Abstract
One of the grand challenges faced by neuroscience is to delineate the determinants of interindividual variation in the comprehensive structural and functional connection matrices that comprise the human connectome. At present, this endeavor appears most tractable at the macroanatomic scale, where intrinsic brain activity exhibits robust patterns of synchrony that recapitulate core functional circuits at the individual level. Here, we use a classical twin study design to examine the heritability of intrinsic functional network properties in 101 twin pairs, including network activity (i.e., variance of a network's specific temporal fluctuations) and internetwork coherence (i.e., correlation between networks' specific temporal fluctuations). Five of 7 networks exhibited significantly heritable (23.3–65.2%) network activity, 6 of the 21 internetwork coherences were significantly heritable (25.6–42.0%), and 11 of the 21 internetwork coherences were significantly influenced by common environmental factors (18.0–47.1%). These results suggest that the source of interindividual variation in functional connectome has a modular architecture: individual modules represented by intrinsic connectivity networks are genetic controlled, while environmental factors influence the interplays between the modules. This work further provides network-specific hypotheses for discovery of the specific genetic and environmental factors influencing functional specialization and integration of the human brain.
Keywords: connectome, environmental contribution, heritability, intrinsic connectivity network, twins
Introduction
The neuroscience community has embraced the challenges of delineating functional interactions within the human connectome (Kelly et al. 2012; Van Essen et al. 2013). From one individual to the next, strikingly similar patterns of temporally correlated spontaneous activity (i.e., functional connectivity) are noted within the connectome, along with substantial variation (Biswal et al. 2010; Tomasi and Volkow 2010; Zuo et al. 2012). In attempting to explain the potential origins of such variation, researchers are increasingly highlighting the need for twin and family study designs, which were successfully used in the morphometric imaging literature to differentiate between genetic and environmental effects (Bartley et al. 1997; Chiang et al. 2009; Kaymaz and van Os 2009; Kochunov et al. 2010, 2011; Chiang et al. 2011; Blokland et al. 2012; Jahanshad et al. 2013).
Initial resting state fMRI-based (rfMRI) investigations have revealed genetic contributions to interindividual variation. First, focusing on the brain's proposed default network (Raichle et al. 2001; Buckner et al. 2008; Andrews-Hanna et al. 2010), an rfMRI study of 333 Mexican American individuals from 29 extended pedigrees found functional connectivity within the default network to be significantly heritable (estimated heritability = 42%). While age was accounted for in their analyses, the authors noted that this value was likely an underestimate due to the wide age range of their participants (Glahn et al. 2010). Using graph theory approaches, in a small sample of twins (N = 58, 16 monozygotic [MZ], 13 dizygotic [DZ] pairs), Fornito et al. (2011) observed highly heritable (60%) cost-efficiency balance of the functional connectome. Most recently, the global efficiency of intrinsic brain networks was also found to be significantly heritable (N = 86, 21 MZ, 22 DZ pairs) in children (van den Heuvel et al. 2013).
However, a burgeoning literature has expanded our appreciation of the intrinsic brain architecture (Smith et al. 2009; Bressler and Menon 2010; Zhang and Raichle 2010) beyond the default network, to include a growing number of large-scale functional modules in the brain connectome (Sporns 2013; van den Heuvel and Sporns 2013)—commonly referred to as intrinsic connectivity networks (ICNs). Disentangling whether genetic and/or environmental factors influence the activities of ICNs and the interplays among ICNs is essential to understand the intrinsic functional architecture of the brain and the neural substrates of individual variations in both behavioral and mental states.
Here, we use a substantial rfMRI dataset obtained in twins (N = 272: 78 MZ pairs; 58 DZ pairs) to examine genetic and environmental contributions to individual variability in fluctuation of ICNs and coherence between ICNs. We first generated a set of ICN templates consisting of 7 commonly identified networks in a separate sample (N = 105) for whom rfMRI was acquired on the same scanner. Applying the ICN templates to the data from 272 twins, we obtained ICN-specific representative time series for the 7 ICNs, based on which we characterized the fluctuations of ICNs using temporal variance (Kaneoke et al. 2012) and characterized the inter-ICN coherences using correlation coefficients between the ICN-specific representative time series. Cross-twin correlation and structural equation modeling (SEM) were performed to examine the genetic and environmental effects on fluctuation of each ICN and coherence between each pair of ICNs. Age and sex were carefully controlled in the analyses.
Methods
Participants
A total of 200 twin pairs, including 86 monozygotic (MZ) and 114 dizygotic (DZ) pairs, were recruited from unrelated Australian families as part of the Queensland Twin Imaging (QTIM) study (de Zubicaray et al. 2008; Chiang et al. 2009; Blokland et al. 2011; Couvy-Duchesne et al. 2014). The participants were originally recruited from the primary and secondary schools in South East Queensland, Australia, and had participated in the Brisbane Twin Cognition study at 16 years of age (Wright and Martin 2004). Zygosity of the twins was assessed objectively with 9 independent DNA microsatellite polymorphisms and crosschecked with blood group (ABO, MNS, and Rh) as well as phenotypic data (hair, skin, and eye color). An overall probability of correct assignment on zygosity is >99.99%. All twins were screened to exclude cases of pathology known to affect brain structure and reported no significant head injury, neurological or psychiatric conditions, or history of substance abuse/dependence or had a first-degree relative with a psychiatric disorder. Informed written consent was obtained from the twins. Ethical approval was obtained from the Human Research Ethics Committee, Queensland Institute of Medical Research.
To avoid the potential confound from the gender difference in brain structure (Ruigrok et al. 2014), we selected only same sex, adult twin pairs who also met an upper limit on head motion measures (see Quality Control Procedure below) from the initial sample of 400 individuals. This criterion required co-twins for each twin pair to have little head motion, so that if one of the co-twins had excessive head motion, the other was excluded. As a result, we included 272 twins in the main analyses (referred to as main dataset, mean age = 22, range 18–28, 75% female), consisting of 78 MZ and 58 DZ pairs. From among those excluded, we used 105 individuals (referred to as template dataset, mean age = 22, range 19–29, 51% female) with mean frame-wise displacement <0.2 mm to generate a set of ICN templates. Eighty of these were opposite-sex DZ twins (40 pairs), the remainder were nonpaired twins (individuals). The following image acquisition, preprocessing, and quality control procedures were identical to both datasets.
MRI Image Acquisition
All twins completed 5′15″ rfMRI scans on a 4 T Bruker Medspec scanner (Bruker Medical) comprising 150 echo-planar imaging (EPI) volumes of whole-brain T2*-weighted blood oxygenation level-dependent (BOLD) contrast: echo time = 30 ms, repetition time = 2100 ms, flip angle = 90, field of view = 230 mm, slice thickness = 3.6 mm, acquisition matrix = 64 × 64 × 36. High-resolution T1-weighted images were also acquired for each pair of twins using the magnetization-prepared rapid gradient echo (MP-RAGE) sequence (echo time = 3.83 ms, repetition time = 2500 ms, inversion time = 1500 ms, flip angle = 15°, slice thickness = 0.9 mm, acquisition matrix = 256 × 256 × 256).
Image Preprocessing
Image processing steps were performed using the Connectome Computation System (CCS, Xu et al. 2015; https://github.com/zuoxinian/CCS), which integrates the functionalities of AFNI (Cox 1996), FSL (Jenkinson et al. 2012), and FreeSurfer (Fischl 2012) with shell and MATLAB scripts. The structural images were first de-noised using a spatially adaptive nonlocal means filter (Xing et al. 2011; Zuo and Xing 2011) and further processed to reconstruct the brain cortical surface (Dale et al. 1999; Fischl et al. 1999, 2001; Segonne et al. 2004, 2007). The rfMRI functional images were preprocessed with the following steps: 1) deleting the first 5 EPI volumes; 2) de-spiking time series; 3) correcting slice timing; 4) aligning each volume to the mean EPI image; 5) normalizing the 4D global mean intensity (10 000); 6) band-pass (0.01–0.1 Hz) filtering the time series; 7) estimating a rigid transformation from individual functional images to the individual anatomical image using the GM-WM boundary-based registration (BBR) algorithm (Greve and Fischl 2009), and 8) a nonlinear transformation between individual anatomical space to the MNI152 template (FNIRT in FSL, 1 mm isotropic).
Quality Control Procedure
CCS includes a quality control procedure to verify the quality of images processed, which produces various screenshots of brain extraction, brain tissue segmentation, pial and white surface reconstruction, and BBR-based functional image registration for visual inspections. The following quantitative measures of head motion and registration quality were computed: 1) the maximum distance of translational head movement (maxTran), 2) the maximum degree of rotational head movement (maxRot), 3) the mean frame-wise displacement (meanFD) (Power et al. 2012), 4) the minimal cost of the BBR co-registration (mcBBR), and 4) the error of the FNIRT spatial normalization (errFNIRT). All participants with poor brain extraction, tissue segmentation, and surface construction were excluded from subsequent analysis. Further, all datasets entering into the subsequent analysis met the following criteria: 1) maxTran ≤2 mm, 2) maxRot ≤2°, 3) meanFD ≤ 0.2 mm, and 4) errFNIRT < 0.2. More details on these steps can be found at the CCS website (http://lfcd.psych.ac.cn/QC.html). Among the original 400 subjects, 21 had excessive head motion (meanFD > 0.2 mm) and 57% of them were females.
Generation of Template ICNs
The analytic pipelines are illustrated in Figure 1. First, generalized ranking and averaging independent component analysis by reproducibility (gRAICAR) (Yang et al. 2008, 2012) was applied to the template dataset (N = 105), with all participants having a mean FD < 0.2 mm. This provided a set of unbiased templates that reflected common ICNs that would be detectable in the main dataset. In this way, the registration accuracy of the ICNs between the templates and the main dataset was maximized, since the 2 datasets were acquired using the same imaging parameters such as field strength and resolution. Specifically, after preprocessing and quality control procedures described above, spatial independent component analysis was applied to the rfMRI dataset using the MELODIC module of FSL software (Beckmann and Smith 2004). The numbers of components were automatically estimated by MELODIC runs. The resultant spatial components from individual participants were pooled, and gRAICAR matched the components from different participants and allocated them into clusters based on their spatial similarity measured using mutual information. For each cluster, a participant could contribute at most one component. A total intersubject spatial similarity among the member component maps (each representing a subject) was calculated to indicate the cross-subject consistency of the cluster. The member component maps in the cluster were weighted averaged to generate a cluster-wise component map. The weights in the averaging were determined by the degree centrality of the individual component maps in the cluster. The degree centrality significance was determined using permutation tests in which components from individual participants were randomly matched, yielding probability values that reflect contributions of individual participants to the clusterwise components. The effectiveness of gRAICAR has been demonstrated in multiple studies (Yang, Chang et al. 2014; Yang, Xu et al. 2014; Kyathanahally et al. 2015). Ranking the clusterwise component maps by their cross-subject consistency, we identified 7 ICNs that represented distributed areas in gray matter and were significantly contributed to by at least 60% of the template subjects.
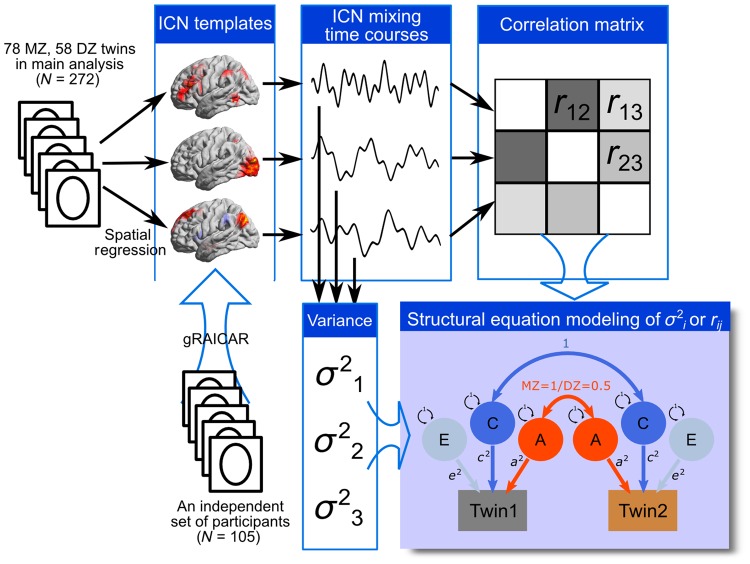
Figure 1.
A flowchart of the analysis pipeline. The analysis pipeline consists of 5 steps. (1) Before the main analysis, an independent dataset containing 105 participants, who had same imaging protocol as in the main analysis, was used to generate common ICN templates (unthresholded spatial maps representing ICNs); (2) For each participant in the main analysis (N = 272), the rfMRI dataset was projected to the ICN spatial templates to yield mixing time courses of the ICNs; (3) The temporal variance of the mixing time courses of the ICNs were computed to represent the level of fluctuations for individual ICNs; (4) The mixing time courses of the ICNs were correlated to generate inter-ICN coherence metrics, as the upper-triangle elements in the inter-ICN correlation matrix; (5) Structural equation modeling was applied to estimate genetic and environmental influence on the each of the ICN fluctuations (variance values) and inter-ICN coherences (correlation coefficients). The capital letters “A,” “C,” and “E” in the model represent additive genetic effect, shared (common) environmental effect, and un-shared (unique) environmental effect, respectively.
Spatial Regression of ICN Maps
Spatial regression was used to project the rfMRI data of the main dataset (N = 272) to the ICN template maps, yielding estimated time-varying contributions of the ICNs to the fMRI data. This approach fulfills our assumption that the ICNs estimated from ICA can overlap in space. An example in this study is that the precuneus network (also referred to as posterior default mode network) and the default mode network overlap in posterior cingulate cortex (see Fig. 2, also observed in Zuo et al. 2010; Jones et al. 2011; Kim and Lee 2011; Knyazev 2012; Yang, Chang et al. 2014). Compared with the mean time series within ROIs, spatial regression not only enables us to disentangle the time series representing the spatially overlapped ICNs detected by ICA, which has been shown to be more sensitive to network changes in aging study (Koch et al. 2010), but also overcomes potential limitations of using seed ROIs to extract representative time series within regions showing highly functional heterogeneity (Jiang et al. 2015; Jiang and Zuo 2015) by taking the whole-brain spatial variability into consideration in the spatial regression model. It has been used as the first half of the “dual regression” model (Beckmann et al. 2009; Zuo et al. 2010). In details, the fMRI dataset was reformed into a Matrix X with v rows and t columns, where v is the number of voxels and t the number of volumes. The 7 ICN template maps plus a constant map (in which all voxels are set to 1 to represent the constant term in the regression model) were formed into a Matrix S, where each of the 8 columns represents a template map (a vector of length v). The intensity of the 7 template maps was standardized so that they had zero mean and unit variance across all voxels. The data matrix X can then be expressed using a linear model: X = SM. The matrix M represents how the ICN template maps, S, are mixed to form X. M is an 8 × t matrix, and each row of M is the representative time series for an ICN. Solving M using a maximal-likelihood approach, we obtained the representative time series for each ICN, reflecting time-varying contribution of the ICN to a given participant (as illustrated in Fig. 1). In Supplementary Figure 1, we showed that the representative time courses were highly correlated with the mean time series from the core regions of the ICNs, supporting the validity of the representative time course.
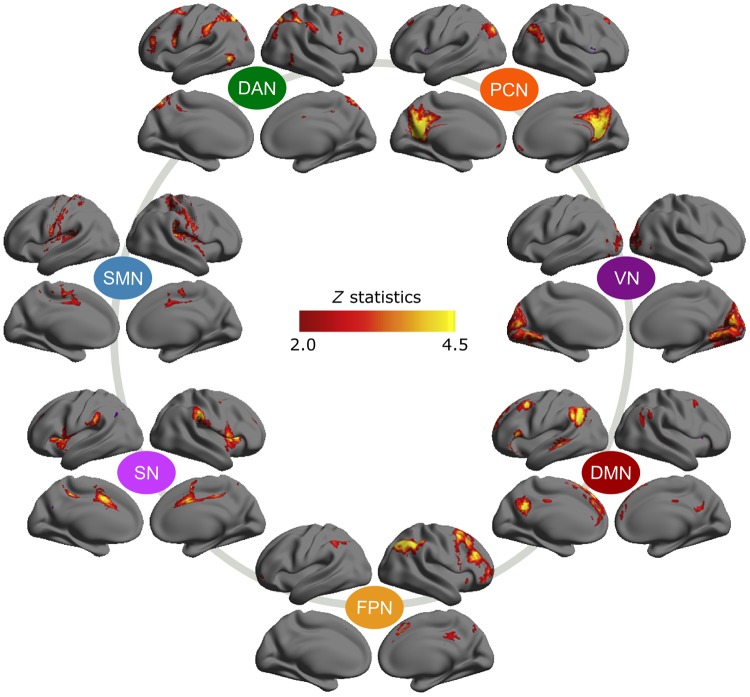
Figure 2.
Template maps of 7 common ICNs. Spatial maps showing the 7 common ICNs detected in 105 individuals. For visualization purpose, the spatial maps are threshold at |Z| > 2.0, but in the following analysis, these spatial maps were not thresholded. Clockwise, the ICNs are the precuneus-dorsal posterior cingulate network (PCN, some studies referred to as posterior default mode network), the visual network (VN), the default mode network (DMN), the fronto-parietal network (FPN), the salience network (SN), the somatosensory-motor network (SMN), and the dorsal attention network (DAN).
Genetic and Environmental Influence on ICN Fluctuations
As demonstrated in Figure 1, the fluctuation level of individual ICNs was quantified using variance of the corresponding representative time courses. Temporal variance has been demonstrated to be closely relevant to the functioning of ICNs (Garrett et al. 2010; Samanez-Larkin et al. 2010; Kaneoke et al. 2012). For each of the ICNs, we first calculated the cross-twin correlation of the variance metrics for MZ and DZ twin pairs separately. The resultant cross-twin correlation coefficients were compared to provide an indication of the relative contribution of genetic and environmental factors to the ICN fluctuations (Fulker and Jinks 1970; Plomin et al. 2000).
Further, we used an ACE model to quantify the genetic and environmental contributions to the ICN fluctuations. In a standard ACE model, the variance of a trait can be decomposed into additive genetic (A), shared or common (C) environment, and nonshared (unique) environmental (E) effects, the latter of which includes measurement error (Neale and Cardon 1992). This model takes advantage of the differences in genetic relatedness between MZ twins who share 100% of their genes, and DZ twins who share, on average, 50% of their genes (Silberg et al. 1990; Rijsdijk and Sham 2002). Based on these assumptions, one can derive the contributions of A and C effects using SEM, as implemented in OpenMx (Boker et al. 2011), with covariates of age and sex modeled as regressions or deviation effects on the mean. In SEM, A, C, and E are represented by latent variables, and their relative variances (denoted as a2, c2, and e2, respectively) reflect the proportion of their contributions to the cross-subject covariance of the observed variables (i.e., variance metrics of ICNs).
Since the variance metric of ICNs may not follow a normal distribution, we used a nonparametric approach to examine the significance of A and C effects (Fornito et al. 2011). The first step is model identification, which is used to examine the significance of the A and C contributions in the model. Besides the ACE model, the path coefficients for 2 submodels, AE and CE, were estimated. The differences in likelihoods between the full model and the 2 submodels were calculated. A significant likelihood difference between ACE and CE models indicated that the A factor is critical in the full model, while a significant likelihood difference between ACE and AE model suggested a significant contribution of C factor.
Null distributions of the likelihood difference were generated by permuting the MZ/DZ labels and the composition of the twins with constraints on exchangeability. Specifically, if a twin from twin pair A was randomly paired with a twin from twin pair B, then the other twins from twin pairs A and B were paired. In addition, the sex and age of the 2 twins forming a twin pair were constrained to be the same, so that age and sex could still be included as covariates in the ACE model. This permutation was repeated 5000 times, and the ACE model, as well as AE and CE submodels, was estimated. The likelihood differences between the full model and submodels were collected to form 2 null distributions, one for ACE-AE and the other for ACE-CE.
The significance of A and C factors for the original dataset was then evaluated within the null distributions. Under the rule of parsimony in model identification, if the C factor was significant but A was not, we used the CE model to estimate the C effect; if the A factor was significant but C was not, we used the AE model to evaluate the A effect. If neither A nor C was significant in these tests, we used the ACE model. For each of the 7 ICNs, we separately conducted the above procedure for model identification. The threshold for significance was P < 0.005 (single-tail test). Once the model was selected, the relative variance of A, C, and E factors (a2, c2, and e2) was evaluated from the identified model.
Genetic and Environmental Influence on Individual Inter-ICN Coherences
Using the same methodology, we investigated genetic and environmental influence on individual inter-ICN coherences. The inter-ICN coherence was characterized using temporal correlation between the representative time courses from a pair of ICNs. For each of the inter-ICN coherence metrics, we compared cross-twin correlations for MZ and DZ twins, and further quantified the contribution of genetic and environmental factors using the ACE model (see above section for details).
Results
ICN Templates
Using 105 individuals whose scan parameters were identical to those in the main analysis, we generated 7 spatial templates, each representing a commonly observed ICN. Figure 2 depicts the spatial ICN template maps. These maps were not thresholded in subsequent spatial regression analysis. Ranked by cross-subject consistency (the sum of spatial similarity among the member component maps, each representing a subject), these 7 ICNs were the precuneus-dorsal posterior cingulate network (PCN, some studies referred to this as posterior default mode network), the visual network (VN), the default mode network (DMN), the fronto-parietal network (FPN), the salience network (SN), the somatosensory-motor network (SMN), and the dorsal attention network (DAN). These networks have been commonly reported in previous studies (Power et al. 2011; Yeo et al. 2011), proved to be reliable (Zuo et al. 2010), and have been associated with behavioral and cognitive taxonomy (Laird et al. 2013).
Genetic and Environmental Influence on ICN Fluctuations
Table 1 presents cross-twin correlation coefficients for MZ and DZ twins. PCN, VN, DMN, FPN, and DAN showed significant cross-twin correlation in MZ twins (P < 0.05 for DMN, P < 0.005 for the other ICNs) and close-to-zero cross-twin correlation in DZ twins, suggesting a major influence of genetic factors. SMN showed significant cross-twin correlation in both MZ and DZ twins (P < 0.005), and the correlation coefficient was similar in the 2 types of twins, suggesting that the level of fluctuations of this ICN is significantly influenced by common environmental factors. SN showed nonsignificant and relatively low cross-twin correlation in both MZ and DZ twins, indicating lack of evidence for genetic and common environmental effects on it.
Table 1.
Cross-twin correlation of ICN fluctuations
| Inter-ICN coherence | MZ |
DZ |
||
|---|---|---|---|---|
| r | 95% CI | r | 95% CI | |
| PCN | 0.478** | 0.286/0.633 | 0.111 | −0.151/0.359 |
| VN | 0.410** | 0.206/0.579 | 0.049 | −0.212/0.304 |
| DMN | 0.248* | 0.026/0.445 | 0.063 | −0.198/0.316 |
| FPN | 0.448** | 0.251/0.610 | 0.107 | −0.155/0.356 |
| SN | 0.139 | −0.086/0.351 | 0.222 | −0.039/0.454 |
| SMN | 0.342** | 0.129/0.525 | 0.409** | 0.168/0.603 |
| DAN | 0.360** | 0.149/0.539 | 0.074 | −0.188/0.326 |
Note: * indicates P < 0.05; ** indicates P < 0.005.
Results from the SEM analyses echoed the above indications. Table 2 presents estimations of genetic (a2), common environmental (c2), and unique environmental effects (e2) from the ACE, AE, and CE models. PCN, VN, DMN, FPN, and DAN showed significant A effect (p(A) < 0.005) and nonsignificant C effect (p(C) > 0.05), and thus, we used AE model to derive the relative variance of A and E. As a result, we obtained a2 values of 51.0%, 34.9%, 23.3%, 65.2%, and 32.7% for the fluctuation levels of PCN, VN, DMN, FPN, and DAN, respectively. On the other hand, SMN showed significant C effect but not A effect. The CE model was then used to derive a c2 value of 34.5% for the fluctuation level of this ICN.
Table 2.
Results of ACE modeling on ICN fluctuations
| ACE |
AE |
CE |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| a2 (%) | c2 (%) | e2 (%) | p(A) | a2 (%) | e2 (%) | p(C) | c2 (%) | e2 (%) | |
| PCN | 51.0 | 0.0 | 49.0 | 0.000 | 51.0* | 49.0 | 0.210 | 18.6 | 81.4 |
| VN | 34.9 | 0.0 | 65.1 | 0.000 | 34.9* | 65.1 | 0.254 | 24.2 | 75.8 |
| DMN | 23.3 | 0.0 | 76.7 | 0.003 | 23.3* | 76.7 | 0.289 | 11.4 | 88.6 |
| FPN | 65.2 | 0.0 | 34.8 | 0.000 | 65.2* | 34.8 | 0.316 | 10.3 | 89.7 |
| SN | 17.7 | 4.2 | 78.1 | 0.047 | 23.4 | 76.6 | 0.071 | 15.5 | 84.5 |
| SMN | 2.2 | 33.0 | 64.8 | 0.132 | 41.3 | 58.7 | 0.003 | 34.5* | 65.5 |
| DAN | 32.7 | 0.0 | 67.3 | 0.000 | 32.7* | 67.3 | 0.218 | 19.0 | 81.0 |
Note: p(A) < 0.005 or p(C) < 0.005 indicates that the A or C factor has a significant contribution in fitting the data. For cases with significant A but nonsignificant C factors, we used AE submodel to estimate the variance explained by A factor (a2); for cases with significant C but nonsignificant C factors, we used CE submodel to estimate the variance explained by C factor (c2). The models selected for the estimation of a2 or c2 are highlighted using bold font.
*Corresponding a2 or c2 is significant at P < 0.005. All P values in this table are single-tailed statistics.
Genetic and Environmental Influences on Individual Inter-ICN Coherences
The results of cross-twin correlation for MZ and DZ twins are summarized in Table 3. All inter-ICN coherences showed significant cross-twin correlation in MZ twins (P < 0.05 for PCN-DAN, DMN-DAN, FPN-SN, FPN-SMN, and SMN-DAN, P < 0.005 for the other pairs). The inter-ICN coherences PCN-VN, PCN-DMN, PCN-FPN, VN-DMN, VN-SN, and VN-DAN showed significant cross-twin correlation in DZ twins at P < 0.005, and PCN-SN, PCN-DAN, VN-FPN, DMN-FPN, DMN-SN, and DMN-DAN showed significant cross-twin correlations in DZ twins at P < 0.05. For these inter-ICN coherences, the cross-twin correlations in MZ twins were less than 2 times of the correlation in DZ twins, indicating influence from both genetic and common environmental factors. On the other hand, the inter-ICN coherences PCN-SMN, VN-SMN, DMN-SMN, FPN-SN, FPN-SMN, FPN-DAN, SN-SMN, SN-DAN, and SMN-DAN showed nonsignificant cross-twin correlations in DZ twins (P > 0.05), indicating that these inter-ICN coherence could be influenced mainly by genetic factors.
Table 3.
Cross-twin correlation of inter-ICN coherences
| Inter-ICN coherence | MZ |
DZ |
||
|---|---|---|---|---|
| r | 95% CI | r | 95% CI | |
| PCN-VN | 0.511** | 0.326/0.659 | 0.436** | 0.200/0.624 |
| PCN-DMN | 0.521** | 0.338/0.666 | 0.391** | 0.148/0.590 |
| PCN-FPN | 0.369** | 0.160/0.547 | 0.434** | 0.198/0.622 |
| PCN-SN | 0.360** | 0.149/0.539 | 0.294* | 0.038/0.513 |
| PCN-SMN | 0.317** | 0.102/0.504 | 0.201 | −0.061/0.436 |
| PCN-DAN | 0.274* | 0.055/0.468 | 0.344* | 0.094/0.553 |
| VN-DMN | 0.418** | 0.216/0.586 | 0.453** | 0.221/0.637 |
| VN-FPN | 0.368** | 0.159/0.546 | 0.256* | 0.002/0.483 |
| VN-SN | 0.449** | 0.252/0.611 | 0.366** | 0.119/0.570 |
| VN-SMN | 0.324** | 0.109/0.510 | 0.108 | −0.155/0.356 |
| VN-DAN | 0.453** | 0.256/0.614 | 0.365** | 0.118/0.570 |
| DMN-FPN | 0.415** | 0.212/0.584 | 0.336* | 0.085/0.547 |
| DMN-SN | 0.336** | 0.123/0.520 | 0.245* | −0.015/0.473 |
| DMN-SMN | 0.437** | 0.238/0.601 | 0.069 | −0.193/0.322 |
| DMN-DAN | 0.272* | 0.053/0.466 | 0.299* | 0.044/0.517 |
| FPN-SN | 0.287* | 0.069/0.479 | 0.140 | −0.123/0.384 |
| FPN-SMN | 0.260* | 0.040/0.456 | −0.207 | −0.442/0.054 |
| FPN-DAN | 0.441** | 0.242/0.604 | 0.223 | −0.038/0.455 |
| SN-SMN | 0.402** | 0.197/0.573 | 0.029 | −0.231/0.285 |
| SN-DAN | 0.356** | 0.145/0.536 | 0.120 | −0.142/0.367 |
| SMN-DAN | 0.284* | 0.066/0.477 | 0.149 | −0.113/0.392 |
Note: * indicates P < 0.05; ** indicates P < 0.005.
Results from the further SEM analyses are summarized in Table 4. The inter-ICN coherences DMN-SMN, FPN-DAN, and SN-SMN showed significant A effect but nonsignificant C effect in model selection (P < 0.005). The a2 for these inter-ICN coherences estimated from the AE model was 39.4%, 42.0%, and 29.3%, respectively. The inter-ICN coherences PCN-VN, PCN-DMN, PCN-FPN, PCN-DAN, VN-DMN, and VN-SN showed significant C effect but nonsignificant A effect in model selection. The c2 for these inter-ICN coherences estimated from the CE model was 45.3%, 47.1%, 38.1%, 29.0%, 41.7%, and 39.7%, respectively. The other inter-ICN coherences did not survive the P < 0.005 threshold for adopting AE or CE submodels, and their a2 and c2 were estimated using the ACE model. Among these inter-ICN coherences, PCN-SN showed significant c2 = 30.8%, PCN-SMN showed significant c2 = 18.0%, VN-DAN showed significant c2 = 28.5%, and DMN-DAN showed significant c2 = 25.9%, while VN-FPN showed significant a2 = 25.6%, VN-SMN showed significant a2 = 30.3%, DMN-FPN showed significant a2 = 23.4%, SN-DAN showed significant a2 = 31.1%, and SMN-DAN showed significant a2 = 29.0%. No inter-ICN coherence showed both significant A and C effects. Figure 3 provides a summary of the SEM analyses on both ICN fluctuations and inter-ICN interplays.
Table 4.
Results of ACE modeling on ICN fluctuations
| ACE |
AE |
CE |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| a2 (%) | c2 (%) | e2 (%) | p(A) | a2 (%) | e2 (%) | p(C) | c2 (%) | e2 (%) | |
| PCN-VN | 10.5 | 37.4 | 52.1 | 0.066 | 52.2 | 47.8 | 0.000 | 45.3* | 54.7 |
| PCN-DMN | 21.4 | 30.2 | 48.4 | 0.024 | 53.7 | 46.3 | 0.003 | 47.1* | 52.9 |
| PCN-FPN | 0 | 38.1 | 61.9 | 0.791 | 39.8 | 60.2 | 0.000 | 38.1* | 61.9 |
| PCN-SN | 0 | 30.8* | 69.2 | 0.779 | 33.2 | 66.8 | 0.019 | 30.8 | 69.2 |
| PCN-SMN | 11.8 | 18.0* | 70.2 | 0.123 | 31.5 | 68.5 | 0.058 | 27.8 | 72.2 |
| PCN-DAN | 0 | 29.0 | 71.0 | 0.527 | 31.7 | 68.3 | 0.000 | 29.0* | 71.0 |
| VN-DMN | 0 | 41.7 | 58.3 | 0.476 | 45.3 | 54.7 | 0.000 | 41.7* | 58.3 |
| VN-FPN | 25.6* | 11.7 | 62.7 | 0.027 | 38.5 | 61.5 | 0.081 | 32.3 | 67.7 |
| VN-SN | 0 | 39.7 | 60.3 | 0.281 | 42.8 | 57.2 | 0.001 | 39.7* | 60.3 |
| VN-SMN | 30.3* | 0 | 69.7 | 0.014 | 30.3 | 69.7 | 0.719 | 24.8 | 75.2 |
| VN-DAN | 21.7 | 24.1* | 54.2 | 0.028 | 48.4 | 51.6 | 0.011 | 40.7 | 59.3 |
| DMN-FPN | 23.4* | 17.3 | 59.2 | 0.027 | 42.4 | 57.6 | 0.049 | 36.1 | 63.9 |
| DMN-SN | 0 | 28.5* | 71.5 | 0.251 | 30.9 | 69.1 | 0.015 | 28.5 | 71.5 |
| DMN-SMN | 39.4 | 0 | 60.6 | 0.001 | 39.4* | 60.6 | 0.782 | 30.3 | 69.7 |
| DMN-DAN | 1.3 | 25.9* | 72.9 | 0.235 | 31.4 | 68.6 | 0.010 | 26.8 | 73.2 |
| FPN-SN | 14.9 | 9.5 | 75.6 | 0.060 | 25.6 | 74.4 | 0.080 | 21.5 | 78.5 |
| FPN-SMN | 19.3 | 0 | 80.7 | 0.006 | 19.3 | 80.7 | 0.772 | 9.8 | 90.2 |
| FPN-DAN | 42.0 | 0 | 58.0 | 0.001 | 42.0* | 58.0 | 0.279 | 32.7 | 67.3 |
| SN-SMN | 29.3 | 0 | 70.7 | 0.004 | 29.3* | 70.7 | 0.275 | 21.8 | 78.2 |
| SN-DAN | 31.1* | 0 | 68.9 | 0.013 | 31.1 | 68.9 | 0.486 | 24.3 | 75.7 |
| SMN-DAN | 29.0* | 0 | 71.0 | 0.014 | 29.0 | 71.0 | 0.985 | 21.5 | 78.5 |
Note: p(A) < 0.005 or p(C) < 0.005 indicates that the A or C factor has a significant contribution in fitting the data. For cases with significant A but nonsignificant C factors, we used AE submodel to estimate the variance explained by A factor (a2); for cases with significant C but nonsignificant C factors, we used CE submodel to estimate the variance explained by C factor (c2). The models selected for the estimation of a2 or c2 are highlighted using bold font.
*Corresponding a2 or c2 is significant at P < 0.005. All P values in this table are single-tailed statistics.
Figure 3.
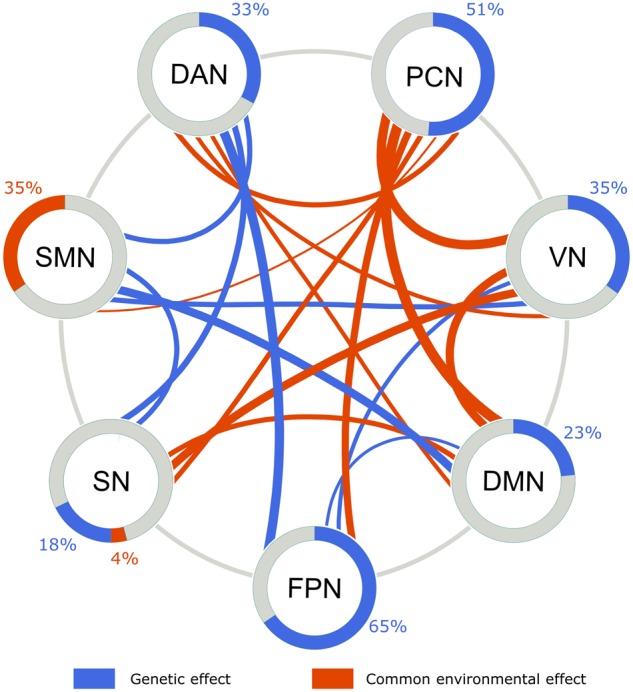
Summary of SEM results for ICN fluctuation and inter-ICN coherence. The genetic (blue) and common environmental (red) contributions to the fluctuation of individual ICNs (a2 and c2 values in Table 2) are visualized using areas of the rings. The genetic (blue) and common environmental (red) effects on the inter-ICN coherences are visualized using the belts linking individual ICNs. The width of the belts represents the contributions (a2 or c2 values in Table 4).
Discussion
Systematic investigation of the intrinsic functional networks in the largest twin sample to date quantified genetic and environmental influence on the functional connectivity architecture of the human brain. We examined the activity level of individual ICNs and the interplays between ICNs, providing a full picture of the genetic and environmental effects on large-scale brain networks (Fig. 3). This study extends previous studies on task-based brain activations (Koten et al. 2009; Blokland et al. 2011), because ICNs and their interplays reflect more fundamental or intrinsic architecture of brain function (Fox and Raichle 2007), and it is hypothesized that the ICNs are recruited and combined to facilitate various tasks (Deco and Corbetta 2011; Buckner et al. 2013). Among the 7 ICNs explored, 5 showed significant genetic effects on the amplitude of fluctuations, and genetic factors were estimated to explain up to 65% of the individual variability. In contrast, 11 of the 21 inter-ICN coherences exhibited significant common environmental effects, and up to 47.1% of the individual variability in inter-ICN coherences can be explained by common environmental factors. These observations suggest that genetic factors play an important role in the activity of individual ICNs, and most of the environmental influence are reflected on the interplays between ICNs. This implication outlines a highly modular organization of the functional connectivity architecture, in which the individual modules represented by ICNs are genetically controlled, while the interplays between the modules are influenced by environmental factors.
Genetic Factors Control Modules of the Connectivity Architecture
ICNs have been hypothesized to be subsystems in the functional brain network (Damoiseaux et al. 2006; Biswal et al. 2010) and have been associated to various cognitive components (Smith et al. 2009; Laird et al. 2011). Our findings that the PCN, VN, DMN, FPN, and DAN are sensitive to genetic mediation indicate that major portions of the subsystems in the intrinsic organization of the brain are controlled by genetic factors. The heritability of the activity level of the individual ICNs supports that ICNs detected with rfMRI are useful mediators to connect brain function to genes. Furthermore, our findings of the intermediate to high heritability (23.3–65.2%) of the ICNs support the hypothesis that the ICNs are more fundamental functional modules that are recruited and combined to perform tasks (Deco and Corbetta 2011), and further provide critical information for associating genes to various cognitive functions. For instance, our results showed that genetic factors explain 65.2% of the interindividual variance in the activity level of FPN. Given that previous studies have associated high-level cognitive tasks such as working memory, reasoning, inhibition, and attention to FPN (Laird et al. 2011), our results predict the heritage of these cognitive control functions, which has been reported separately (Fan et al. 2001; Blokland et al. 2011; Schachar et al. 2011; Mosing et al. 2012). Combining previous findings and our results, it is likely that the high-level cognitive control functions are facilitated by the same FPN network. More importantly, it is reasonable to hypothesize that the same set of genes controlling the activity of FPN is the common source of the individual variability of these high-level cognitive control functions.
Similarly, the significant genetic influences on the PCN, VN, DMN, and DAN suggest that their associated brain functions are heritable. In particular, as the central region of the PCN, precuneus, is thought to be a key connectome hub (Hagmann et al. 2008; van den Heuvel and Kahn 2011; Zuo et al. 2012) that is highly functional heterogeneous and is commonly considered to play a central role in the integration of auditory, somatosensory-motor, and visual information, as well as spontaneous cognition (Greenwood et al. 1993; Cavanna and Trimble 2006; Margulies et al. 2009; Zhang and Li 2012; Park et al. 2013; Jiang et al. 2015). The high heritability of PCN suggests that the internal mechanism for integrating multi-modal sensory information (does not include the amount and ways of connections to unimodal cortex) is controlled by genetic factors. This argument is further supported by the relevance of this ICN to drug-naive, first-episode early-onset schizophrenia (Yang, Xu et al. 2014) and to healthy aging (Yang, Chang et al. 2014).
Comparing to previous findings on the heritability (42.4%) of DMN (Glahn et al. 2010), our results showed a lower genetic contribution to DMN (23.3%). This difference does not conflict the previous finding, but rather reflects a difference in the definition of DMN. In our study, the PCN was functionally separated from the DMN, while the DMN template in Glahn et al. (2010) includes PCN. The PCN has been referred as posterior DMN in some studies (Jones et al. 2011; Kim and Lee 2011; Knyazev 2012), exhibited different cross-lifespan dynamics than DMN (Jones et al. 2011; Yang, Chang et al. 2014), and has been related to high-order memory function termed as a parietal memory network (Gilmore et al. 2015). In fact, based on the connectional anatomy, Buckner et al. (2008) have suspected the involvement of precuneus in DMN. Echoing these evidences, our ICN templates generated using a fully data-driven approach revealed separate PCN and DMN. Given the difference in heritability estimates for DMN and PCN, we speculate that the activity level of both DMN and PCN is influenced by genetic factors, but the activity of PCN may receive more genetic control than that of DMN. This speculation explains the 42.4% heritability of DMN estimated by Glahn et al. (2010), since in the current study, the mean heritability of DMN and PCN is around 40%.
Environmental Factor Influence Interplays Between Modules
Plasticity is an essential property of our brain that allows us to learn. Our results enrich our understanding of the plasticity in brain networks by mapping the environmental influence on specific coherences between ICNs. We showed that a large portion (11 of 21) of the interplays between ICNs are modulated by common environmental effects, highlighting the contribution of environment in the functional architecture of the brain connectivity.
Six inter-ICN coherences between PCN and the other ICNs are significantly influenced by common environmental effects. These observations generated an important hypothesis that helps to disentangle the main source of the plasticity in the neural system, that is, the interplays between PCN and other ICNs, instead of the PCN per se, underpin the brain network plasticity due to environment. This hypothesis explains the observed behaviors of the precuneus. This region has been reported to involve in a wide range of high-level cognitive processing, such as visuospatial imagery (Hanakawa et al. 2003), episodic memory retrieval (Henson et al. 1999), and self-processing (Kircher et al. 2000). These behaviors exhibit strong relationships to environment. For instance, studies have indicated the relationship between spatial imagery and visual experience (Gandhi et al. 2014), the relationship between episodic memory and specific training (Brehmer et al. 2007), and the relationship between self-perception and teaching strategy (Penick 1982). Importantly, when involving in various functions, the precuneus never activates alone, but is accompanied by regions in other systems, such as visual, auditory, and motor systems, as well as prefrontal cortex (Cavanna and Trimble 2006). These behaviors make the precuneus a functional “router” in the brain—highly flexible processing of various information flow by connecting to the corresponding brain networks (Cole et al. 2013). Given this context, our results provide an interpretation, at the large-scale neural network level, of the plasticity of precuneus-relevant behaviors and further hypothesize that the main sources of the plasticity originated from the interplays between PCN and other ICNs.
The other inter-ICN coherences showing significant environmental influences are from either DMN or VN. The visual system processes the major portion of the external information, and DMN is the largest consumer of the “dark energy” in the brain. Recent studies on brain network reorganizations after meditation (Kemmer et al. 2015) and arm amputation (Makin et al. 2015) have echoed the plasticity of the functional connectivity between these networks and other ICNs, although more investigations are needed to verify the plasticity of the inter-ICN coherence and its cause. It should be noted that here we discuss the environmental influence based on significant influence from common environmental effects, but the unique environmental effect could also likely attribute to environmental influence. Thus, the above discussions do not exclude the environmental contribution to the inter-ICN coherences that did not exhibit significant common environmental effect.
Genetic Factors Influence Interplays Between Modules
We found 8 inter-ICN coherences that exhibit significant genetic effect, and the estimated genetic contribution ranged between 25.6% and 42.0%. These findings call for an expansion of the search space for heritable ICN-derived endophenotypes for neuropsychiatric illness. A growing literature has implicated abnormal network properties in neuropsychiatric illness and suggested their potential utility as endophenotypes (Hasler and Northoff 2011). Our findings regarding the heritability of inter-ICN coherence provide support for this notion, and further shed light on interplays between ICNs as promising endophenotype candidates. While it is possible that a pathologic process can specifically impact a single functional system (e.g., stroke), neuropsychiatric illnesses appear to be associated with more widespread abnormalities, affecting multiple networks and their interactions (Jafri et al. 2008; Stephan et al. 2009; Park and Thakkar 2010; Menon 2011). Recent studies have highlighted inter-ICN coherence as an important feature in brain abnormalities underlying various mental disorders (Kaiser and Feng 2015; Nomi and Uddin 2015). For instance, Kaiser and Feng (2015) reported that teenagers with autism spectrum disorder did not differ from typical developing subjects in within-ICN connectivity, but showed significantly decreased inter-ICN coherence (). Heritability is a critical property for inter-ICN coherence features to be considered as potential endophenotypes representing genetic vulnerability to neuropsychiatric illness. Our findings provide evidence for this possibility. The estimation of heritability for the individual inter-ICN coherences further provides a reference for screening potential endophenotypes for neuropsychiatric illness.
The FPN-DAN coherence is the most heritable inter-ICN interplay according to our estimation. The FPN is relevant to executive control, memory encoding, and speech (Miller and Cohen 2001), and the DAN is associated with top-down orienting of attention (Fox et al. 2006). The interplay between these 2 ICNs may serve to provide greater specificity in efforts to develop diagnostic indices or biomarkers than individual ICN properties themselves. A meta-analysis containing 25 studies provided strong support for this argument by highlighting that decreased coherence between the FPN and DAN is a remarkable deficit in major depressive disorder patients (Kaiser and Feng 2015). Another study also reported decreased FPN-DAN coherence in Parkinson's disease (Baggio et al. 2015). The intermediate heritability (0.42) in FPN-DAN coherence observed in our study adds support for this inter-ICN coherence to be considered as an effective endophenotype of mental disorders. At the system neuroscience level, heritable FPN-DAN coherence complements our understanding of the prefrontal cortex. The prefrontal cortex is generally believed to be shaped largely by environmental influences (Cerqueira et al. 2007; McEwen and Morrison 2013). Studies have emphasized the impact of stress and environmental enrichment, as well as suggested the ability of cognitive training to impact prefrontal function (Olesen et al. 2004; Klingberg 2010; Soderqvist et al. 2012). Our finding that FPN-DAN coherence is substantially heritable provides support for genetic contribution of prefrontal functioning and bolsters recent discussions indicating that the functions of prefrontal cortex are shaped by a combination of genetic and environmental factors (Colizzi et al. 2015).
The SN-SMN, DMN-SMN, and VN-FPN represent interplay between unimodal (SMN and VN) and cross-modal cognitive networks (SN, DMN, and FPN). Functional parcellation studies commonly show that unimodal ICNs and cross-modal cognitive networks are separable (Power et al. 2011; Yeo et al. 2011), indicating that the functions of these networks may be divergent. However, this does not mean that the individual variability in inter-ICN interplay is not due to genetic factors. With the increasing attention to inter-ICN interactions (Steffener et al. 2012; Cerliani et al. 2015; Diez et al. 2015), we expect that future studies investigating inter-ICN interactions in psychiatric disorders will likely detect biomarkers based on the interplay between unimodal ICNs and cross-modal cognitive networks. Similarly, those examining age-related genetic influences (Lenroot et al. 2009; Gao et al. 2014) will likely find development-related changes in such inter-ICN interactions.
Limitations
In considering the magnitude of genetic or common environmental effects in the present work, it is important to note the impact of the sample size and data quality. Our results are conservative in that they are based on a moderate sample size for a classical twin study. Additionally, the quality of the data has limited the power to detect significant genetic and environmental effects. This is relevant to the estimates from the full ACE models, where we failed to find an inter-ICN coherence that showed both significant genetic and common environmental effects, as a large amount of variance was explained by the nonshared environmental effects and measurement error (E). We are optimistic that future studies using newly optimized data acquisition protocols (e.g., the Human Connectome Project, Van Essen et al. 2013), which have been demonstrated to have satisfactory reliability (Zuo and Xing 2014), will have the power to reveal whether an even greater proportion of the variance in the connectome is due to genetic factors or others interfering both reliability and reproducibility (Zuo et al. 2014), than has been possible in the present work. In addition, the imaging sample used in this study is from Australia, and the generalization of the findings to eastern population should be cautious and can be further tested using easterner twins in future.
Conclusions
Systematic investigation of the activity level of individual ICNs and the interplays between ICNs provides a full characteristic picture of the genetic and environmental effects on large-scale brain networks. While the activity levels of most ICNs are influenced by genetic factors, large portions of the interplays between ICNs are modulated by common environmental effect. These findings outline a highly modular organization of the functional connectivity architecture, in which the individual modules represented by ICNs are genetically controlled, while most of the interplays between the modules could be influenced by environmental factors. These findings initiate the process of detecting the genetic and environmental sources of individual variability of the human connectome.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
This work was supported by the National Basic Research (973) Program (grant number: 2015CB351702), the Natural Science Foundation of China (grant numbers: 81270023, 81571756, 81278412, 81171409, 81000583, 81471740, 81220108014), the Beijing Nova Program for Science and Technology (XXJH2015B079 to Z.Y.), the Outstanding Young Investigator Award of Institute of Psychology, Chinese Academy of Sciences (to Z.Y.), the Foundation of Beijing Key Laboratory of Mental Disorders (2014JSJB03 to Z.Y.), the Key Research Program and the Hundred Talents Program of the Chinese Academy of Sciences (KSZD-EW-TZ-002 to X.N.Z), NIH (U01MH099059, R01MH083246, R01HD050735), NIMH BRAINS (R01MH094639-01 to M.P.M), and gifts from Phyllis Green and Randolph Cowen to F.X.C. and M.P.M. M.P.M and X.N.Z are members of an international collaboration team supported by the Chinese Academy of Sciences K.C. Wong Education Foundation. Funding to pay the Open Access publication charges for this article was provided by the National Basic Research (973) Program of China.
Supplementary Material
Notes
Conflict of Interest: None declared.
References
- Andrews-Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL. 2010. Functional-anatomic fractionation of the brain's default network. Neuron. 65:550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggio HC, Segura B, Sala-Llonch R, Marti MJ, Valldeoriola F, Compta Y, Tolosa E, Junque C. 2015. Cognitive impairment and resting-state network connectivity in parkinson's disease. Hum Brain Mapp. 36:199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley AJ, Jones DW, Weinberger DR. 1997. Genetic variability of human brain size and cortical gyral patterns. Brain. 120:257–269. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Mackay CE, Filippini N, Smith SM. 2009. Group comparison of resting-state fMRI data using multi-subject ica and dual regression. Neuroimage. 47:S148. [Google Scholar]
- Beckmann CF, Smith SM. 2004. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 23:137–152. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S et al. 2010. Toward discovery science of human brain function. Proc Natl Acad Sci USA. 107:4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokland GA, de Zubicaray GI, McMahon KL, Wright MJ. 2012. Genetic and environmental influences on neuroimaging phenotypes: a meta-analytical perspective on twin imaging studies. Twin Res Hum Genet. 15:351–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokland GA, McMahon KL, Thompson PM, Martin NG, de Zubicaray GI, Wright MJ. 2011. Heritability of working memory brain activation. J Neurosci. 31:10882–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T, Spies J, Estabrook R, Kenny S, Bates T et al. 2011. OpenMX: an open source extended structural equation modeling framework. Psychometrika. 76:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehmer Y, Li S-C, Muller V, von Oertzen T, Lindenberger U. 2007. Memory plasticity across the lifespan: uncovering children's latent potential. Develop Psych. 43:465–478. [DOI] [PubMed] [Google Scholar]
- Bressler SL, Menon V. 2010. Large-scale brain networks in cognition: emerging methods and principles. Trends Cogn Sci. 14:277–290. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. 2008. The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 1124:1–38. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Yeo BT. 2013. Opportunities and limitations of intrinsic functional connectivity MRI. Nat Neurosci. 16:832–837. [DOI] [PubMed] [Google Scholar]
- Cavanna AE, Trimble MR. 2006. The precuneus: a review of its functional anatomy and behavioural correlates. Brain. 129:564–583. [DOI] [PubMed] [Google Scholar]
- Cerliani L, Mennes M, Thomas RM, Di Martino A, Thioux M, Keysers C. 2015. Increased functional connectivity between subcortical and cortical resting-state networks in autism spectrum disorder. JAMA Psychiatry. 72:767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira JJ, Mailliet F, Almeida OF, Jay TM, Sousa N. 2007. The prefrontal cortex as a key target of the maladaptive response to stress. J Neurosci. 27:2781–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, Avedissian C, Barysheva M, Toga AW, McMahon KL, de Zubicaray GI, Wright MJ, Thompson PM. 2009. Extending genetic linkage analysis to diffusion tensor images to map single gene effects on brain fiber architecture. Med Image Comput Comput Assist Interv. 12:506–513. [DOI] [PubMed] [Google Scholar]
- Chiang MC, Barysheva M, Toga AW, Medland SE, Hansell NK, James MR, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ et al. 2011. BDNF gene effects on brain circuitry replicated in 455 twins. Neuroimage. 55:448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Reynolds JR, Power JD, Repovs G, Anticevic A, Braver TS. 2013. Multi-task connectivity reveals flexible hubs for adaptive task control. Nat Neurosci. 16:1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colizzi M, Fazio L, Ferranti L, Porcelli A, Masellis R, Marvulli D, Bonvino A, Ursini G, Blasi G, Bertolino A. 2015. Functional genetic variation of the cannabinoid receptor 1 and cannabis use interact on prefrontal connectivity and related working memory behavior. Neuropsychopharmacology. 40:640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvy-Duchesne B, Blokland GA, Hickie IB, Thompson PM, Martin NG, de Zubicaray GI, McMahon KL, Wright MJ. 2014. Heritability of head motion during resting state functional mri in 462 healthy twins. Neuroimage. 102:424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW. 1996. Afni: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 29:162–173. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. 1999. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 9:179–194. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. 2006. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA. 103:13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Corbetta M. 2011. The dynamical balance of the brain at rest. Neuroscientist. 17:107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zubicaray GI, Chiang MC, McMahon KL, Shattuck DW, Toga AW, Martin NG, Wright MJ, Thompson PM. 2008. Meeting the challenges of neuroimaging genetics. Brain Imaging Behav. 2:258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez I, Erramuzpe A, Escudero I, Mateos B, Cabrera A, Marinazzo D, Sanz-Arigita EJ, Stramaglia S, Cortes Diaz JM. 2015. Information flow between resting-state networks. Brain Connect. 5:554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Wu Y, Fossella JA, Posner MI. 2001. Assessing the heritability of attentional networks. BMC Neurosci. 2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. 2012. Freesurfer. Neuroimage. 62:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Liu A, Dale AM. 2001. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging. 20:70–80. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. 1999. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 9:195–207. [DOI] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Bassett DS, Meunier D, Ellison-Wright I, Yucel M, Wood SJ, Shaw K, O'Connor J, Nertney D et al. 2011. Genetic influences on cost-efficient organization of human cortical functional networks. J Neurosci. 31:3261–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. 2006. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc Natl Acad Sci USA. 103:10046–10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 8:700–711. [DOI] [PubMed] [Google Scholar]
- Fulker DW, Jinks JL. 1970. Comparative analyses of twin data. Acta Genet Med Gemellol (Roma). 19:140. [DOI] [PubMed] [Google Scholar]
- Gandhi TK, Ganesh S, Sinha P. 2014. Improvement in spatial imagery following sight onset late in childhood. Psychol Sci. 25:693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Elton A, Zhu H, Alcauter S, Smith JK, Gilmore JH, Lin W. 2014. Intersubject variability of and genetic effects on the brain's functional connectivity during infancy. J Neurosci. 34:11288–11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett DD, Kovacevic N, McIntosh AR, Grady CL. 2010. Blood oxygen level-dependent signal variability is more than just noise. J Neurosci. 30:4914–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AW, Nelson SM, McDermott KB. 2015. A parietal memory network revealed by multiple MRI methods. Trends Cogn Sci. 19:534–543. [DOI] [PubMed] [Google Scholar]
- Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, Curran JC, Olvera RL, Laird AR, Smith SM et al. 2010. Genetic control over the resting brain. Proc Natl Acad Sci USA. 107:1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood PM, Parasuraman R, Haxby JV. 1993. Changes in visuospatial attention over the adult lifespan. Neuropsychologia. 31:471–485. [DOI] [PubMed] [Google Scholar]
- Greve DN, Fischl B. 2009. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 48:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann CF, Robertson NJ, Leung WC, Chong KW, Chitty LS. 2008. Foetal brain imaging: Ultrasound or mri. A comparison between magnetic resonance imaging and a dedicated multidisciplinary neurosonographic opinion. Acta Paediatr. 97:414–419. [DOI] [PubMed] [Google Scholar]
- Hanakawa T, Immisch I, Toma K, Dimyan MA, Van Gelderen P, Hallett M. 2003. Functional properties of brain areas associated with motor execution and imagery. J Neurophysiol. 89:989–1002. [DOI] [PubMed] [Google Scholar]
- Hasler G, Northoff G. 2011. Discovering imaging endophenotypes for major depression. Mol Psychiatry. 16:604–619. [DOI] [PubMed] [Google Scholar]
- Henson RNA, Rugg MD, Shallice T, Josephs O, Dolan RJ. 1999. Recollection and familiarity in recognition memory: an event-related functional magnetic resonance imaging study. J Neurosci. 19:3962–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri MJ, Pearlson GD, Stevens M, Calhoun VD. 2008. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. Neuroimage. 39:1666–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Kochunov PV, Sprooten E, Mandl RC, Nichols TE, Almasy L, Blangero J, Brouwer RM, Curran JE, de Zubicaray GI et al. 2013. Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: a pilot project of the enigma-dti working group. Neuroimage. 81:455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. 2012. FSL. Neuroimage. 62:782–790. [DOI] [PubMed] [Google Scholar]
- Jiang L, Xu T, He Y, Hou XH, Wang J, Cao XY, Wei GX, Yang Z, He Y, Zuo XN. 2015. Toward neurobiological characterization of functional homogeneity in the human cortex: regional variation, morphological association and functional covariance network organization. Brain Struct Funct. 220:2485–2507. [DOI] [PubMed] [Google Scholar]
- Jiang L, Zuo XN. 2015. Regional homogeneity: a multi-modal, multi-scale neuroimaging marker of the human connectome. Neuroscientist. doi:10.1177/1073858415595004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Machulda MM, Vemuri P, McDade EM, Zeng G, Senjem ML, Gunter JL, Przybelski SA, Avula RT, Knopman DS et al. 2011. Age-related changes in the default mode network are more advanced in Alzheimer disease. Neurology. 77:1524–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T, Feng G. 2015. Modeling psychiatric disorders for developing effective treatments. Nat Med. 21:979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneoke Y, Donishi T, Iwatani J, Ukai S, Shinosaki K, Terada M. 2012. Variance and autocorrelation of the spontaneous slow brain activity. PLoS One. 7:e38131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaymaz N, van Os J. 2009. Heritability of structural brain traits an endophenotype approach to deconstruct schizophrenia. Int Rev Neurobiol. 89:85–130. [DOI] [PubMed] [Google Scholar]
- Kelly C, Biswal BB, Craddock RC, Castellanos FX, Milham MP. 2012. Characterizing variation in the functional connectome: promise and pitfalls. Trends Cogn Sci. 16:181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmer PB, Guo Y, Wang Y, Pagnoni G. 2015. Network-based characterization of brain functional connectivity in Zen practitioners. Front Psychol. 6:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Lee JH. 2011. Are posterior default-mode networks more robust than anterior default-mode networks? Evidence from resting-state fMRI data analysis. Neurosci Lett. 498:57–62. [DOI] [PubMed] [Google Scholar]
- Kircher TTJ, Senior C, Phillips ML, Benson PJ, Bullmore ET, Brammer M, Simmons A, Williams SC, Bartels M, David AS. 2000. Towards a functional neuroanatomy of self-processing: effects of faces and words. Cogn Brain Res. 10:133–144. [DOI] [PubMed] [Google Scholar]
- Klingberg T. 2010. Training and plasticity of working memory. Trends Cogn Sci. 14:317–324. [DOI] [PubMed] [Google Scholar]
- Knyazev G. 2012. Extraversion and anterior vs. posterior DMN activity during self-referential thoughts. Front Hum Neurosci. 6:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W, Teipel S, Mueller S, Buerger K, Bokde AL, Hampel H, Coates U, Reiser M, Meindl T. 2010. Effects of aging on default mode network activity in resting state fMRI: does the method of analysis matter? Neuroimage. 51:280–287. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Lancaster J, Winkler A, Kent JW Jr, Olvera RL, Cole SA, Dyer TD, Almasy L, Duggirala R et al. 2010. Whole brain and regional hyperintense white matter volume and blood pressure: overlap of genetic loci produced by bivariate, whole-genome linkage analyses. Stroke. 41:2137–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Nichols TE, Winkler AM, Hong EL, Holcomb HH, Stein JL, Thompson PM, Curran JE, Carless MA et al. 2011. Genetic analysis of cortical thickness and fractional anisotropy of water diffusion in the brain. Front Neurosci. 5:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koten JW Jr, Wood G, Hagoort P, Goebel R, Propping P, Willmes K, Boomsma DI. 2009. Genetic contribution to variation in cognitive function: an FMRI study in twins. Science. 323:1737–1740. [DOI] [PubMed] [Google Scholar]
- Kyathanahally SP, Jia H, Pustovyy OM, Waggoner P, Beyers R, Schumacher J, Barrett J, Morrison EE, Salibi N, Denney TS et al. 2015. Anterior-posterior dissociation of the default mode network in dogs. Brain Struct Funct. 220:1063–1076. [DOI] [PubMed] [Google Scholar]
- Laird AR, Eickhoff SB, Rottschy C, Bzdok D, Ray KL, Fox PT. 2013. Networks of task co-activations. Neuroimage. 80:505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Eickhoff SB, Turner JA, Ray KL, McKay DR, Glahn DC, Beckmann CF, Smith SM, Fox PT. 2011. Behavioral interpretations of intrinsic connectivity networks. J Cogn Neurosci. 23:4022–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, Kendler KS, Evans AC, Giedd JN. 2009. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Hum Brain Mapp. 30:163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makin TR, Filippini N, Duff EP, Henderson Slater D, Tracey I, Johansen-Berg H. 2015. Network-level reorganisation of functional connectivity following arm amputation. Neuroimage. 114:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Vincent JL, Kelly C, Lohmann G, Uddin LQ, Biswal BB, Villringer A, Castellanos FX, Milham MP, Petrides M. 2009. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci USA. 106:20069–20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Morrison JH. 2013. The brain on stress: vulnerability and plasticity of the prefrontal cortex over the life course. Neuron. 79:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V. 2011. Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn Sci. 15:483–506. [DOI] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. 2001. An integrative theory of prefrontal cortex function. Annu Rev Neurosci. 24:167–202. [DOI] [PubMed] [Google Scholar]
- Mosing MA, Mellanby J, Martin NG, Wright MJ. 2012. Genetic and environmental influences on analogical and categorical verbal and spatial reasoning in 12-year old twins. Behav Gene. 42:722–731. [DOI] [PubMed] [Google Scholar]
- Neale MC, Cardon LR. 1992. Methodology for genetic studies of twins and families. Dordrecht, the Netherlands: Kluwer Academic. [Google Scholar]
- Nomi JS, Uddin LQ. 2015. Developmental changes in large-scale network connectivity in autism. Neuroimage Clin. 7:732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen PJ, Westerberg H, Klingberg T. 2004. Increased prefrontal and parietal activity after training of working memory. Nat Neurosci. 7:75–79. [DOI] [PubMed] [Google Scholar]
- Park H, Kennedy KM, Rodrigue KM, Hebrank A, Park DC. 2013. An fMRI study of episodic encoding across the lifespan: changes in subsequent memory effects are evident by middle-age. Neuropsychologia. 51:448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Thakkar KN. 2010. “Splitting of the mind” revisited: recent neuroimaging evidence for functional dysconnection in schizophrenia and its relation to symptoms. Am J Psychiatry. 167:366–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penick JE. 1982. Self-perceptions in science, cognitive development, and teaching strategy. J Exp Edu. 51:75–80. [Google Scholar]
- Plomin R, DeFries JC, McClearn GE, McGuffin P. 2000. Behavioral genetics. New York, NY: Worth Publishers. [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL et al. 2011. Functional network organization of the human brain. Neuron. 72:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. 2001. A default mode of brain function. Proc Natl Acad Sci USA. 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijsdijk FV, Sham PC. 2002. Analytic approaches to twin data using structural equation models. Brief Bioinform. 3:119–133. [DOI] [PubMed] [Google Scholar]
- Ruigrok AN, Salimi-Khorshidi G, Lai M-C, Baron-Cohen S, Lombardo MV, Tait RJ, Suckling J. 2014. A meta-analysis of sex differences in human brain structure. Neurosci Behav Rev. 39:34–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanez-Larkin GR, Kuhnen CM, Yoo DJ, Knutson B. 2010. Variability in nucleus accumbens activity mediates age-related suboptimal financial risk taking. J Neurosci. 30:1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachar RJ, Forget-Dubois N, Dionne G, Boivin M, Robaey P. 2011. Heritability of response inhibition in children. J Int Neuropsych Soc. 17:238–247. [DOI] [PubMed] [Google Scholar]
- Segonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B. 2004. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 22:1060–1075. [DOI] [PubMed] [Google Scholar]
- Segonne F, Pacheco J, Fischl B. 2007. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans Med Imaging. 26:518–529. [DOI] [PubMed] [Google Scholar]
- Silberg JL, Heath AC, Kessler R, Neale MC, Meyer JM, Eaves LJ, Kendler KS. 1990. Genetic and environmental effects on self-reported depressive symptoms in a general population twin sample. J Psychiatr Res. 24:197–212. [DOI] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR et al. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci USA. 106:13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderqvist S, Bergman Nutley S, Peyrard-Janvid M, Matsson H, Humphreys K, Kere J, Klingberg T. 2012. Dopamine, working memory, and training induced plasticity: implications for developmental research. Dev Psychol. 48:836–843. [DOI] [PubMed] [Google Scholar]
- Sporns O. 2013. Network attributes for segregation and integration in the human brain. Curr Opin Neurobiol. 23:162–171. [DOI] [PubMed] [Google Scholar]
- Steffener J, Habeck CG, Stern Y. 2012. Age-related changes in task related functional network connectivity. PLoS One. 7:e44421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Friston KJ, Frith CD. 2009. Dysconnection in schizophrenia: from abnormal synaptic plasticity to failures of self-monitoring. Schizophr Bull. 35:509–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND. 2010. Functional connectivity density mapping. Proc Natl Acad Sci USA. 107:9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Kahn RS. 2011. Abnormal brain wiring as a pathogenetic mechanism in schizophrenia. Biol Psychiatry. 70:1107–1108. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. 2013. Network hubs in the human brain. Trends Cogn Sci. 17:683–696. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, van Soelen IL, Stam CJ, Kahn RS, Boomsma DI, Hulshoff Pol HE. 2013. Genetic control of functional brain network efficiency in children. Eur Neuropsychopharmacol. 23:19–23. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, The WU-Minn HCP Consortium. 2013. The wu-minn human connectome project: an overview. Neuroimage. 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MJ, Martin NG. 2004. Brisbane adolescent twin study: Outline of study methods and research projects. Aust J Psychol. 56:65–78. [Google Scholar]
- Xing XX, Zhou YL, Adelstein JS, Zuo XN. 2011. Pde-based spatial smoothing: a practical demonstration of impacts on MRI brain extraction, tissue segmentation and registration. Magn Reson Imaging. 29:731–738. [DOI] [PubMed] [Google Scholar]
- Xu T, Yang Z, Jiang L, Xing XX, Zuo XN. 2015. A connectome computation system for discovery science of brain. Sci Bull. 60:86–95. [Google Scholar]
- Yang Z, Chang C, Xu T, Jiang L, Handwerker DA, Castellanos FX, Milham MP, Bandettini PA, Zuo XN. 2014. Connectivity trajectory across lifespan differentiates the precuneus from the default network. Neuroimage. 89:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, LaConte S, Weng X, Hu X. 2008. Ranking and averaging independent component analysis by reproducibility (RAICAR). Hum Brain Mapp. 29:711–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Xu Y, Xu T, Hoy CW, Handwerker DA, Chen G, Northoff G, Zuo XN, Bandettini PA. 2014. Brain network informed subject community detection in early-onset schizophrenia. Sci Rep. 4:5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zuo XN, Wang P, Li Z, LaConte SM, Bandettini PA, Hu XP. 2012. Generalized RAICAR: discover homogeneous subject (sub)groups by reproducibility of their intrinsic connectivity networks. Neuroimage. 63:403–414. [DOI] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Raichle ME. 2010. Disease and the brain's dark energy. Nat Rev Neurol. 6:15–28. [DOI] [PubMed] [Google Scholar]
- Zhang S, Li CS. 2012. Functional connectivity mapping of the human precuneus by resting state fmri. Neuroimage. 59:3548–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J, Breitner JC, Buckner RL, Calhoun VD, Castellanos FX et al. 2014. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data. 1:140049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, Milham MP. 2012. Network centrality in the human functional connectome. Cereb Cortex. 22:1862–1875. [DOI] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Adelstein JS, Klein DF, Castellanos FX, Milham MP. 2010. Reliable intrinsic connectivity networks: test-retest evaluation using ICA and dual regression approach. Neuroimage. 49:2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Xing XX. 2011. Effects of non-local diffusion on structural mri preprocessing and default network mapping: statistical comparisons with isotropic/anisotropic diffusion. PLoS One. 6:e26703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Xing XX. 2014. Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: a systems neuroscience perspective. Neurosci Biobehav Rev. 45:100–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.