SUMMARY
Nitrogen permease regulator-like 2 (NPRL2) is a component of a conserved complex that inhibits mTORC1 (mammalian Target Of Rapamycin Complex 1) in response to amino acid insufficiency. Here we show that NPRL2 is required for mouse viability and its absence significantly compromises fetal liver hematopoiesis in developing embryos. Moreover, NPRL2 KO embryos have significantly reduced methionine levels and exhibit phenotypes reminiscent of cobalamin (vitamin B12)-deficiency. Consistent with this idea, NPRL2 KO liver and mouse embryonic fibroblasts (MEFs) show defective processing of the cobalamin-transport protein transcobalamin 2, along with impaired lysosomal acidification and lysosomal gene expression. NPRL2 KO MEFs exhibit a significant defect in the cobalamin-dependent synthesis of methionine from homocysteine, which can be rescued by supplementation with cyanocobalamin. Taken together, these findings demonstrate a role for NPRL2 and mTORC1 in the regulation of lysosomal-dependent cobalamin processing, methionine synthesis and maintenance of cellular re-methylation potential, which are important during hematopoiesis.
INTRODUCTION
The mTORC1 pathway regulates cellular growth by sensing growth factors and nutrients, and relaying these signals to downstream effectors through its kinase activity (Dibble and Manning, 2013; Shimobayashi and Hall, 2014). Multicellular eukaryotes rely on growth factor signaling as a means to communicate energy availability between tissues and cells and significant progress has been made defining regulators of the pathways that contribute to mTORC1 activity, including TSC1/2, AKT, and PTEN (Inoki et al., 2005; Laplante and Sabatini, 2012). Activation of mTORC1 results in the phosphorylation of targets such as S6 Kinase and 4EBP1, which stimulate translation and growth. In response to growth factor or nutrient insufficiency, mTORC1 is inhibited by upstream negative regulators that act on small GTPases that are important for mTORC1 function. The TSC1/2 complex is one such negative regulator whose loss leads to hyperactive mTORC1 signaling (Inoki et al., 2002; Manning et al., 2002; Tee et al., 2002). Mutations in TSC1/2 are associated with tuberous sclerosis and various forms of tumorigenesis, phenotypes which are consistent with mTORC1 dysregulation in tumor formation (Guertin and Sabatini, 2007; Inoki et al., 2005).
Genetic studies in yeast revealed the existence of additional upstream negative regulators of TORC1. An evolutionarily conserved complex consisting of Npr2p, Npr3p and Iml1p (NPRL2, NPRL3, and DEPDC5 in mammals, respectively) was identified to inhibit mTORC1 activity and induce autophagy in response to specific nutrient limitations (Dokudovskaya et al., 2011; Neklesa and Davis, 2009; Wu and Tu, 2011). Biochemical studies of the Npr2-complex, termed SEACIT in yeast and GATOR1 in mammals, have shown that it inhibits TORC1 activity by functioning as a GTPase-activating protein (GAP) toward the Rag family of small GTPases (Bar-Peled et al., 2013; Kira et al., 2014; Panchaud et al., 2013). Consistent with these observations, mutants lacking Npr2, Npr3, or Iml1 fail to induce autophagy and exhibit unchecked growth under specific nutrient limitations (Sutter et al., 2013; Wu and Tu, 2011). The presence of the Npr2-complex, but not TSC orthologs in single-cell eukaryotes, suggests the NPRL2-complex might have a more ancestral role in modulating mTORC1 activity in response to amino acid availability. Loss of a genomic locus containing Nprl2 is frequently associated with lung and other cancers (Bar-Peled et al., 2013; Lerman and Minna, 2000; Li et al., 2004), suggesting it might have tumor suppressive functions.
While a multitude of mTORC1 regulators contribute to diverse physiological outcomes, as reviewed elsewhere (Laplante and Sabatini, 2012), the in vivo function of NPRL2 in mammals has not yet been addressed. To determine the physiological role of NPRL2, we created a global Nprl2 knockout mouse. Here we show that NPRL2 KO animals have impaired fetal liver hematopoiesis and a methionine synthesis deficit. We further show that loss of NPRL2 produces an apparent “folate-trap” and implicate mTORC1 as a regulator of cobalamin (vitamin B12)-dependent processes and cellular re-methylation potential. These findings reveal a previously unrecognized mechanism whereby a negative regulator of mTORC1 contributes to hematopoiesis.
RESULTS
Defective Hematopoiesis in NPRL2 KO Embryos
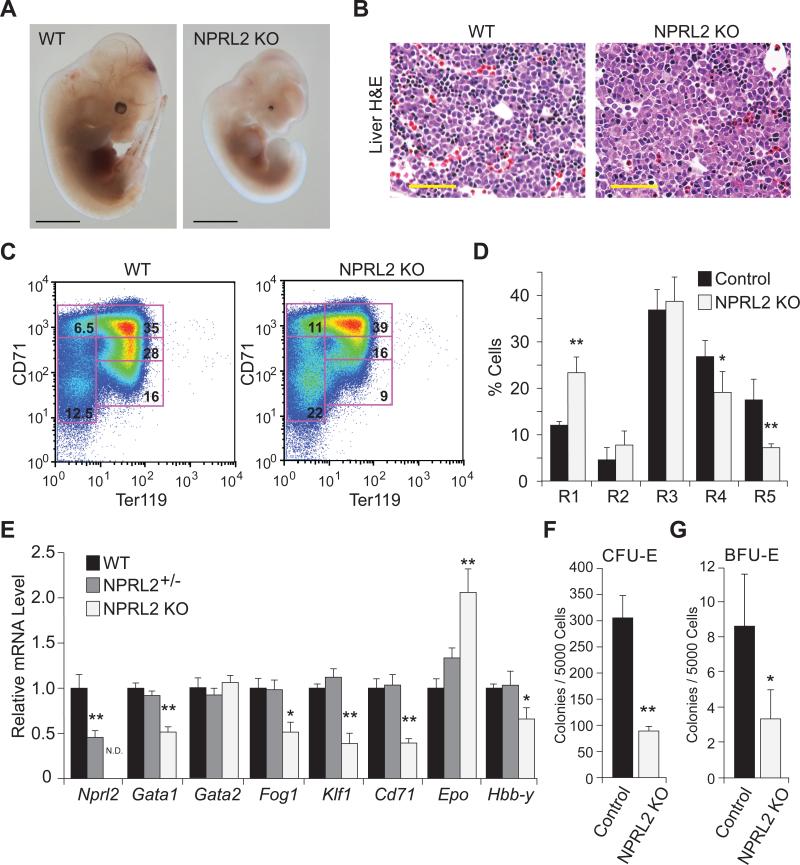
To determine the function of NPRL2 in vivo, we created a global knockout of Nprl2 in mice (Figure S1). Breeding heterozygous animals did not produce NPRL2 KO pups, but E12.5 embryos were obtained for analysis. Gross phenotypic observation showed NPRL2 KO embryos were smaller than wild type (WT) in size, and displayed microphthalmia, occasional unilateral anopthalmia, and notably pale liver (Figure 1A). Histological examination of the fetal livers showed reduced hematopoietic precursor cells in the NPRL2 KO compared to WT littermates (Figure 1B). Flow cytometry of isolated fetal livers targeting cell surface markers CD71 and Ter119 showed impaired erythroid differentiation in the NPRL2 KO, with increased percentages of early progenitors and reduced percentages of late progenitors (Figure 1C,D). Quantitative RT-PCR of erythroid-related genes, including Gata1, Fog1, Klf1, and Cd71, showed they were significantly decreased in the NPRL2 KO compared to Nprl2+/− and WT (Figure 1E). Consistent with a defect in fetal liver hematopoiesis, fetal hemoglobin Hbb-y mRNA was significantly reduced in the NPRL2 KO liver, whereas the expression of Epo was significantly increased, possibly due to feedback regulation resulting from inefficient erythropoiesis. To determine if hematopoiesis was defective in the early differentiation cascade, fetal livers were isolated and used to determine the capacity of hematopoietic stem cells to differentiate into erythroid-colony forming units (CFU-e) and more immature erythroid-blast forming units (BFU-e) (Figure 1F,G). Both types of colonies formed in WT and NPRL2 KO cultures; however, the number of colonies was significantly reduced in NPRL2 KO. These data suggest NPRL2 KO embryos have an early defect in hematopoiesis, resulting in reduced numbers of erythroid progenitor cells. Analysis of Nprl2+/− and WT blood from 20-week old mice did not show any hematological differences by complete blood count with differential analysis (Figure S1), suggesting that one functional Nprl2 allele is sufficient for normal production of blood cells.
Figure 1. Loss of NPRL2 reduces fetal liver hematopoiesis.
(A) Representative image of WT and NPRL2 KO embryos at E12.5, scale bar is 0.5 mm. Note: microphthalmia and pale liver in NPRL2 KO embryo. Scale bar is 2 mm.
(B) Haematoxylin and eosin staining of fetal liver sections of WT and NPRL2 KO embryos at E12.5 show decreased number of small, darkly-stained cells in the NPRL2 KO indicative of reduced hematopoiesis. Scale bar is 50 μM.
(C) Flow cytometric analysis of freshly isolated WT and NPRL2 KO fetal livers at E12.5 labeled with FITC-CD71 and APC-Ter119 antibodies show decreased abundance of mature erythrocytes in NPRL2 KO.
(D) Quantification of flow cytometry data (n=6/group).
(E) mRNA levels of Nprl2 and erythroid-related genes, including: Gata1, Gata2, Fog1, Klf1, Cd71 Epo, and Hbb-y in WT, Nprl2+/−, and NPRL2 KO liver at E12.5 (n=7, 10 and 6; respectively. N.D.=not detectable).
(F-G) Erythroid colony-forming units (CFU-E) and erythroid burst-forming units (BFU-E) assays were performed using isolated E12.5 livers and show NPRL2 KO cells have reduced colony formation (Control n=4; NPRL2 KO n=3). *, p < 0.05 vs control; **, p < 0.01 vs control.
Altered Metabolism in NPRL2 KO Embryos
Previous metabolic characterization of yeast npr2Δ mutants suggested that disruption of mammalian Nprl2 may alter amino acid metabolism in the developing embryo (Laxman et al., 2014). We obtained WT, Nprl2+/−, and NPRL2 KO embryos at E12.5 and performed targeted LC-MS/MS analysis of whole embryo metabolite extracts. Intriguingly, metabolomic analysis showed markedly lower abundance of methionine and elevated 5-methyltetrahydrofolate (5-Me-THF) in NPRL2 KO embryos, compared to WT (Table 1). These metabolic differences suggest the presence of a “folate-trap” (Herbert and Zalusky, 1962; Scott and Weir, 1981), a hallmark of defective methionine synthase (MTR) and cobalamin-deficient anemia (Koury and Ponka, 2004). We also analyzed metabolites of betaine-homocysteine methyltransferase (BHMT), an alternative pathway of methionine synthesis that uses betaine as a methyl donor. We observed a decrease in N,N-dimethylglycine and reduced BHMT mRNA levels in the NPRL2 KO (Table 1, Figure S2A), suggesting no significant compensation by this pathway. The abundance of several other amino acids was reduced in the NPRL2 KO, including leucine, isoleucine, and glutamine, while most others remained largely unchanged (Table 1). Importantly, not all metabolites were repressed in the NPRL2 KO as assessed by our targeted detection methods (Table 1, S1). Nucleic acid moieties were significantly increased in the NPRL2 KO, whereas several glycolytic and TCA intermediates were reduced, consistent with altered folate and one-carbon metabolism (Table 1 and S1).
Table 1.
Relative metabolite amount from E12.5 WT and NPRL2 KO embryos determined by targeted LC-MS/MS.
| Metabolite | Q1/Q3 m/z | Wild Type | NPRL2 KO |
|---|---|---|---|
|
Methionine:Folate Cycle:
| |||
| Methionine | 150.1/104.1 | 1.00 ± 0.02 | 0.71 ± 0.03c |
| 150.1/133.0 | 1.00 ± 0.02 | 0.72 ± 0.03c | |
| 5-Me-THF | 460.1/313.3 | 1.00 ± 0.06 | 1.58 ± 0.19b |
| 460.1/180.1 | 1.00 ± 0.07 | 1.50 ± 0.20a | |
| SAM | 399.1/250.2 | 1.00 ± 0.04 | 1.02 ± 0.10 |
| 399.1/136.0 | 1.00 ± 0.03 | 0.92 ± 0.07 | |
| SAH | 385.1/136.2 | 1.00 ± 0.08 | 1.18 ± 0.09 |
| 385.1/134.0 | 1.00 ± 0.09 | 1.19 ± 0.10 | |
| N,N Dimethylglycine | 104.0/58.1 | 1.00 ± 0.02 | 0.78 ± 0.06b |
| 104.0/42.0 | 1.00 ± 0.03 | 0.86 ± 0.03a | |
| Betaine | 118.1/72.2 | 1.00 ± 0.17 | 1.12 ± 0.15 |
|
Amino Acids: | |||
| Isoleucine | 130.0/82.1 | 1.00 ± 0.10 | 0.47 ± 0.07b |
| 130.0/45.2 | 1.00 ± 0.13 | 0.43 ± 0.06b | |
| Leucine | 130.0/84.0 | 1.00 ± 0.08 | 0.51 ± 0.06c |
| Glutamine | 144.9/127.2 | 1.00 ± 0.04 | 0.34 ± 0.05c |
| 144.9/109.0 | 1.00 ± 0.05 | 0.36 ± 0.04c | |
| Glutamate | 148.1/84.1 | 1.00 ± 0.04 | 0.92 ± 0.04 |
| 148.1/130.1 | 1.00 ± 0.04 | 0.88 ± 0.03a | |
| Valine | 118.1/72.1 | 1.00 ± 0.02 | 0.67 ± 0.03c |
| 118.1/55.0 | 1.00 ± 0.02 | 0.66 ± 0.03c | |
| Cysteine | 122.0/58.9 | 1.00 ± 0.12 | 1.18 ± 0.06 |
| 122.0/76.1 | 1.00 ± 0.09 | 1.05 ± 0.10 | |
| Proline | 116.0/70.1 | 1.00 ± 0.06 | 0.89 ± 0.05 |
| 116.0/69.0 | 1.00 ± 0.07 | 0.89 ± 0.04 | |
| Tyrosine | 182.1/136.1 | 1.00 ± 0.04 | 0.74 ± 0.03c |
| 182.1/91.0 | 1.00 ± 0.04 | 0.72 ± 0.03c | |
| Tryptophan | 205.1/188.2 | 1.00 ± 0.05 | 0.86 ± 0.04a |
| 205.1/146.2 | 1.00 ± 0.05 | 0.84 ± 0.03a | |
| Arginine | 175.1/70.1 | 1.00 ± 0.04 | 0.68 ± 0.04c |
| 175.1/116.0 | 1.00 ± 0.04 | 0.67 ± 0.05c | |
| His tidine | 156.1/110.0 | 1.00 ± 0.02 | 0.81 ± 0.06a |
| 156.1/83.2 | 1.00 ± 0.04 | 0.69 ± 0.04c | |
| Phenylalanine | 166.1/120.1 | 1.00 ± 0.03 | 0.80 ± 0.05b |
| 166.1/103.2 | 1.00 ± 0.04 | 0.75 ± 0.04b | |
p<0.05 vs WT
p<0.01 vs WT
p<0.001 vs WT. (n=4-7)
The significant changes in methionine levels prompted us to examine the expression of genes pertaining to methionine, folate, and cobalamin metabolism, as well as the transsulfuration pathway in embryonic liver (Figure S2A). Folate-related genes (Dhfr, Mthfr) were unchanged in NPRL2 KO versus WT; however Mtr and the lysosomal transporter for cobalamin (Lmbrd1) were moderately induced in the NPRL2 KO. Expression of Cth, part of the transsulfuration pathway, was induced in the NPRL2 KO. These data indicate that expression of genes involved in sulfur amino acid metabolism is influenced by the loss of NPRL2 despite no apparent increase in pS6K and pmTOR in liver at E12.5 (Figure S2B).
mTORC1 and Lysosomal Dysfunction in NPRL2 KO cells
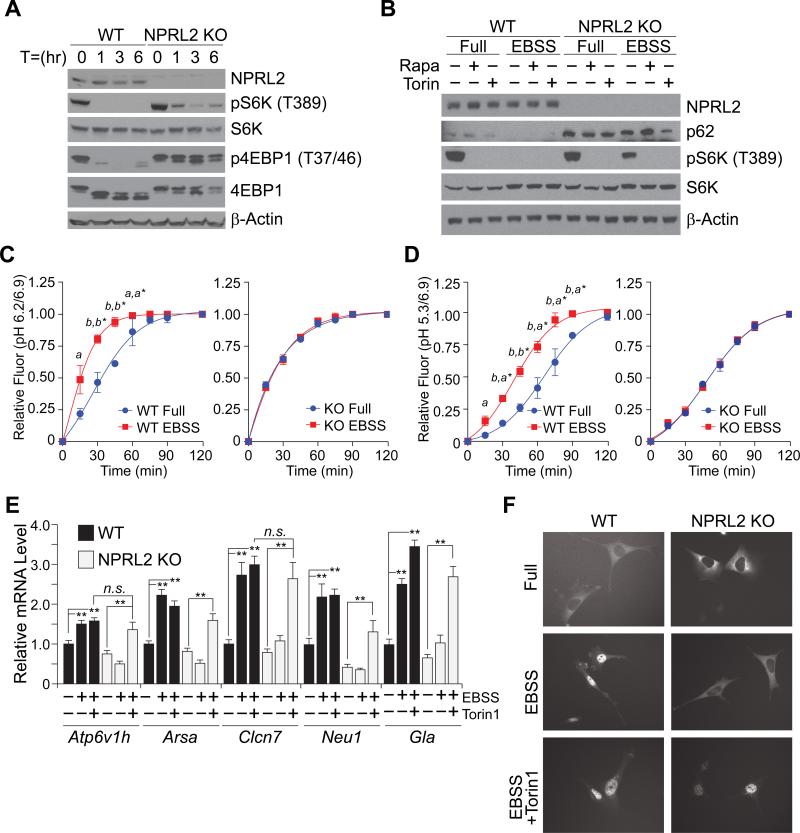
To test if complete loss of NPRL2 altered mTORC1-dependent pathways, mouse embryonic fibroblasts (MEF) cells were generated from E12.5 embryos. Consistent with previous NPRL2 knockdown studies (Bar-Peled et al., 2013), NPRL2 KO MEF cells were unable to repress mTORC1 activity during starvation in Earle's buffered saline solution (EBSS) and retained phosphorylation of S6K and 4EBP1, compared to WT (Figure 2A). To determine if the phosphorylation of S6K in the NPRL2 KO cells was due to dysregulated mTORC1 activity, NPRL2 KO cells were treated with either rapamycin or torin 1 during EBSS incubation. Both pharmacological mTORC1 inhibitors reduced S6K phosphorylation in NPRL2 KO cells, suggesting that NPRL2 regulates mTORC1 activity (Figure 2B). Moreover, the amount of p62, a ubiquitin-binding protein that is degraded by autophagy, was increased in the NPRL2 KO MEF cells and not degraded during EBSS stimulation suggesting defective autophagy or lysosomal function (Figure 2B). These data collectively indicate that NPRL2 KO MEFs exhibit hallmarks of active mTORC1 signaling despite starvation, consistent with its role as a negative regulator of TORC1.
Figure 2. Dysregulation of lysosomal-related function in NPRL2 KO MEF cells.
(A) WT and NPRL2 KO MEF cells cultured in Earle's buffered saline solution (EBSS) for 1, 3, and 6 h. Western blot analysis was used to determine the abundance of NPRL2, pS6K (T389), total S6K, p4EBP1 (T37/46), 4EBP1, and β-actin.
(B) WT and NPRL2 KO MEF cells were cultured in either complete media (Full) or EBSS for 1 h in the presence of 10 μM rapamycin or 250 μM torin 1. Western blot analysis was performed to determine the abundance of NPRL2, p62, pS6K (T389), total S6K and β-actin.
(C-D) Confocal microscopy of WT and NPRL2 KO MEF cells treated with 100 μg/mL hybrid ultra pH-sensitive nanoprobes was used to determine the intracellular vesicle pH of cells cultured in full media or EBSS over a 120 minute period. Relative fluorescence of nanoprobes at pH 6.2/6.9 indicating early endosome acidification and pH 5.3/6.9 indicating late endosome and lysosomal acidification was determined. (Similar data was obtained from replicate experiments with 2-5 cells per group. a, p<0.02; b, p<0.001 WT EBSS vs WT full; *WT EBSS vs KO EBSS).
(E) Quantitative RT-PCR analysis of TFEB target genes, including: Atp6v1h, Arsa, Clcn7, Neu1 and Gla in WT and NPRL2 KO MEF cells incubated for 6 h in EBSS in the presence or absence of torin1. ** p<0.01; n.s.=not significant.
(F) WT and NPRL2 KO MEF cells expressing TFEB-GFP were analyzed by fluorescent microscopy to determine TFEB localization in complete media, and after 2 h incubation in EBSS in the presence and absence of torin 1.
Since mTORC1 inhibition is necessary for the acidification of lysosomes during autophagy (Zhou et al., 2013), we tested if NPRL2 KO MEF cells had altered rates of lysosomal acidification. We utilized hybrid ultra pH-sensitive nanoprobes to quantitate lysosomal acidification over a time course with EBSS starvation (Ma et al., 2014; Zhou et al., 2011). WT MEF cells showed a significant and rapid increase in early-endosome and late-endosome/lysosome acidification over the starvation period (Figure 2C, D and S4A-D). In contrast, NPRL2 KO MEF cells did not show compartmental pH difference between nutrient-rich or starved conditions, consistent with impaired lysosomal-function (Mindell, 2012). Moreover, WT MEF cells overexpressing the GTP-locked RagA Q66L mutant had similar defects in acidification (Figure S3E), which supports the reported function of the NPRL2-containing complex as a GAP for the Rag GTPases (Bar-Peled et al., 2013; Panchaud et al., 2013). In addition, pretreatment of Nprl2 KO cells with torin1 increased acidification similar to WT cells in EBSS (Figure S3F). These data suggest particular lysosomal activities requiring low pH may be impaired during starvations in the NPRL2 KO cells.
We next investigated whether the regulation of genes involved in lysosomal biogenesis was altered by loss of NPRL2. Quantitative RT-PCR of lysosomal gene targets of the transcription factor TFEB (Sardiello et al., 2009), including Atp6v1h, Arsa, Clcn7, Neu1, and Gla showed that their induction by EBSS was dependent on NPRL2 expression and mTORC1 inhibition (Figure 2E). We then transfected WT and NPRL2 KO MEF cells with TFEB-GFP to determine if the nuclear localization of TFEB in response to starvation might be compromised in the absence of NPRL2. In nutrient-rich media, TFEB retained its cytoplasmic localization in both cell types, but localized to the nucleus during EBSS incubation in WT MEF cells, not NPRL2 KO (Figure 2F). Since the localization of TFEB to the nucleus is dependent on mTORC1 repression (Sardiello et al., 2009), we treated WT and NPRL2 KO cells with torin 1 during the switch to EBSS. Treatment of NPRL2 KO cells with torin 1 caused TFEB to localize to the nucleus, consistent with induction of TFEB target genes, suggesting lysosomal gene expression is dependent on mTORC1 inhibition by NPRL2.
Defective Cobalamin-Dependent Metabolism in NPRL2 KO cells
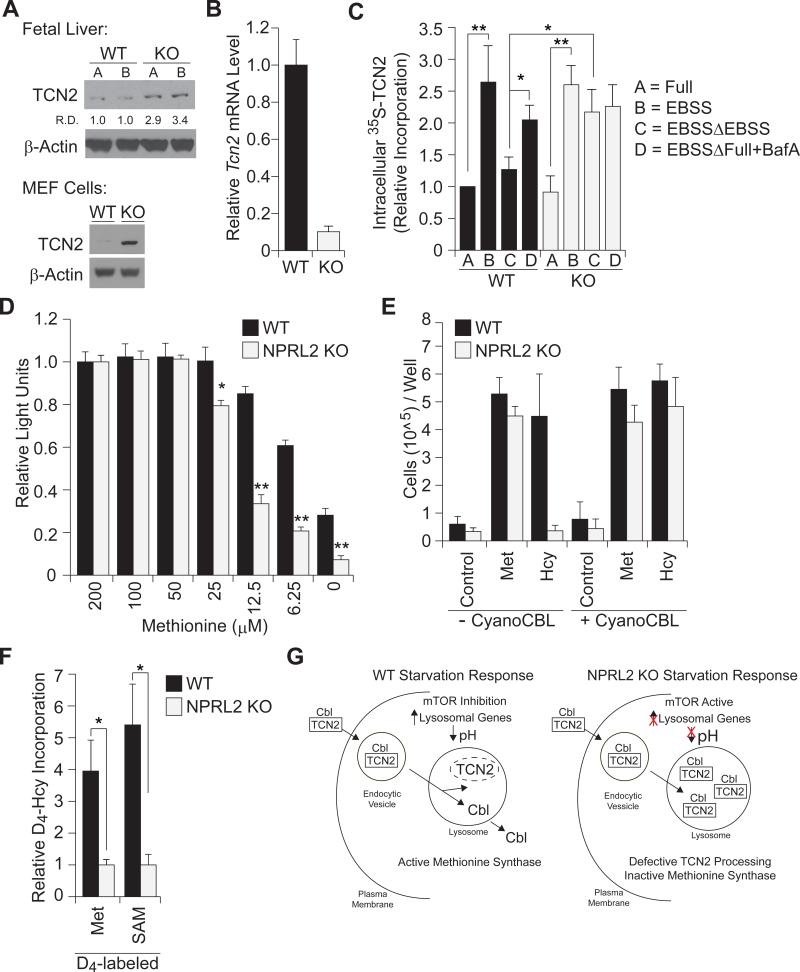
We realized that defects in lysosomal function might help explain the reduced methionine amounts in NPRL2 KO embryos. The methionine synthase enzyme is dependent on cobalamin (vitamin-B12), which is carried in plasma bound to the cobalamin-transport protein, called TCN2, and transported into cells by the endocytic-lysosomal pathway (Banerjee and Matthews, 1990; Rosenblatt et al., 1985; Youngdahl-Turner et al., 1978). Western blot analysis showed increased abundance of TCN2 in both NPRL2 KO embryonic liver and MEF cells, compared with WT (Figure 3A). These data suggested a cell-autonomous regulation of cobalamin metabolism could impact methionine synthesis. Notably, NPRL2 KO MEF cells expressed significantly lower levels of Tcn2 mRNA, compared to WT, suggesting a negative feedback due to high TCN2 protein uptake (Figure 3B). We assayed TCN2 uptake in WT and NPRL2 KO MEF cells using cobalamin-loaded, 35S-labeled TCN2. Both WT and NPRL2 KO cells took up the protein and EBSS stimulated the effect (Figure 3C). Pulse-chase experiments with 35S-labeled TCN2 showed that continued incubation of cells in unlabeled media reduced intracellular 35S-TCN2 in WT MEF cells to a greater extent than NPRL2 KO cells. Since NPRL2 KO cells display impaired lysosomal function, these data could explain the increased TCN2 content in NPRL2 KO (Figure 3A). Treating WT cells with the lysosomal V-ATPase inhibitor, bafilomycin A1, during the incubation period caused retention of TCN2 protein, indicating TCN2 is indeed processed by lysosomes.
Figure 3. Defective TCN2 processing, cobalamin-deficiency, and impaired methionine synthesis in NPRL2 KO MEF cells.
(A) Western blot analysis of TCN2 and β-actin show increased TCN2 protein content in NPRL2 KO fetal liver and MEFs cells, relative to WT. (R.D.=relative densitometry)
(B) Quantitative RT-PCR analysis of TCN2 mRNA show decreased expression in the NPRL2 KO MEF cells, relative to WT MEF cells. (n=3/group)
(C) 35S-TCN2 assays were performed in WT and NPRL2 KO MEF cells incubated with labeled-TCN2 for 2 h in (A) complete media, (B) EBSS starvation media, (C) EBSS followed by a 4 h switch to non-labeled media, or (D) EBSS followed by a 4 h switch to complete media with 100 nM bafilomycin A. Phosphorimager measurements of 35S-TCN2 in cell lysates were normalized to total protein. Data represent 3 independent experiments. * p<0.05; ** p<0.01.
(D) WT and NPRL2 KO MEF cells were cultured for 4 days in methionine-deficient media, supplemented with increasing concentrations of methionine. Cell viability was determined by cell-titer glo analysis. n=3/group, * p<0.01.
(E) WT and NPRL2 KO MEF cells (20,000/well) were cultured for 4 days in methionine-deficient media with 1% HI FBS (control) in the presence or absence of 200 μM L-methionine or 200 μM L-homocysteine, and 250 nM cyanocobalamin. In contrast to WT cells, NPRL2 KO cells die when media is supplemented with L-homocysteine alone, but are able to proliferate with L-homocysteine and cyanocobalamin supplementation for proliferation. Cell number was determined with trypan-blue exclusion and automated cell counting. Data represent two independent experiments performed in duplicate.
(F) WT and NPRL2 KO MEF cells were cultured in methionine-deficient media with 200 μM homocysteine-3,3,4,4-D4 for 24 h. Metabolite extracts were analyzed by LC-MS/MS to determine the synthesis of heavy-labeled [D4]-methionine and [D4]-SAM. Data were normalized against NPRL2 KO. * p<0.05.
(G) Model depicting NPRL2-regulated functions during the starvation response.
To assess whether the defects in methionine synthesis in NPRL2 KO embryos could be recapitulated in a cell culture model, WT and NPRL2 KO MEF cells were cultured in various nutrient conditions. Although similar proliferation rates of WT and NPRL2 KO MEF cells were observed in complete media (Figure S4A), NPRL2 KO cell viability was more sensitive to methionine depletion, compared to WT cells (Figure 3D). We tested if NPRL2 KO cells might exhibit altered sensitivity to anti-folate treatments, using a dose-response with methotrexate, but found no difference (Figure S4B). We then measured proliferation of low-passage fibroblasts in methionine-deficient media, in the presence and absence of methionine or homocysteine supplementation. The synthesis of methionine from homocysteine requires the cobalamin-dependent enzyme methionine synthase. WT MEF cells were able to proliferate in media containing 1% HI-FBS and either methionine or homocysteine. In striking contrast, NPRL2 KO MEF cells only grew in media containing methionine (Figure 3E). These data are consistent with defective cobalamin metabolism in the NPRL2 KO cells and suggest NPRL2 is necessary for maintaining the cellular demand of methionine synthesis from homocysteine. We next asked if the NPRL2 KO phenotype could be rescued by exogenous cobalamin treatment. Remarkably, supplementation of cyanocobalamin permitted growth of NPRL2 KO MEF cells in homocysteine, at rates comparable to WT cells (Figure 3E). To directly test whether NPRL2 KO MEF cells harbored a defect in the cobalamin-dependent synthesis of methionine from homocysteine, we quantitated the conversion of homocysteine to methionine using LC-MS/MS. WT and NPRL2 KO MEF cells were incubated in methionine-deficient media and supplemented with the labeled homocysteine-3,3,4,4-d4 for a 24 h period. WT MEF cells synthesized approximately 4-fold more methionine and 5-fold more SAM from labeled-homocysteine, compared to NPRL2 KO cells (Figure 3F). Treatment of WT cells with torin1 for 4 hours slightly increased methionine synthesis from labeled homocysteine, in contrast to NPRL2 KO cells (Figure S4C). Collectively, these data strongly suggest that loss of NPRL2 produces cobalamin-deficiency, and thus methionine-deficiency, perhaps due to defective lysosomal-dependent TCN2 processing (Figure 3G).
DISCUSSION
In this report, we show an important role for NPRL2 in the regulation of amino acid metabolism and fetal hematopoiesis. Loss of NPRL2 leads to low methionine and high 5-Me-THF amounts in embryos, suggesting a “folate-trap” that can occur by cobalamin-deficiency (Herbert and Zalusky, 1962). We further observe that lysosomal processing of cobalamin is downstream of NPRL2 and establish mTORC1 as a regulator of cobalamin-processing and the cobalamin-dependent enzyme, methionine synthase. Comprehensive studies of human inborn-errors of cobalamin metabolism have uncovered multiple proteins in this pathway that regulate the availability of cobalamin in cells, but thus far have not examined mTORC1 as a regulatory pathway (Watkins and Rosenblatt, 2013). Our observations suggest that mTORC1 repression is essential for lysosomal processing of TCN2 for the bioavailability of cobalamin for methionine synthase (Banerjee and Matthews, 1990). This enzyme resides at the intersection of the methionine and folate cycle and influences flux through these pathways. Defects in either pathway can present with similar hematological pathologies, but distinct metabolic profiles (Carmel et al., 2003). Collectively, our findings reveal that NPRL2, a negative regulator of mTORC1, controls cobalamin availability, methionine homeostasis, and re-methylation potential, which are important metabolic pathways for hematopoiesis (Klee, 2000; Koury and Ponka, 2004).
Previous studies have shown that growth factor-dependent regulators of mTORC1, including PTEN and TSC1, contribute to hematopoiesis (Chen et al., 2008; Gan et al., 2008; Knight et al., 2014; Yilmaz et al., 2006). A surprising result from our studies is that NPRL2 KO animals do not phenocopy RagA GTPase-deficient mice, which do not present with malformations of the eye, nor pallor (Efeyan et al., 2013). In contrast, the constitutively GTP-bound RagA mutant mice are born alive, but succumb shortly after birth. These phenotypic differences may be explained by genetic background differences, compensatory effects of the other Rag proteins, or the possibility that NPRL2 has additional functions in vivo. Our observations, however, are consistent with the idea that dynamic regulation of mTORC1 is required for red blood cell development, which has been observed in other genetically-engineered mouse models of dysregulated mTORC1 (Chen et al., 2008; Gan et al., 2008; Knight et al., 2014; Yilmaz et al., 2006). The combined early lethality and hematopoietic phenotype of NPRL2 KO embryos is consistent with additional lysosomal pathway defects, including the failure to activate lysosomal gene transcription in response to starvation. Striking microphtalmia in the NPRL2 KO embryos is consistent with a dysfunction of micropthalmia-associated transcription factor, another known lysosomal-regulated transcription factor (Roczniak-Ferguson et al., 2012; Steingrimsson et al., 1994). The consequence of dysregulated lysosomal gene transcription likely contributes to extra-hematologic deficiencies and will be of interest in future studies.
Our finding that NPRL2 KO embryos have reduced hematopoietic progenitor cells is consistent with a special requirement for methionine metabolism in the maintenance and proliferation of embryonic stem cells (Shiraki et al., 2014). As starvation-induced lysosomal acidification is impaired in NPRL2 KO cells, these data are consistent with the observation that mTORC1 inactivation is required for lysosomal acidification (Zhou et al., 2013), which if compromised could result in defective TCN2 processing (Rosenblatt et al., 1985; Youngdahl-Turner et al., 1978). Interestingly, complete loss of RagA/B also results in defective lysosomal acidification (Kim et al., 2014), similar to NPRL2 KO cells that harbor RagA/B in the GTP-locked state. These observations suggest that cycling of the Rag GTPases between GTP- and GDP-bound states may be required for proper lysosomal acidification.
NPRL2 functions in a complex with two other proteins, NPRL3 and DEPDC5, which are each indispensable for proper nutrient regulation of mTORC1 (Bar-Peled et al., 2013; Kira et al., 2014; Neklesa and Davis, 2009; Wu and Tu, 2011). Although loss of NPRL3 has been studied in mice by deleting the Nprl3 promoter region (Kowalczyk et al., 2012), the mice do not delete NPRL3 expression in erythroid cells making comparisons to our global NPRL2 KO model challenging. Intriguingly, the NPRL3 gene locus is proximal to α-globin cluster in mice, indicating cis-elements in this region may contribute to NPRL3 and α-globin expression. While no reports have directly investigated the function of DEPDC5, recent studies have shown DEPDC5 mutations are associated with epilepsy in humans (Dibbens et al., 2013). These observations are intriguing since cobalamin deficiencies may contribute to seizure activity (Akaike et al., 1993; Kumar, 2004) and indicate that the NPRL2-complex may contribute to neurological pathologies that are sensitive to cobalamin (Stabler, 2013).
Methionine-dependence of cancer cells is a hallmark of many tumors (Agrawal et al., 2012; Glaudemans et al., 2013). Our observation that loss of Nprl2 causes stricter methionine-dependence and remethylation deficits suggests that cells lacking NPRL2 may consequently exhibit increased methionine uptake and dependency. Consistent with this idea, yeast npr2Δ mutants have significantly higher levels of SAM, which may promote their unchecked growth under particular nutrient starvations (Laxman et al., 2013; Sutter et al., 2013). Although budding yeast do not utilize cobalamin for the synthesis of methionine, the similar phenotype linked to methionine metabolism compared to mammalian cells suggests an ancient, conserved function for this complex and TORC1 centered on methionine and SAM homeostasis. Future work will determine if loss of Nprl2, or other negative regulators of mTORC1, increases the demand for methionine in tumors and whether methionine limitation might represent a therapeutic strategy to compromise their growth.
In summary, we describe an unanticipated role for NPRL2 in the regulation of lysosomal processing of cobalamin in mammals, which is critical during hematopoiesis. These results provide mechanistic support for an intimate link between the mTORC1 signaling pathway and its proper regulation required during hematopoiesis (Knight et al., 2014; Yilmaz et al., 2006). Our study suggests a key metabolic requirement during blood cell formation could be the proper coordination of re-methylation potential with the demands of DNA nucleotide synthesis. We conclude that NPRL2 coordinates a cellular response to amino acid starvation by orchestrating the uptake and bioavailability of cobalamin, which in turn is required for the synthesis of the sentinel metabolites methionine and SAM that play critical roles in the regulation of cell growth and homeostasis.
EXPERIMENTAL PROCEDURES
Generation of NPRL2 KO Mice
The Nprl2 targeting vector was created using the pGKNeoF2L2DTA (Addgene, #13445). A 5-kb long arm harboring the Nprl2 promoter, a 2.9-kb fragment harboring exons 1-9, and a 1.6-kb short arm harboring exons 10 and 11 of the Nprl2 gene were generated by high-fidelity polymerase amplification (PfuUltra®, Stratagene). The Nprl2 targeting vector was linearized with AloI (Thermo Scientific) and injected into ES cells. ES clones were isolated, screened for homologous recombination by PCR, and verified by Southern blot analysis. Validated homologous recombined clones were injected into 3.5-day C57BL/6 blastocysts, and the resulting chimeras were crossed with C57BL/6 females to obtain germ-line transmission of the targeted Nprl2 allele (Nprl2neo-loxP/+). The Nprl2 gene was deleted by crossing Nprl2loxP/+ mice with female Zp3-cre (C57BL/6) to generate the Nprl2+/− mice. Removal of the Zp3-cre allele and deletion of the Nprl2 gene were confirmed by PCR genotyping with primers listed in Table S3. Nprl2+/− mice were backcrossed to C57BL/6 mice at least 4 generations and subsequent heterozygous crosses, resulting in littermates of Nprl2+/+, Nprl2+/−, Nprl2−/− (NPRL2 KO), were used in the studies. See also Figure S1.
Flow Cytometry Analysis of Erythropoiesis
Nprl2+/− mice were crossed, and embryonic livers were collected at E12.5 by decapitation of the dam and excision of the fetal liver into sterile PBS with 2% FBS and analyzed as previously described (Zhang et al., 2003). Briefly, cells were isolated by trituration, passed through a 70 μM nylon mesh cell strainer (Falcon), immuno-labeled at 4°C with FITC-CD71 (1:200 dilution; BD, #553266) and APC-Ter119 (1:200 dilution; BD, #557909) antibodies for 15 minutes, pelleted, and re-suspended in PBS with 2% FBS and 3 ug/mL propidium iodide. Flow cytometry was carried out on a BD FACSCalibur (Franklin Lakes, NJ) and analyzed using FlowJo V8.8.4.
Colony Assays
For CFU-E and mature BFU-E assays, E12.5 fetal liver cells were isolated and plated in Methocult™ media with erythropoietin (M3334, Stem Cell Technology) following the manufactures protocol. Cells were cultured at 37°C and 5% CO2 in a humidified incubator and colonies were scored on day 2 (CFU-E) and day 4 (BFU-E).
Analysis of Methionine Synthesis
WT and NPRL2 KO MEF cells were incubated in methionine-deficient media, consisting of DMEM (Gibco®, #21013-024), 1× GlutaMAX™ (Gibco®, 35050-061), 200 μM L-cysteine (Sigma), 5% FBS (Gibco®, #10082-147) and 200 μM DL-Homocysteine-3,3,4,4-d4 (CDN Isotopes, D-6452), for 24 h. Metabolites were extracted from cells in ice-cold extraction buffer with 0.3% formic acid, and processed for LC-MS/MS. Synthesis of metabolites from labeled-homocysteine was calculated relative to total ion counts for each sample.
TCN2 Uptake Assay
TCN2 was cloned from duodenum of C57/B6 and inserted in the pCI vector (Promega). 35S-incoporation was performed by using the TNT® Quick Coupled Transcription/Translation kit (Promega), according to the manufacturer's protocol. The 35S-TCN2 labeling reaction was loaded with 1 ug/mL cyanocobalamin for 15 minutes and dialyses in PBS overnight at 4°C in a 10,000 MWCO Slide-A-Lyzer® cassette (Thermo Scientific). Cells were treated with 35S-TCN2 as described in the figure legends, washed twice with PBS, and harvested in 2X Laemmli sample buffer. Protein lysates were separated by SDS-PAGE and exposed to a phosphorimager screen for 48 h and imaged with a Typhoon scanner (Amersham Biosciences).
Statistical Analysis
Statistical analysis was performed by two-tailed Student's t-test using Microsoft Excel 2010. A p-value <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank J. Shelton, J. Richardson, and the UTSW Molecular Pathology Core Laboratory for histology and analysis; R. Hammer and the UTSW Transgenic Core Facility; C. Sephton for imaging assistance; M. Roth, Y. Yu, and D. Rosenblatt for helpful discussions. This work was funded by a Damon Runyon-Rachleff Innovation Award (B.P.T.), a Packard Fellowship (B.P.T.), a Cancer Prevention Research Institute of Texas (CPRIT) individual investigator award, (B.P.T.), a research fellowship from CPRIT (P.A.D.), and a NIH grant R01HL089966 (L.J.H.).
REFERENCES
- Agrawal V, Alpini SE, Stone EM, Frenkel EP, Frankel AE. Targeting methionine auxotrophy in cancer: discovery & exploration. Expert opinion on biological therapy. 2012;12:53–61. doi: 10.1517/14712598.2012.636349. [DOI] [PubMed] [Google Scholar]
- Akaike A, Tamura Y, Sato Y, Yokota T. Protective effects of a vitamin B12 analog, methylcobalamin, against glutamate cytotoxicity in cultured cortical neurons. European journal of pharmacology. 1993;241:1–6. doi: 10.1016/0014-2999(93)90925-8. [DOI] [PubMed] [Google Scholar]
- Banerjee RV, Matthews RG. Cobalamin-dependent methionine synthase. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1990;4:1450–1459. doi: 10.1096/fasebj.4.5.2407589. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel R, Green R, Rosenblatt DS, Watkins D. Update on cobalamin, folate, and homocysteine. Hematology / the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2003:62–81. doi: 10.1182/asheducation-2003.1.62. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. The Journal of experimental medicine. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, de Vries B, Donatello S, Heron SE, Hodgson BL, Chintawar S, Crompton DE, Hughes JN, Bellows ST, Klein KM, et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nature genetics. 2013;45:546–551. doi: 10.1038/ng.2599. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nature cell biology. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, Williams R, Salamero J, Chait BT, Sali A, et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Molecular & cellular proteomics : MCP. 2011;10:M110 006478. doi: 10.1074/mcp.M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, Williams DA, Kwiatkowski DJ, DePinho RA. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19384–19389. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaudemans AW, Enting RH, Heesters MA, Dierckx RA, van Rheenen RW, Walenkamp AM, Slart RH. Value of 11C-methionine PET in imaging brain tumours and metastases. European journal of nuclear medicine and molecular imaging. 2013;40:615–635. doi: 10.1007/s00259-012-2295-5. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Herbert V, Zalusky R. Interrelations of vitamin B12 and folic acid metabolism: folic acid clearance studies. The Journal of clinical investigation. 1962;41:1263–1276. doi: 10.1172/JCI104589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nature genetics. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature cell biology. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Kim YC, Park HW, Sciarretta S, Mo JS, Jewell JL, Russell RC, Wu X, Sadoshima J, Guan KL. Rag GTPases are cardioprotective by regulating lysosomal function. Nature communications. 2014;5:4241. doi: 10.1038/ncomms5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kira S, Tabata K, Shirahama-Noda K, Nozoe A, Yoshimori T, Noda T. Reciprocal conversion of Gtr1 and Gtr2 nucleotide-binding states by Npr2-Npr3 inactivates TORC1 and induces autophagy. Autophagy. 2014;10:1565–1578. doi: 10.4161/auto.29397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee GG. Cobalamin and folate evaluation: measurement of methylmalonic acid and homocysteine vs vitamin B(12) and folate. Clinical chemistry. 2000;46:1277–1283. [PubMed] [Google Scholar]
- Knight ZA, Schmidt SF, Birsoy K, Tan K, Friedman JM. A critical role for mTORC1 in erythropoiesis and anemia. eLife. 2014:e01913. doi: 10.7554/eLife.01913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury MJ, Ponka P. New insights into erythropoiesis: the roles of folate, vitamin B12, and iron. Annual review of nutrition. 2004;24:105–131. doi: 10.1146/annurev.nutr.24.012003.132306. [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, et al. Intragenic enhancers act as alternative promoters. Molecular cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Kumar S. Recurrent seizures: an unusual manifestation of vitamin B12 deficiency. Neurology India. 2004;52:122–123. [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Shi L, Tu BP. Npr2 inhibits TORC1 to prevent inappropriate utilization of glutamine for biosynthesis of nitrogen-containing metabolites. Science signaling. 2014;7:ra120. doi: 10.1126/scisignal.2005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell. 2013;154:416–429. doi: 10.1016/j.cell.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer research. 2000;60:6116–6133. [PubMed] [Google Scholar]
- Li J, Wang F, Haraldson K, Protopopov A, Duh FM, Geil L, Kuzmin I, Minna JD, Stanbridge E, Braga E, et al. Functional characterization of the candidate tumor suppressor gene NPRL2/G21 located in 3p21.3C. Cancer research. 2004;64:6438–6443. doi: 10.1158/0008-5472.CAN-03-3869. [DOI] [PubMed] [Google Scholar]
- Ma X, Wang Y, Zhao T, Li Y, Su LC, Wang Z, Huang G, Sumer BD, Gao J. Ultra-pH-sensitive nanoprobe library with broad pH tunability and fluorescence emissions. Journal of the American Chemical Society. 2014;136:11085–11092. doi: 10.1021/ja5053158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Molecular cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- Mindell JA. Lysosomal acidification mechanisms. Annual review of physiology. 2012;74:69–86. doi: 10.1146/annurev-physiol-012110-142317. [DOI] [PubMed] [Google Scholar]
- Neklesa TK, Davis RW. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS genetics. 2009;5:e1000515. doi: 10.1371/journal.pgen.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud N, Peli-Gulli MP, De Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Science signaling. 2013;6:ra42. doi: 10.1126/scisignal.2004112. [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Science signaling. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt DS, Hosack A, Matiaszuk NV, Cooper BA, Laframboise R. Defect in vitamin B12 release from lysosomes: newly described inborn error of vitamin B12 metabolism. Science. 1985;228:1319–1321. doi: 10.1126/science.4001945. [DOI] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Scott JM, Weir DG. The methyl folate trap. A physiological response in man to prevent methyl group deficiency in kwashiorkor (methionine deficiency) and an explanation for folic-acid induced exacerbation of subacute combined degeneration in pernicious anaemia. Lancet. 1981;2:337–340. doi: 10.1016/s0140-6736(81)90650-4. [DOI] [PubMed] [Google Scholar]
- Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nature reviews Molecular cell biology. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell metabolism. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Stabler SP. Vitamin B12 deficiency. The New England journal of medicine. 2013;368:2041–2042. doi: 10.1056/NEJMc1304350. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D'Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nature. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Sutter BM, Wu X, Laxman S, Tu BP. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell. 2013;154:403–415. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins D, Rosenblatt DS. Lessons in biology from patients with inborn errors of vitamin B12 metabolism. Biochimie. 2013;95:1019–1022. doi: 10.1016/j.biochi.2013.01.013. [DOI] [PubMed] [Google Scholar]
- Wu X, Tu BP. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Molecular biology of the cell. 2011;22:4124–4133. doi: 10.1091/mbc.E11-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Youngdahl-Turner P, Rosenberg LE, Allen RH. Binding and uptake of transcobalamin II by human fibroblasts. The Journal of clinical investigation. 1978;61:133–141. doi: 10.1172/JCI108911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- Zhou J, Tan SH, Nicolas V, Bauvy C, Yang ND, Zhang J, Xue Y, Codogno P, Shen HM. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell research. 2013;23:508–523. doi: 10.1038/cr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Wang Y, Huang X, Luby-Phelps K, Sumer BD, Gao J. Tunable, ultrasensitive pH-responsive nanoparticles targeting specific endocytic organelles in living cells. Angewandte Chemie. 2011;50:6109–6114. doi: 10.1002/anie.201100884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.