Abstract
Studies in mice genetically lacking natural killer T (NKT) cells show that these lymphocytes make important contributions to both innate and adaptive immune responses. However, the usefulness of murine models to study human NKT cells is limited by the many differences between mice and humans, including that their NKT cell frequencies, subsets and distribution are dissimilar. A more suitable model may be swine that share many metabolic, physiological and growth characteristics with humans and are also similar for NKT cells. Thus, we analyzed genetically modified pigs made deficient for CD1d that is required for the development of Type I invariant NKT (iNKT) cells that express a semi-invariant T cell receptor (TCR) and Type II NKT cells that use variable TCRs. Peripheral blood analyzed by flow cytometry and interferon-γ (IFNγ) enzyme-linked immuno spot (ELISPOT) assays demonstrated that CD1d-knockout pigs completely lack iNKT cells while other leukocyte populations remain intact. CD1d and NKT cells have been shown to be involved in shaping the composition of the commensal microbiota in mice. Therefore, we also compared the fecal microbiota profile between pigs expressing and lacking NKT cells. However, no differences were found between pigs lacking or expressing CD1d. Our results are the first to show that knocking-out CD1d prevents the development of iNKT cells in a non-rodent species. CD1d-deficient pigs should offer a useful model to more accurately determine the contribution of NKT cells for human immune responses. They also have potential for understanding how NKT cells impact the health of commercial swine.
Introduction
CD1 molecules are a family of highly conserved antigen presenting glycoproteins that present lipid antigens to CD1-restricted T cells. In humans, the CD1 family is comprised of five members (CD1a-e) encoded by CD1A-E (Park and Bendelac 2000). Of these, CD1d has been the subject of much interest following the discovery that the molecule is the only member conserved between mice and humans, although mice express two copies of the CD1d gene (Park and Bendelac 2000). CD1d molecules are predominantly found on hematopoietic cell types where they present lipid antigens to a specialized subset of immunoregulatory T cells known as natural killer T (NKT) cells (Van Kaer et al. 2011). NKT cells are comprised of two main subsets; Type I and Type II. Most Type I NKT cells express a semi-invariant T cell receptor (TCR) and are referred to as invariant NKT (iNKT) cells. They also react to the prototypic antigen α-galactosylceramide (α-GalCer). Type II NKT cells recognize different antigens using an oligoclonal TCR repertoire (Godfrey et al. 2010). Both Type I and Type II NKT cells are capable of profound effects on the innate and adaptive immune system, primarily through their rapid secretion of both pro- and anti-inflammatory cytokines (Kumar and Delovitch 2014).
Mice deficient for either the Jα18 TCR segment or CD1d, which respectively lack Type I and both Type I and Type II NKT cells, have demonstrated that NKT cells have effects that promote as well as suppress a variety of immune responses. In general, murine NKT cells suppress autoimmune responses [reviewed in (Kumar and Delovitch 2014)] and protect against graft-versus-host disease (Pillai et al. 2007), probably through the anti-inflammatory cytokines they secrete, while their pro-inflammatory responses participate in protective immunity against tumors [reviewed in (Robertson et al. 2014)] and a wide range of infectious agents, including viral, bacterial, fungal and parasitic pathogens [reviewed in (Kinjo et al. 2013)]. NKT cells also exacerbate various mouse models of inflammatory disease such as allergic airway reactivity, hepatitis, ischemia-reperfusion injury, colitis, sickle-cell disease and atherosclerosis [reviewed in (Van Kaer et al. 2011)]. Furthermore, CD1d knockout (KO) mice have demonstrated that NKT cells are important for shaping the bacterial colonization of the intestine (Nieuwenhuis et al. 2009) and for the development of medullary thymic epithelial cells that control negative selection of αβ T cells (White et al. 2014). These and other studies support that NKT cells likely contribute to wide range of immune responses in people. However, mouse-based discoveries have been difficult to translate to humans due to considerable differences in NKT cell frequencies, subsets, cytokine secretion profiles and tissue distribution patterns between these two species (Bendelac et al. 2007; Van Kaer et al. 2011). Hence, there is a need for better animal models to establish how NKT cells contribute to immune responses in people. The current manuscript describes our recently generated CD1d KO pigs (Whitworth et al. 2014) that were generated for this purpose.
The advantage of pigs is that porcine and human NKT cells appear far more similar for frequency, tissue distribution and subsets compared to murine NKT cells (Artiaga et al. 2014; Thierry et al. 2012). Furthermore, pigs like humans express all five classes of CD1 molecules, each capable of presenting lipid antigens to CD1-resticted T cells (Eguchi-Ogawa et al. 2007). Therefore, eliminating CD1d is likely to affect the glycolipid response of pigs similarly to humans and differently than mice. CD1d KO pigs will allow the evaluation of NKT cells in a species that closely matches humans for anatomy, physiology, growth and metabolism. Also, adaptive and innate immune cell subsets are highly homologous between pigs and humans, which likely accounts for why both species are susceptible to many of the same infectious agents. This may present opportunities to better define which human pathogens are subject to the immunomodulatory effects of NKT cells. Finally, pigs lacking CD1d could provide important information about how NKT cells participate in immune responses against pathogens that threaten commercial swine.
Materials and methods
Pigs
All animal procedures were performed with an approved University of Missouri and University of Florida Institutional Animal Care and Use (ACUC) protocol. The National Swine Resource and Research Center (NSRRC) generated pigs carrying various targeted disruptions of CD1D using the CRISPR/Cas9 system. Modifications were made to somatic cells followed by somatic cell nuclear transfer or by directly altering in vitro produced zygotes as previously described (Whitworth et al. 2014). The resulting mutants included seven pigs (158-1, 158-4, 158-5, 159-1, 159-4, 166-1, 167-1) with various CD1D gene disruptions in both alleles that are fully described in Whitworth et al. 2014. These are hereafter designated as CD1d KO. For controls, samples were collected at the NSRRC from two pigs respectively heterozygous (166-2) and homozygous (166-3) for non-deleterious CD1D mutations as well as from four CD1D intact pigs (164-3, 164-6, 164-9, 165-1). For analysis of PB NKT cells, additional wild-type animals were sampled from a crossbred herd maintained by the University of Florida’s Animal Sciences department.
Blood collection and preparation
Blood samples were collected from the jugular vein in vacutainer plasma tubes coated with sodium heparin (BD Biosciences, San Jose, CA) according to the recommendations of the United States Department of Agriculture regulations, the National Research Council’s Guide for the Care and Use of Laboratory Animals, as well as all relevant state and federal regulations and policies. Isolation of peripheral blood mononuclear cells (PBMCs) was performed using Ficoll-Paque TM PREMIUM (GE Healthcare Bio-Sciences Corp., Uppsala, Sweden) as previously described (Artiaga et al. 2014). To obtain total blood leukocytes, 200 μl of whole blood was incubated with an ammonium chloride based buffer to lyse red blood cells.
Flow cytometry and reagents
Immune cell populations from total blood leukocytes were characterized by flow cytometry using a BD Accuri C6 flow cytometer. Cell suspensions were Fc receptor blocked with rat IgG from Sigma-Aldrich (Saint Louis, MO) and stained with the indicated fluorochrome-conjugated antibodies for 30 minutes at 4°C (Table 1). To identify iNKT cells, samples stained with anti-CD3ε and α-galactosylceramide (α-GalCer) analog PBS57-loaded mouse CD1d (mCD1d) tetramer reagent were compared to cells stained with anti-CD3ε and unloaded tetramer. αβ T lymphocyte subsets were identified using anti-CD3ε, anti-CD8α, and anti-CD4. γδ T lymphocytes were identified by co-staining leukocytes with anti-CD3ε and anti-TCRδ antibodies. Regulatory T lymphocytes (Tregs) were identified by surface staining with anti-CD3ε and anti-CD4 and intracellular staining with anti-FoxP3 according to the FoxP3 staining kit from eBioscience (San Diego, CA). B-lymphocytes and monocytes were identified according to surface staining with anti-MHC class II (MHCII), anti-CD172α and intracellular staining with anti-CD79α according to BD the Cytofix/Cytoperm kit from BD Biosciences (San Jose, CA). Cells identified as MHCII+CD172−CD79α+ and MHCII+CD172+CD79α− were respectively classified as B cells and monocytes. Granulocytes were distinguished based on size and complexity of this cell population after gating on total leukocytes. Data were analyzed using FlowJo software (Treestar, Palo Alto, CA).
Table 1.
Reagents used for flow cytometry analysis of surface markers and intracellular FoxP3 and CD79α
| Antigen | Clone | Isotype | Fluorochrome conjugation |
Source |
|---|---|---|---|---|
| CD1d tetramer | N/A | N/A | PE | NIH Tetramer Core Facility |
| Anti-CD3ε | BB23-8E6-8C8 | Mouse IgG2aκ | FITC/PE | BD Biosciences |
| Anti-CD8 | 76-2-11 | Mouse IgG2aκ | PE | BD Biosciences |
| Anti-CD4 | 74-12-4 | Mouse IgG2bκ | Alexa647 | Southern Biotech |
| Anti-FoxP3 | FJK-16s | Rat IgG2aκ | PE | eBioscience |
| Anti-TCRδ | PGBL22A | Mouse IgG1 | Alexa647 | Washington State University |
| Anti-CD172α | 74-22-15A | Mouse IgG2bκ | Alexa488 | BD Biosciences |
| Anti-CD79α | HM47 | Mouse IgG1κ | PE | BD Biosciences |
| Anti-MHC class II | H42A | Mouse IgG2a | Alexa647 | Accurate |
IFNγ enzyme-linked immunosorbent spot assays
PBMCs were plated in triplicate at 0.5 and 1×106 cells per well into MultiScreen HTS plates (Millipore, Billerica, MA) pre-coated with purified anti-IFNγ (P2G10, BD Biosciences) with or without 1 μg/ml α-GalCer from Toronto Research Chemicals (Toronto, ON, Canada). α-GalCer stock solutions were prepared as previously described (Artiaga et al. 2014). Plates were incubated for 3 days at 5% CO2 and 37°C after which IFNγ production was detected using biotin conjugated anti-IFNγ (P2C11) detection antibody, streptavidin-HRP and the BD Cytokine ELISPOT AEC substrate set all supplied by BD Biosciences. After development, spots were read using an automated ELISPOT reader (AID EliSpot High-Resolution Reader System ELHR03). Data are presented as mean number of spots ± SEM/105 viable cells in each of the α-GalCer-stimulated triplicates after subtracting spots counted in unstimulated wells.
Profiling of the fecal bacterial community by 16S rDNA sequencing
To determine the composition of the fecal bacterial community by 16S rDNA sequencing, individual fecal samples were collected from the seven CD1d KO pigs and three control pigs (165-1, 166-2, 166-3) at 6–8 months of age. Fecal samples were frozen immediately after sampling and stored at −80°C. DNA was extracted from samples using the RBB+C method of Yu and Morrison (2004). Briefly, 0.25 g of freshly-thawed samples were subjected to 2 rounds of bead beating in the presence of SDS. Nucleic acids were precipitated by ammonium acetate and isopropanol. Protein and RNA were removed with proteinase K and RNase A. DNA was purified using the columns from the QIAGEN DNA Mini Stool Kit (QIAGEN, Valencia, CA). The amount of DNA was determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Integrity and size of DNA was assessed by agarose (0.8%) gel electrophoresis. All DNA samples were stored at −20°C until further processing.
PCR amplification and MiSeq sequencing were performed by MR DNA (Shallowater, TX, USA). The V1-V3 region of the 16S rRNA gene was PCR amplified using 27F/519R primers (Lane 1991) and a HotStarTaq Plus Master Mix Kit (Qiagen, USA). Forward primers included an 8-bp barcode for sample identification. Amplification included initial denaturation at 94°C for 3 min; 28 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 40 s, and extension at 72°C for 1 min; and a final extension step at 72°C for 5 min. PCR products were assessed by agarose (2%) gel electrophoresis. Samples were pooled in equal proportions based on molecular weight and DNA concentration and then purified using an Agencourt AMPure XP kit (Beckman Coulter, Brea, CA, USA). The MiSeq v. 3 sequencing platform (Illumina, Inc., San Diego, CA, USA) was used to generate 300 bp paired-end reads. Sequences were trimmed and joined using the MR DNA analysis pipeline.
Quantitative Insights Into Microbial Ecology (QIIME; v. 1.8.0) was used for further sequence processing and analysis (Caporaso et al. 2010). Following demultiplexing and primary quality-filtering, operational-taxonomic units (OTUs) were picked using the pick_open_reference_otus.py script. Using this open-reference OTU picking protocol, sequences were clustered at 97% similarity against the GreenGenes reference database [release 13_5; (DeSantis et al. 2006)], and sequences that did not cluster against the reference database were clustered de novo. Chimeras were removed with usearch [v. 6.1.544 (Edgar 2010)] with the usearch61 option. Taxonomy was assigned using the RDP classifier (Wang et al. 2007). We applied secondary quality-filtering with an OTU threshold of 0.005% with the filter_otus_from_otu_table.py script (Bokulich et al. 2013). The core_diversity_analyses.py script was used to compare relative abundance of OTUs, compute the phylogenetic diversity metric of α-diversity, and build principal coordinate analysis (PCoA) plots of unweighted UniFrac distances (Lozupone and Knight 2005).
Statistical analysis
The flow cytometry and ELISPOT assay data were analyzed using a nonparametric Wilcoxon U test from GraphPad Prism, version 6.0d for Macintosh (GraphPad Software, Inc., La Jolla, CA). Relative abundance of OTUs was compared using ANOVA with false-discovery rate control for multiple comparisons. The phylogenetic diversity metric was compared across treatments at highest rarefaction depth using a t-test. The within-treatment UniFrac distances and between-treatment UniFrac distances were compared with a nonparametric Monte Carlo test (no Bonferroni correction applied). Data was considered significant when p- or q-value < 0.05.
Results
CD1d-deficient pigs lack iNKT cells
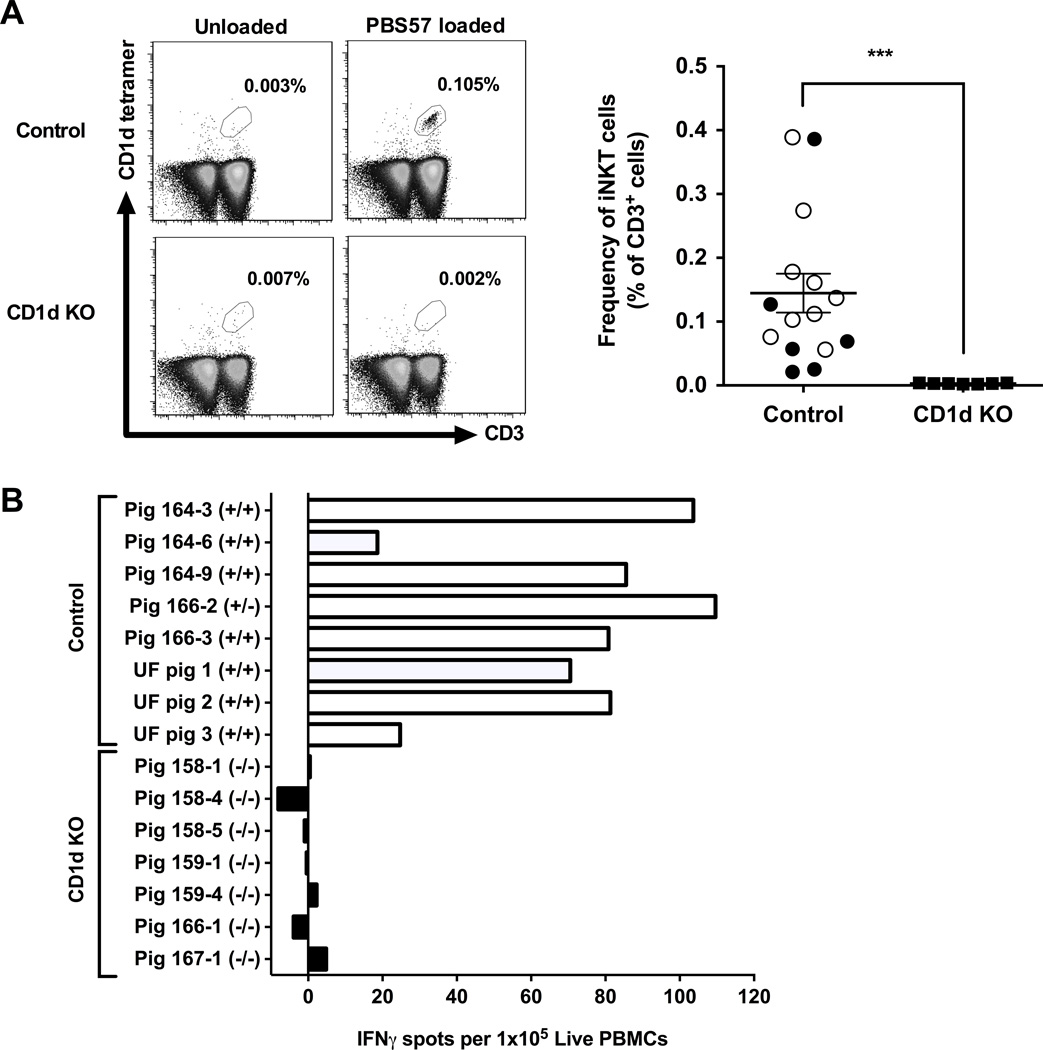
To determine if pigs lacking CD1d fail to develop NKT cells, flow cytometry was used to compare peripheral blood (PB) between seven CD1d KO pigs and 15 control pigs from the NSRCC and UF using anti-CD3ε antibody and PBS57-loaded mouse CD1d-tetramer+ that detects porcine Type I iNKT cells (Artiaga et al. 2014). Additional samples were stained with unloaded CD1d tetramer to determine the extent of non-specific tetramer binding (Fig 1A). No iNKT cells were detected in CD1d KO pigs, while iNKT cell frequencies in control pigs from the NSRRC (0.11% ± 0.06) and UF (0.17% ± 0.04) were similar to what has previously been reported for swine (Artiaga et al. 2014; Thierry et al. 2012) (Fig 1A).
Figure 1.
Comparison of iNKT cells between CD1d KO and control pigs. (A) Frequency of peripheral blood iNKT cells. Left panel describes the gating strategy to identify iNKT cells. Single cell suspensions were membrane-labeled with anti-CD3ε mAb and PBS57-loaded CD1d tetramer. Some samples were also labeled with an unloaded CD1d tetramer to validate the staining of PBS57-loaded CD1d tetramer. Right panel describes the frequency of iNKT cells as a proportion of CD3+ cells. For controls, samples were collected from 6 NSRRC pigs (closed circles) and 9 UF pigs (open circles) (B) iNKT cell responses to α-GalCer as measured by IFNγ-ELISPOT assay in which PBMCs were incubated for 72 h with 1 μg/ml α-GalCer. Results represent mean IFNγ spots per 1×105 PBMCs after subtracting spots in unstimulated samples.
We also assessed pigs for iNKT cells by analyzing their blood for the presence of α-GalCer-reactive cells. For this, IFNγ-ELISPOT assays were performed by incubating PBMCs from each pig for 3 days in the presence or absence of α-GalCer. Frequencies of α-GalCer-responsive cells for each animal was determined after subtracting spots counted in unstimulated wells. Responses to α-GalCer were detected for all control pigs. In contrast, no IFNγ spots were detected above baseline levels for any of the CD1d KO pigs (Fig 1B). Together, these results indicate that pigs genetically lacking CD1D fail to develop iNKT cells. To determine if other immune cells were affected by the ablation of CD1D, PB from the CD1d KO and NSRRC control pigs were compared by flow cytometry for various leukocyte populations, including conventional, regulatory and γδ T lymphocytes, B lymphocytes, monocytes and granulocytes (Table 2). No differences were found in any of the non-iNKT cell populations analyzed. These results suggest that pigs lacking CD1d are deficient for iNKT cells but not other immune cell populations.
Table 2.
Flow cytometric analysis of cell surface proteins from peripheral blood leukocytes obtained from CD1d KO and control pigs
| Immune cell population | CD1d KO | Control | p-value |
|---|---|---|---|
| CD3+ T cells | 83.94 ± 2.06 | 79.03 ± 1.53 | NS |
| CD8+ T cells (% of CD3+) | 30.93 ± 2.77 | 28.93 ± 2.39 | NS |
| CD4+ T cells (% of CD3+) | 16.09 ± 1.69 | 13.25 ± 4.40 | NS |
| CD4+CD8+ T cells (% of CD3+) | 10.34 ± 0.86 | 11.95 ± 1.71 | NS |
| FoxP3+ T cells (% of CD4+) | 8.68 ± 0.53 | 7.35 ± 0.34 | NS |
| (FoxP3 MFI) | 387.86 ± 12.05 | 406 ± 21.39 | NS |
| γδ T cells (% of CD3+) | 55.14 ± 3.01 | 56.70 ± 5.76 | NS |
| B cells | 7.34 ± 0.45 | 8.50 ± 0.38 | NS |
| Monocytes | 32.11 ± 1.78 | 32.57 ± 5.00 | NS |
| Granulocytes | 29.63 ± 1.45 | 28.77 ± 3.59 | NS |
Table values represent the variable mean ± SEM for viable cells. Seven CD1d KO and three control pigs were compared at 6–8 months of age. NS indicates no significant difference (p ≥ 0.05) between CD1d KO and control pigs when analyzed by the Wilcoxon U test. MFI indicates median fluorescence intensity.
Composition of fecal microbiota is unaffected by CD1d-deficiency
From fecal samples of seven CD1d KO and three control pigs, we identified 962 OTUs from 512,662 quality-filtered 16S rDNA sequences. There were no differences in abundance of OTUs across CD1d KO and control pigs (q = 0.815). No differences were apparent for other taxonomic levels (phylum, class, order, family, genus; data not shown).
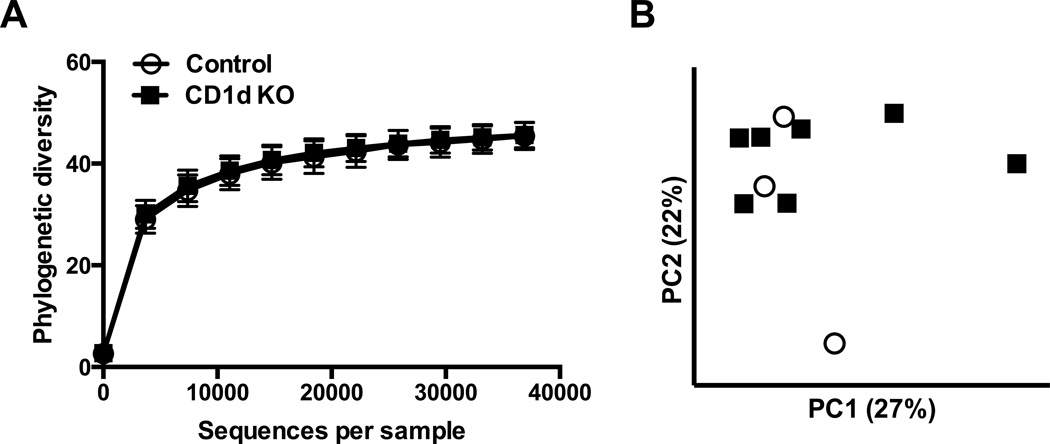
The within-sample (α) diversity was similar between CD1d KO and control pigs when measured using the phylogenetic diversity metric. This was apparent both visually from the rarefaction plot (Fig. 2A) and statistically when tested at the highest rarefaction level (p=0.916). When between-sample (β) diversity was measured using unweighted UniFrac distances and visualized with PCoA plots, samples did not appear to group by CD1d KO or control treatment (Fig. 2B). In support of this visual finding, within-treatment distances and between-treatment distances did not differ (p = 0.297). Results were similar when using weighted UniFrac distances (data not shown). Together, these analyses reveal no pronounced compositional differences in the bacterial communities across CD1d KO and control pigs, though our conclusions must be qualified by our small sample size.
Figure 2.
Profiling of the fecal bacterial community of CD1d KO and control pigs by 16S rDNA sequencing. (A) Rarefaction plot for phylogenetic diversity metric of α-diversity for bacterial communities of fecal samples from CD1d KO (n = 7) and control pigs (n = 3). The highest rarefaction level corresponds to the number of sequences of the sample with fewest sequences. Values are mean ± SEM. (B) Principal coordinates analysis (PCoA) plot of unweighted UniFrac distances between bacterial communities of fecal samples from CD1d KO and control pigs.
Discussion
Our results demonstrate that porcine iNKT cells require CD1d for their development. However, CD1d-deficiency in pigs did not appear to affect other immune cell populations, which is consistent with what has been reported for CD1d and Jα18 deficient mice (Cui et al. 1997; Mendiratta et al. 1997). We also compared the fecal microbiota profile between pigs expressing and lacking NKT cells as CD1d and NKT cells have been shown to be involved in shaping the composition of the commensal microbiota in mice. For example, CD1d deficiency is associated with increased quantities of adherent bacteria as a consequence of defects in CD1d-mediated release of antimicrobial peptides from small intestinal Paneth cells (Nieuwenhuis et al. 2009). We did not detect any differences in the composition of fecal bacterial by 16S rDNA sequencing for pigs. Notably, fecal samples did not group by CD1d KO or control treatments in PCoA plots of UniFrac distances. However, in a similar analysis but with murine fecal samples, Nieuwenhuis et al. (2009) found that samples grouped strongly by treatment, even with a modest sample size (n = 5 each for KO and WT mice). Future experiments should analyze the community composition of the small intestine, not just the feces, because KO mice were more susceptible to pathogen colonization in the small intestine (Nieuwenhuis et al. 2009).
In conclusion, CD1d-deficient pigs may offer an important tool with which to establish how NKT cells contribute to immune responses in a species more biologically similar to humans than mice. This model may be particularly useful for determining the significance of NKT cells for microbial immunity in a species susceptible to many of the same pathogens as humans that also share human NKT cell characteristics. Additional applications could include using CD1d knockout pigs to understand the role of NKT cells in tumor immunity, transplantation tolerance, inflammatory diseases and autoimmunity. Finally, CD1d-deficient pigs may provide useful information about the importance of NKT cells for the immune health of commercial pigs.
Acknowledgments
This work was supported by the National Swine Resource and Research Center, UF Research Opportunity Seed Fund and IFAS Early Career Seed Fund awards. The National Institutes of Health Tetramer Core Facility provided the CD1d tetramers. G.Y is supported by a UF Animal Molecular and Cellular Biology graduate fellowship.
References
- Artiaga BL, Whitener RL, Staples CR, Driver JP. Adjuvant effects of therapeutic glycolipids administered to a cohort of NKT cell-diverse pigs. Vet Immunol Immunopathol. 2014;162:1–13. doi: 10.1016/j.vetimm.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annual review of immunology. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Eguchi-Ogawa T, Morozumi T, Tanaka M, Shinkai H, Okumura N, Suzuki K, Awata T, Uenishi H. Analysis of the genomic structure of the porcine CD1 gene cluster. Genomics. 2007;89:248–261. doi: 10.1016/j.ygeno.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, Kitano N, Kronenberg M. The role of invariant natural killer T cells in microbial immunity. J Infect Chemother. 2013;19:560–570. doi: 10.1007/s10156-013-0638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Delovitch TL. Different subsets of natural killer T cells may vary in their roles in health and disease. Immunology. 2014;142:321–336. doi: 10.1111/imm.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997;6:469–477. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis EE, Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, Simons-Oosterhuis Y, Brugman S, Yamaguchi K, Ishikawa H, Aiba Y, Koga Y, Samsom JN, Oshima K, Kikuchi M, Escher JC, Hattori M, Onderdonk AB, Blumberg RS. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. The Journal of clinical investigation. 2009;119:1241–1250. doi: 10.1172/JCI36509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Bendelac A. CD1-restricted T-cell responses and microbial infection. Nature. 2000;406:788–792. doi: 10.1038/35021233. [DOI] [PubMed] [Google Scholar]
- Pillai AB, George TI, Dutt S, Teo P, Strober S. Host NKT cells can prevent graft-versus-host disease and permit graft antitumor activity after bone marrow transplantation. J Immunol. 2007;178:6242–6251. doi: 10.4049/jimmunol.178.10.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson FC, Berzofsky JA, Terabe M. NKT cell networks in the regulation of tumor immunity. Front Immunol. 2014;5:543. doi: 10.3389/fimmu.2014.00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A, Robin A, Giraud S, Minouflet S, Barra A, Bridoux F, Hauet T, Touchard G, Herbelin A, Gombert JM. Identification of invariant natural killer T cells in porcine peripheral blood. Vet Immunol Immunopathol. 2012;149:272–279. doi: 10.1016/j.vetimm.2012.06.023. [DOI] [PubMed] [Google Scholar]
- Van Kaer L, Parekh VV, Wu L. Invariant natural killer T cells: bridging innate and adaptive immunity. Cell Tissue Res. 2011;343:43–55. doi: 10.1007/s00441-010-1023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AJ, Jenkinson WE, Cowan JE, Parnell SM, Bacon A, Jones ND, Jenkinson EJ, Anderson G. An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. J Immunol. 2014;192:2659–2666. doi: 10.4049/jimmunol.1303057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, Samuel MS, Mao J, O'Gorman C, Walters EM, Murphy CN, Driver J, Mileham A, McLaren D, Wells KD, Prather RS. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014;91:78. doi: 10.1095/biolreprod.114.121723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques. 2004;36:808–812. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]