may become a truly high-throughput compliment to bottom-up MS in near future. Until then it is sensible to take the advantage of hybrid approaches such as middle down MS.
Acknowledgments
This work was supported by funding from the NIH grants R01GM110174 and DP2OD007447 to BA Garcia and National Cancer Institute grants 5R01CA149566 and 5R21CA185365 to M Carroll.
Footnotes
Financial & competing interests disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Feinberg AP, Tycko B. The history of cancer epigenetics. Nature reviews. Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 2.Laird PW. The power and the promise of DNA methylation markers. Nature reviews. Cancer. 2003;3(4):253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 3.Mattick JS, Makunin IV. Non-coding RNA. Human molecular genetics. 2006;15:R17–29. doi: 10.1093/hmg/ddl046. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends in cell biology. 2009;19(5):207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Developmental cell. 2003;4(4):497–508. doi: 10.1016/s1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 8.Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nature genetics. 2005;37(1):41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 9.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(17):6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santaguida S, Musacchio A. The life and miracles of kinetochores. The EMBO journal. 2009;28(17):2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosgrove MS. Histone proteomics and the epigenetic regulation of nucleosome mobility. Expert review of proteomics. 2007;4(4):465–478. doi: 10.1586/14789450.4.4.465. [DOI] [PubMed] [Google Scholar]
- 12.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 13.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nature biotechnology. 2010;28(10):1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]; * The first epigenetic mechanism to be linked to cancer development.
- 15.Lee JV, Carrer A, Shah S, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell metabolism. 2014;20(2):306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nature reviews. Molecular cell biology. 2012;13(3):153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 17.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]; * Recent review describes the role of epigtenetics in oncogenesis.
- 18.Baylin SB, Hoppener JW, de Bustros A, Steenbergh PH, Lips CJ, Nelkin BD. DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer research. 1986;46(6):2917–2922. [PubMed] [Google Scholar]
- 19.Greger V, Passarge E, Hopping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Human genetics. 1989;83(2):155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 20.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews. Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Nagaraja S, Delcuve GP, Hendzel MJ, Davie JR. Effects of histone acetylation, ubiquitination and variants on nucleosome stability. The Biochemical journal. 1993;296(Pt 3):737–744. doi: 10.1042/bj2960737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(23):13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Sprung R, Tang Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Molecular & cellular proteomics : MCP. 2007;6(5):812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(1):60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Comprehensive catalog of histone PTM sites in mammalian cells.
- 26.Boulikas T. DNA strand breaks alter histone ADP-ribosylation. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(10):3499–3503. doi: 10.1073/pnas.86.10.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(46):19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidoli S, Cheng L, Jensen ON. Proteomics in chromatin biology and epigenetics: Elucidation of post-translational modifications of histone proteins by mass spectrometry. Journal of proteomics. 2012;75(12):3419–3433. doi: 10.1016/j.jprot.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 29.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 30.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Molecular cell. 2007;28(5):730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 31.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature genetics. 2005;37(4):391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]; * Comprehensive examination of the modification patterns of histone H4 in human normal tissues and cancer cells.
- 32.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 33.van Zutven LJ, Onen E, Velthuizen SC, et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes, chromosomes & cancer. 2006;45(5):437–446. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- 34.Wang GG, Song J, Wang Z, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reader JC, Meekins JS, Gojo I, Ning Y. A novel NUP98-PHF23 fusion resulting from a cryptic translocation t(11;17)(p15;p13) in acute myeloid leukemia. Leukemia. 2007;21(4):842–844. doi: 10.1038/sj.leu.2404579. [DOI] [PubMed] [Google Scholar]
- 36.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Molecular cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 37.Van Aller GS, Reynoird N, Barbash O, et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics : official journal of the DNA Methylation Society. 2012;7(4):340–343. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beguelin W, Popovic R, Teater M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer cell. 2013;23(5):677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen X, Liu Y, Hsu YJ, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24(2):268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 42.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Albadine R, Magheli A, et al. Increased EZH2 protein expression is associated with invasive urothelial carcinoma of the bladder. Urologic oncology. 2012;30(4):428–433. doi: 10.1016/j.urolonc.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Lewis PW, Muller MM, Koletsky MS, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Mutation of lysine to methionine in position 27 of histone H3 (H3K27M) observed in pediatric brain tumors inhibits the enzymatic H3K27 tri- and di-methyltransferase activity of the PRC2 by blocking free PRC2 in the cells.
- 45.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nature structural & molecular biology. 2007;14(11):997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 46.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128(4):721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 47.Albig W, Kioschis P, Poustka A, Meergans K, Doenecke D. Human histone gene organization: nonregular arrangement within a large cluster. Genomics. 1997;40(2):314–322. doi: 10.1006/geno.1996.4592. [DOI] [PubMed] [Google Scholar]
- 48.Pusarla RH, Bhargava P. Histones in functional diversification. Core histone variants. The FEBS journal. 2005;272(20):5149–5168. doi: 10.1111/j.1742-4658.2005.04930.x. [DOI] [PubMed] [Google Scholar]
- 49.Dhillon N, Oki M, Szyjka SJ, Aparicio OM, Kamakaka RT. H2A.Z functions to regulate progression through the cell cycle. Molecular and cellular biology. 2006;26(2):489–501. doi: 10.1128/MCB.26.2.489-501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Molecular cell. 2012;48(5):723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** The first selective dual inhibitor of EZH2 and EZH1 enzymes.
- 51.Larochelle M, Gaudreau L. H2A.Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. The EMBO journal. 2003;22(17):4512–4522. doi: 10.1093/emboj/cdg427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marques M, Laflamme L, Gervais AL, Gaudreau L. Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics : official journal of the DNA Methylation Society. 2010;5(4):267–272. doi: 10.4161/epi.5.4.11520. [DOI] [PubMed] [Google Scholar]
- 53.O'Donnell A, Yang SH, Sharrocks AD. PARP1 orchestrates variant histone exchange in signal-mediated transcriptional activation. EMBO reports. 2013;14(12):1084–1091. doi: 10.1038/embor.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarcinella E, Zuzarte PC, Lau PN, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Molecular and cellular biology. 2007;27(18):6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112(5):725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 56.Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nature structural & molecular biology. 2004;11(7):650–655. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
- 57.Adam M, Robert F, Larochelle M, Gaudreau L. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Molecular and cellular biology. 2001;21(18):6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henikoff S, Smith MM. Histone variants and epigenetics. Cold Spring Harbor perspectives in biology. 2015;7(1):a019364. doi: 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunican DS, McWilliam P, Tighe O, Parle-McDermott A, Croke DT. Gene expression differences between the microsatellite instability (MIN) and chromosomal instability (CIN) phenotypes in colorectal cancer revealed by high-density cDNA array hybridization. Oncogene. 2002;21(20):3253–3257. doi: 10.1038/sj.onc.1205431. [DOI] [PubMed] [Google Scholar]
- 60.Baptista T, Graca I, Sousa EJ, et al. Regulation of histone H2A.Z expression is mediated by sirtuin 1 in prostate cancer. Oncotarget. 2013;4(10):1673–1685. doi: 10.18632/oncotarget.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim K, Punj V, Choi J, et al. Gene dysregulation by histone variant H2A.Z in bladder cancer. Epigenetics & chromatin. 2013;6(1):34. doi: 10.1186/1756-8935-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hua S, Kallen CB, Dhar R, et al. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Molecular systems biology. 2008;4:188. doi: 10.1038/msb.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kapoor A, Goldberg MS, Cumberland LK, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468(7327):1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brandt WF, von Holt C. The determination of the primary structure of histone F3 from chicken erythrocytes by automatic Edman degradation. 2. Sequence analysis of histone F3. European journal of biochemistry / FEBS. 1974;46(2):419–429. doi: 10.1111/j.1432-1033.1974.tb03635.x. [DOI] [PubMed] [Google Scholar]
- 65.Chicoine LG, Schulman IG, Richman R, Cook RG, Allis CD. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena. Evidence for functionally distinct H4 acetylation sites. The Journal of biological chemistry. 1986;261(3):1071–1076. [PubMed] [Google Scholar]
- 66.Bock I, Dhayalan A, Kudithipudi S, Brandt O, Rathert P, Jeltsch A. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics : official journal of the DNA Methylation Society. 2011;6(2):256–263. doi: 10.4161/epi.6.2.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheung P. Generation and characterization of antibodies directed against di-modified histones, and comments on antibody and epitope recognition. Methods in enzymology. 2004;376:221–234. doi: 10.1016/S0076-6879(03)76015-7. [DOI] [PubMed] [Google Scholar]
- 68.Byrum SD, Raman A, Taverna SD, Tackett AJ. ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell reports. 2012;2(1):198–205. doi: 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soldi M, Bonaldi T. The ChroP approach combines ChIP and mass spectrometry to dissect locus-specific proteomic landscapes of chromatin. Journal of visualized experiments : JoVE. 2014;(86) doi: 10.3791/51220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ji X, Dadon DB, Abraham BJ, et al. Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(12):3841–3846. doi: 10.1073/pnas.1502971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith LM, Kelleher NL. Consortium for Top Down P. Proteoform: a single term describing protein complexity. Nature methods. 2013;10(3):186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang K, Williams KE, Huang L, et al. Histone acetylation and deacetylation: identification of acetylation and methylation sites of HeLa histone H4 by mass spectrometry. Molecular & cellular proteomics : MCP. 2002;1(7):500–508. doi: 10.1074/mcp.m200031-mcp200. [DOI] [PubMed] [Google Scholar]
- 73.Zhang K, Tang H, Huang L, et al. Identification of acetylation and methylation sites of histone H3 from chicken erythrocytes by high-accuracy matrix-assisted laser desorption ionization-time-of-flight, matrix-assisted laser desorption ionization-postsource decay, and nanoelectrospray ionization tandem mass spectrometry. Analytical biochemistry. 2002;306(2):259–269. doi: 10.1006/abio.2002.5719. [DOI] [PubMed] [Google Scholar]
- 74.Garcia BA, Mollah S, Ueberheide BM, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nature protocols. 2007;2(4):933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith CM. Quantification of acetylation at proximal lysine residues using isotopic labeling and tandem mass spectrometry. Methods. 2005;36(4):395–403. doi: 10.1016/j.ymeth.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 76.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Analytical biochemistry. 2003;316(1):23–33. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 77.Karczmarski J, Rubel T, Paziewska A, et al. Histone H3 lysine 27 acetylation is altered in colon cancer. Clinical proteomics. 2014;11(1):24. doi: 10.1186/1559-0275-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cuomo A, Moretti S, Minucci S, Bonaldi T. SILAC-based proteomic analysis to dissect the “histone modification signature” of human breast cancer cells. Amino acids. 2011;41(2):387–399. doi: 10.1007/s00726-010-0668-2. [DOI] [PubMed] [Google Scholar]
- 79.Jaffe JD, Wang Y, Chan HM, et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nature genetics. 2013;45(11):1386–1391. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]; * 42 distict histone modification of histone H3 lysine methylation monitored across 115 cancer cell lines by using quantitative mass spectrometric approach.
- 80.Beck HC, Nielsen EC, Matthiesen R, et al. Quantitative proteomic analysis of post-translational modifications of human histones. Molecular & cellular proteomics : MCP. 2006;5(7):1314–1325. doi: 10.1074/mcp.M600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Drogaris P, Villeneuve V, Pomies C, et al. Histone deacetylase inhibitors globally enhance h3/h4 tail acetylation without affecting h3 lysine 56 acetylation. Scientific reports. 2012;2:220. doi: 10.1038/srep00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Drogaris P, Wurtele H, Masumoto H, Verreault A, Thibault P. Comprehensive profiling of histone modifications using a label-free approach and its applications in determining structure-function relationships. Analytical chemistry. 2008;80(17):6698–6707. doi: 10.1021/ac800739d. [DOI] [PubMed] [Google Scholar]
- 83.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & cellular proteomics : MCP. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 84.Scharf AN, Barth TK, Imhof A. Establishment of histone modifications after chromatin assembly. Nucleic acids research. 2009;37(15):5032–5040. doi: 10.1093/nar/gkp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. The Journal of biological chemistry. 2010;285(5):3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leroy G, Dimaggio PA, Chan EY, et al. A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics & chromatin. 2013;6(1):20. doi: 10.1186/1756-8935-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 88.Tu S, Bulloch EM, Yang L, et al. Identification of histone demethylases in Saccharomyces cerevisiae. The Journal of biological chemistry. 2007;282(19):14262–14271. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelleher NL. Top-down proteomics. Analytical chemistry. 2004;76(11):197A–203A. [PubMed] [Google Scholar]
- 90.Loo JA, Edmonds CG, Smith RD. Primary sequence information from intact proteins by electrospray ionization tandem mass spectrometry. Science. 1990;248(4952):201–204. doi: 10.1126/science.2326633. [DOI] [PubMed] [Google Scholar]
- 91.Ginter JM, Zhou F, Johnston MV. Generating protein sequence tags by combining cone and conventional collision induced dissociation in a quadrupole time-of-flight mass spectrometer. Journal of the American Society for Mass Spectrometry. 2004;15(10):1478–1486. doi: 10.1016/j.jasms.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 92.Zubarev RAK, N.L., McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 93.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Compton PD, Strukl JV, Bai DL, Shabanowitz J, Hunt DF. Optimization of electron transfer dissociation via informed selection of reagents and operating parameters. Analytical chemistry. 2012;84(3):1781–1785. doi: 10.1021/ac202807h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Udeshi ND, Shabanowitz J, Hunt DF, Rose KL. Analysis of proteins and peptides on a chromatographic timescale by electron-transfer dissociation MS. The FEBS journal. 2007;274(24):6269–6276. doi: 10.1111/j.1742-4658.2007.06148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Banks GC, Deterding LJ, Tomer KB, Archer TK. Hormone-mediated dephosphorylation of specific histone H1 isoforms. The Journal of biological chemistry. 2001;276(39):36467–36473. doi: 10.1074/jbc.M104641200. [DOI] [PubMed] [Google Scholar]
- 97.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird's eye view. Journal of proteome research. 2006;5(2):240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 98.Boyne MT, 2nd, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. Journal of proteome research. 2006;5(2):248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 99.Siuti N, Roth MJ, Mizzen CA, Kelleher NL, Pesavento JJ. Gene-specific characterization of human histone H2B by electron capture dissociation. Journal of proteome research. 2006;5(2):233–239. doi: 10.1021/pr050268v. [DOI] [PubMed] [Google Scholar]
- 100.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Analytical chemistry. 2006;78(13):4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 101.Vellaichamy A, Tran JC, Catherman AD, et al. Size-sorting combined with improved nanocapillary liquid chromatography-mass spectrometry for identification of intact proteins up to 80 kDa. Analytical chemistry. 2010;82(4):1234–1244. doi: 10.1021/ac9021083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bloom GC, Eschrich S, Zhou JX, Coppola D, Yeatman TJ. Elucidation of a protein signature discriminating six common types of adenocarcinoma. International journal of cancer. Journal international du cancer. 2007;120(4):769–775. doi: 10.1002/ijc.22041. [DOI] [PubMed] [Google Scholar]
- 103.Zhou G, Li H, DeCamp D, et al. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Molecular & cellular proteomics : MCP. 2002;1(2):117–124. doi: 10.1074/mcp.m100015-mcp200. [DOI] [PubMed] [Google Scholar]
- 104.Sarg B, Faserl K, Kremser L, Halfinger B, Sebastiano R. Lindner HH. Comparing and combining capillary electrophoresis electrospray ionization mass spectrometry and nano-liquid chromatography electrospray ionization mass spectrometry for the characterization of post-translationally modified histones. Molecular & cellular proteomics : MCP. 2013;12(9):2640–2656. doi: 10.1074/mcp.M112.024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meng F, Cargile BJ, Patrie SM, Johnson JR, McLoughlin SM, Kelleher NL. Processing complex mixtures of intact proteins for direct analysis by mass spectrometry. Analytical chemistry. 2002;74(13):2923–2929. doi: 10.1021/ac020049i. [DOI] [PubMed] [Google Scholar]
- 106.Taverna SD, Ueberheide BM, Liu Y, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(7):2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garcia BA. What does the future hold for Top Down mass spectrometry? Journal of the American Society for Mass Spectrometry. 2010;21(2):193–202. doi: 10.1016/j.jasms.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 108.Cannon J, Lohnes K, Wynne C, Wang Y, Edwards N, Fenselau C. High-throughput middle-down analysis using an orbitrap. Journal of proteome research. 2010;9(8):3886–3890. doi: 10.1021/pr1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Swatkoski S, Gutierrez P, Wynne C, et al. Evaluation of microwave-accelerated residue-specific acid cleavage for proteomic applications. Journal of proteome research. 2008;7(2):579–586. doi: 10.1021/pr070502c. [DOI] [PubMed] [Google Scholar]
- 110.Lindner H, Sarg B, Meraner C, Helliger W. Separation of acetylated core histones by hydrophilic-interaction liquid chromatography. Journal of chromatography. A. 1996;743(1):137–144. doi: 10.1016/0021-9673(96)00131-8. [DOI] [PubMed] [Google Scholar]
- 111.Mant CT, Hodges RS. Mixed-mode hydrophilic interaction/cation-exchange chromatography (HILIC/CEX) of peptides and proteins. Journal of separation science. 2008;31(15):2754–2773. doi: 10.1002/jssc.200800243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nature methods. 2007;4(6):487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 113.Jung HR, Sidoli S, Haldbo S, et al. Precision mapping of coexisting modifications in histone H3 tails from embryonic stem cells by ETD-MS/MS. Analytical chemistry. 2013;85(17):8232–8239. doi: 10.1021/ac401299w. [DOI] [PubMed] [Google Scholar]
- 114.Sidoli S, Schwammle V, Ruminowicz C, et al. Middle-down hybrid chromatography/tandem mass spectrometry workflow for characterization of combinatorial post-translational modifications in histones. Proteomics. 2014;14(19):2200–2211. doi: 10.1002/pmic.201400084. [DOI] [PubMed] [Google Scholar]
- 115.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. Highthroughput characterization of combinatorial histone codes. Molecular & cellular proteomics : MCP. 2009;8(10):2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]; * First highthroughput automated characterization of histone H3 code.
- 116.Tran JC, Zamdborg L, Ahlf DR, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480(7376):254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gault J, Malosse C, Machata S, et al. Complete posttranslational modification mapping of pathogenic Neisseria meningitidis pilins requires top-down mass spectrometry. Proteomics. 2014;14(10):1141–1151. doi: 10.1002/pmic.201300394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. The New England journal of medicine. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 119.Delpu Y, Cordelier P, Cho WC, Torrisani J. DNA methylation and cancer diagnosis. International journal of molecular sciences. 2013;14(7):15029–15058. doi: 10.3390/ijms140715029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Powrozek T, Krawczyk P, Kucharczyk T, Milanowski J. Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: preliminary report. Medical oncology. 2014;31(4):917. doi: 10.1007/s12032-014-0917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wei SH, Balch C, Paik HH, et al. Prognostic DNA methylation biomarkers in ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12(9):2788–2794. doi: 10.1158/1078-0432.CCR-05-1551. [DOI] [PubMed] [Google Scholar]
- 122.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 123.Liu TF, McCall CE. Deacetylation by SIRT1 Reprograms Inflammation and Cancer. Genes & cancer. 2013;4(3–4):135–147. doi: 10.1177/1947601913476948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Barber MF, Michishita-Kioi E, Xi Y, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487(7405):114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Seligson DB, Horvath S, McBrian MA, et al. Global levels of histone modifications predict prognosis in different cancers. The American journal of pathology. 2009;174(5):1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Grignani F, De Matteis S, Nervi C, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391(6669):815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 127.Fenrick R, Hiebert SW. Role of histone deacetylases in acute leukemia. Journal of cellular biochemistry. Supplement. 1998;30–31:194–202. [PubMed] [Google Scholar]
- 128.Falk H, Connor T, Yang H, et al. An efficient high-throughput screening method for MYST family acetyltransferases, a new class of epigenetic drug targets. Journal of biomolecular screening. 2011;16(10):1196–1205. doi: 10.1177/1087057111421631. [DOI] [PubMed] [Google Scholar]
- 129.Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 130.Chopra M, Bohlander SK. Disturbing the histone code in leukemia: translocations and mutations affecting histone methyl transferases. Cancer genetics. 2014 doi: 10.1016/j.cancergen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 131.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 132.Hayami S, Kelly JD, Cho HS, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. International journal of cancer. Journal international du cancer. 2011;128(3):574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 133.Katz TA, Vasilatos SN, Harrington E, Oesterreich S, Davidson NE, Huang Y. Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast cancer research and treatment. 2014;146(1):99–108. doi: 10.1007/s10549-014-3012-9. [DOI] [PubMed] [Google Scholar]
- 134.Vardabasso C, Hasson D, Ratnakumar K, Chung CY, Duarte LF, Bernstein E. Histone variants: emerging players in cancer biology. Cellular and molecular life sciences : CMLS. 2014;71(3):379–404. doi: 10.1007/s00018-013-1343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Campbell RM, Tummino PJ. Cancer epigenetics drug discovery and development: the challenge of hitting the mark. The Journal of clinical investigation. 2014;124(1):64–69. doi: 10.1172/JCI71605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Barneda-Zahonero B, Parra M. Histone deacetylases and cancer. Molecular oncology. 2012;6(6):579–589. doi: 10.1016/j.molonc.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Friend C, Scher W, Holland JG, Sato T. Hemoglobin synthesis in murine virus-induced leukemic cells in vitro: stimulation of erythroid differentiation by dimethyl sulfoxide. Proceedings of the National Academy of Sciences of the United States of America. 1971;68(2):378–382. doi: 10.1073/pnas.68.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Richon VM, Emiliani S, Verdin E, et al. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(6):3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The first FDA approved epigenetic drug targeting histone modifier enzymes
- 139.DiNardo CD, Cortes JE. New treatment for acute myelogenous leukemia. Expert opinion on pharmacotherapy. 2015;16(1):95–106. doi: 10.1517/14656566.2015.981527. [DOI] [PubMed] [Google Scholar]
- 140.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature chemical biology. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 141.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 142.Qi W, Chan H, Teng L, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(52):21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu B, On DM, Ma A, et al. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2015;125(2):346–357. doi: 10.1182/blood-2014-06-581082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Christophorou MA, Castelo-Branco G, Halley-Stott RP, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507(7490):104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dai L, Peng C, Montellier E, et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nature chemical biology. 2014;10(5):365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]
- 146.Tessarz P, Santos-Rosa H, Robson SC, et al. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature. 2014;505(7484):564–568. doi: 10.1038/nature12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gillet LC, Navarro P, Tate S, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Molecular & cellular proteomics : MCP. 2012;11(6) doi: 10.1074/mcp.O111.016717. O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sidoli S, Lin S, Xiong L, et al. SWATH Analysis for Characterization and Quantification of Histone Post-translational Modifications. Molecular & cellular proteomics : MCP. 2015 doi: 10.1074/mcp.O114.046102. [DOI] [PMC free article] [PubMed] [Google Scholar]