Klebsiella pneumoniae ST258 is the dominant host of KPC enzymes in the UK outside of North-West England and, as multiple UK hospitals have encountered this clone since 2007, there is a need for continued surveillance and implementation of rigorous infection prevention and control measures.
Abstract
Objectives
Klebsiella pneumoniae carbapenemases (KPCs) have been increasingly reported in the UK since 2003. We analysed patient and isolate data for KPC-positive bacteria confirmed by the national reference laboratory from UK laboratories from August 2003 to August 2014, excluding North-West England, where the epidemiology has previously been studied.
Methods
MICs were determined by BSAC agar dilution. Carbapenem-resistant isolates lacking imipenem/EDTA synergy were tested by PCR for blaKPC. MLST and blaKPC sequencing were performed on a subset of isolates. Plasmid analysis was performed by transformation, PCR-based replicon typing and, in some cases, whole-plasmid sequencing. Patient data provided by the sending laboratories were reviewed.
Results
Two hundred and ten isolates with KPC enzymes were submitted from 71 UK laboratories outside North-West England, representing 160 patients. All were Enterobacteriaceae, predominantly K. pneumoniae (82%; 173/210), and most (91%; 191/210) were from hospitalized patients. Analysis of 100 isolates identified blaKPC-2 (62%), blaKPC-3 (30%) and blaKPC-4 (8%). Clonal group (CG) 258 was dominant among K. pneumoniae (64%; 54/84), but 21 unrelated STs were also identified. Plasmid analysis identified a diverse range of plasmids representing >11 different replicon types and found in multiple STs and species. Most (34/35) plasmids with IncFIB/FIIK replicons exhibited >99% sequence identity to pKpQIL.
Conclusions
KPC enzymes are increasingly detected in Enterobacteriaceae in the UK, albeit without the major outbreaks seen in North-West England. K. pneumoniae CG258 are the dominant hosts, but plasmid spread plays a major role in KPC dissemination between other K. pneumoniae STs and enterobacterial species.
Introduction
Klebsiella pneumoniae carbapenemases (KPCs) were first identified in 1996 in a K. pneumoniae isolate obtained from a patient hospitalized in North Carolina, USA.1 Since then, they have disseminated globally, predominantly among Enterobacteriaceae, although there are also reports of production by Acinetobacter spp. and Pseudomonas aeruginosa isolates in the Americas.2–4 They have been reported in all inhabited continents, with numerous outbreaks described, particularly in Greece, Israel, Italy, the USA and China.4 Many of these outbreaks are associated with an internationally disseminated lineage of K. pneumoniae, ST258, and other members of its clonal group (CG) CG258, which comprises ST258, along with its single-locus variants (SLVs), such as ST512, and their SLVs.5 KPC enzymes are typically encoded within the Tn3-based transposon Tn4401, which has five known isoforms (a, b, c, d and e) as defined by insertions or deletions within a polymorphic region immediately upstream of blaKPC.6 The first fully sequenced plasmid encoding a KPC enzyme was an IncFIB/IncFIIK replicon type designated pKpQIL from an ST258 isolate in Israel; highly similar plasmids have since been found in several other countries.7–9 blaKPC has also less frequently been reported to be carried by plasmids of other replicon types including IncI2, IncN, IncL/M and IncX.10–12
Most bacteria with KPC enzymes are multiresistant, harbouring genes whose products compromise non-β-lactam antibiotics [e.g. aac(6′)-Ib, encoding resistance to aminoglycosides except gentamicin],13,14 resulting in a paucity of treatment options. Members of the K. pneumoniae ST258 lineage usually remain susceptible only to colistin, gentamicin and tigecycline, but there have been documented outbreaks of colistin-resistant K. pneumoniae ST258, further reducing therapeutic options, and such variants have become nationally disseminated in Italy.14,15
The first KPC enzyme identified in the UK was a KPC-4, found in 2003 in an Enterobacter cloacae complex blood culture isolate from Scotland,16 whilst the first K. pneumoniae ST258 KPC isolate was found in a urine specimen in 2007, also in Scotland.17 Since then, numbers of isolates with KPC enzymes referred to PHE's Antimicrobial Resistance and Healthcare Associated Infections (AMRHAI) Reference Unit have risen sharply.4 Most (>95%) originate from hospitals in North-West England (defined as the counties of Cheshire, Cumbria, Greater Manchester, Lancashire and Merseyside), where an outbreak, centred in Manchester, has been ongoing since 2010 despite control efforts.4,18 In contrast with most international experience, this outbreak is polyclonal in nature and attributable to the horizontal spread of a pKpQIL-like plasmid amongst multiple strains of multiple species of Enterobacteriaceae.4,18 Similar polyclonal situations have been described recently in other countries, including Spain and Canada.19,20
Here, we describe the first 210 KPC-positive bacteria referred to AMRHAI from 160 infected or colonized UK patients outside North-West England.
Materials and methods
Bacterial isolates, identification and susceptibility testing
Isolates were submitted to PHE's AMRHAI Reference Unit from laboratories across the UK (excluding North-West England) between August 2003 and August 2014 for investigation of ‘unusual’ resistance, including to carbapenems. They were identified using chromogenic agars [CHROMagar™ Orientation (CHROMagar, Paris, France) and Brilliance UTI (Oxoid, Basingstoke, UK)], API-20E tests (bioMérieux, Marcy-l'Étoile, France) or, since August 2012, MALDI-TOF MS (Bruker Microflex LT, Bruker Daltonik, Bremen, Germany).
MICs were determined by BSAC agar dilution21 using AMRHAI's standard Gram-negative antibiotic panel, which includes ertapenem, meropenem and imipenem (with/without 320 mg/L EDTA to detect metallocarbapenemase producers), and interpreted using EUCAST breakpoints.22
Screening for KPC genes
Isolates exhibiting raised cefotaxime and ceftazidime MICs, with no significant clavulanic acid synergy, and resistance, based on EUCAST criteria, to one or more of imipenem, meropenem and ertapenem, but lacking imipenem/EDTA synergy (≥8-fold potentiation of imipenem by 320 mg/L EDTA), were screened by in-house PCR for blaKPC1 and/or with a commercial microarray (Check-Points CT102, Check-Points, Wageningen, The Netherlands).23
WGS
WGS was performed on 40 clinical isolates and 61 blaKPC transformants (see below), chosen to be geographically representative of AMRHAI submissions. Genomes were sequenced using the Nextera sample preparation method and standard 2 × 251 base sequencing protocols on a MiSeq instrument (Illumina, San Diego, CA, USA). Reads were assembled into contigs using VelvetOptimiser (http://bioinformatics.net.au/software.velvetoptimiser.shtml), with k-mer values from 55 to 75. STs and plasmid replicon types were extracted in silico by BLASTn using reference sequences from http://mlst.warwick.ac.uk/mlst/dbs/Ecoli and http://pubmlst.org/plasmid/ databases. The presence of resistance genes was determined as previously described.24
MLST and sequencing of KPC alleles
A subset of 123 isolates (100 K. pneumoniae, 16 E. cloacae complex, 4 Escherichia coli, 2 Klebsiella oxytoca and 1 Citrobacter freundii), geographically representative of submissions and selected from throughout the study period, were chosen for further analysis. STs were determined for K. pneumoniae, E. cloacae complex and E. coli isolates by traditional MLST25–27 or were inferred from WGS data. blaKPC alleles were defined either by sequencing PCR amplicons as previously described1 or from analysis of WGS data.
Plasmid transformation, replicon typing and plasmid sequencing
Transformation of blaKPC plasmids was attempted by electroporation using the subset of 123 isolates and E. coli α-Select recipient cells (Bioline, London, UK). Transformants were selected on LB agar containing 100 mg/L ampicillin and colonies were screened for blaKPC by PCR. A subgroup of 61 transformants was selected and subjected to WGS as above; selection was based on the geographical and temporal distribution, KPC alleles and the species and STs of their parent strains.
PCR-based replicon typing (PBRT) of blaKPC plasmids was performed as described previously28,29 or was inferred from WGS data.
Patient demographic information
Patient data were obtained from the accompanying request forms sent by referring laboratories. A patient was categorized as ‘new’ if they were found to have KPC-positive isolates detected by AMRHAI for the first time and ‘known’ if KPC-positive isolates, irrespective of species, had previously been identified from the patient by AMRHAI.
Data analysis
Data were analysed using Microsoft Excel and Bionumerics software v6.1 (Applied Maths, Sint-Martens-Latem, Belgium).
Results
Demographics of patients affected and distribution
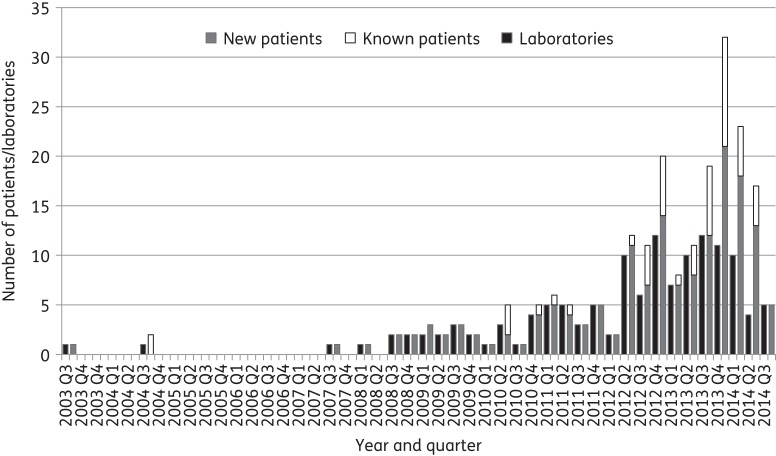
During the study period, AMRHAI confirmed 210 KPC-positive isolates from outside of North-West England. These were submitted by 71 UK laboratories and obtained from 160 patients. Figure 1 illustrates the distribution of these isolates among ‘new’ and ‘known’ patients and among submitting laboratories. The first three KPC-producing organisms, as noted above and previously reported,16 were E. cloacae isolates found in blood specimens from a single patient across consecutive years (one in 2003 and two in 2004) from Scotland; all were found to carry blaKPC-4. The first KPC-producing K. pneumoniae isolate was identified in 2007 in a blood specimen, also from Scotland.17 The numbers of ‘new’ patients increased significantly from 2008 onwards (Figure 1).
Figure 1.
Numbers of new and known affected patients and laboratories sending KPC-positive isolates per quarter during the study period.
KPC-positive isolates were submitted from laboratories across all 11 UK regions. The national distribution of affected patients, as ascertained by AMRHAI referrals from outside North-West England, was as follows: England (n = 124), Scotland (n = 26), Northern Ireland (n = 9) and Wales (n = 1). The greatest number of affected patients was in the Yorkshire and the Humber region (n = 39), followed by London (n = 27) and the West Midlands (n = 20).
Most source patients were hospitalized (86%; 138/160), but a few were outpatients (4%; 6/160), in primary care (6%; 10/160) or in an unknown setting (4%; 6/160). The mean patient age was 60.4 years and most patients were male (58%; 92/160).
Foreign travel history was available for only 51/160 (32%) patients. Of these, 19 patients had documented travel within the previous 6 months to Greece (11/19), Italy (2/19), Bulgaria, Curaçao, India, Israel, Macedonia and Saudi Arabia (1 patient each). Four of these 19, with histories of travel to Curaçao, Greece, Israel and Italy, were known to have been hospitalized whilst abroad. Information on patient transfers between UK regions was very limited, but 5/160 (3%) patients were recognized to have had isolates with KPC enzymes submitted from hospitals across two UK regions. Four KPC-positive patients had previously been hospitalized in North-West England (Manchester) prior to isolations of bacteria with KPC enzymes in Wales, Northern Ireland and Yorkshire and the Humber (two patients) and one KPC-positive patient had previously been hospitalized in Yorkshire and the Humber prior to the East Midlands.
Single KPC-producing isolates were referred from 125/160 (78%) patients and multiple isolates were referred from the remaining 35/160 (22%). Amongst the 35 patients with multiple KPC-producing isolates, 6 (17%) yielded isolates of different species or genera with these enzymes and 14 (40%) had isolates with KPC enzymes from different anatomical sites. The KPC-positive isolates were referred over a period of <14 days in 19/35 (54%) instances and over a period of >6 months from just 1 patient.
The date when the sample was taken was available for 89% (187/210) of isolates and the median duration between this date and the isolate being received at AMRHAI was 8 days (range = 1–49 days).
Microbiology
All KPC-positive isolates were Enterobacteriaceae. The majority were K. pneumoniae (82%; 173/210), followed by E. cloacae complex (11%; 24/210), E. coli (4%; 9/210), K. oxytoca (1%; 3/210) and C. freundii (<1%; 1/210).
If samples, rather than patients, were considered as the denominator, most were taken in hospitals (91%; 191/210), but some were from general practice urines (5%; 11/210) and a few were from an unknown setting (4%; 8/210). The most frequently reported specimen types were urines (33%; 70/210), followed by screening swabs (24%; 51/210). Ten percent (21/210) of isolates were obtained from blood cultures and line tips, 13% (27/210) from tissue and fluid samples and 10% (21/210) from faeces (Table 1).
Table 1.
Sources of the KPC-positive isolates
| Species | Hospital and GP settings |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| urines | screening swabs | blood cultures and line tips | respiratory | tissue and fluid | faeces | not known | GP urines | total | |
| C. freundii | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| E. cloacae complex | 7 | 0 | 4 | 2 | 4 | 2 | 0 | 3 | 22 |
| E. coli | 4 | 1 | 0 | 0 | 0 | 2 | 0 | 1 | 8 |
| Klebsiella spp. | 44 | 47 | 17 | 12 | 22 | 17 | 5 | 7 | 171 |
| Total | 55 | 49 | 21 | 14 | 26 | 21 | 5 | 11 | 202 |
| Species | Unknown setting |
||||||||
| urines | screening swabs | blood cultures and line tips | respiratory | tissue and fluid | faeces | not known | total | ||
| E. cloacae complex | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | |
| E. coli | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Klebsiella spp. | 2 | 1 | 0 | 0 | 1 | 0 | 1 | 5 | |
| Total | 4 | 2 | 0 | 0 | 1 | 0 | 1 | 8 | |
GP, general practice.
KPC alleles and typing of the isolates
One hundred and twenty-three isolates distributed throughout the entire collection period (2003–14) were typed by MLST and their KPC variants were defined (Table 2). After the exclusion of isolates exhibiting the same ST from single patients, 100 isolates remained for analysis, comprising: 84 K. pneumoniae, 12 E. cloacae complex and 4 E. coli. blaKPC-2 was the most prevalent variant found in 62/100 isolates, followed by blaKPC-3 (30/100) and blaKPC-4 (8/100).
Table 2.
MLST and KPC allele characteristics of isolates from 100 patients (one isolate per patient)
| Species | Number of isolates | KPC variant(s) (number) | STs (number if >1) | Number of submitting laboratories | Number of regions |
|---|---|---|---|---|---|
| K. pneumoniae | 84 | KPC-2 (54), KPC-3 (30) | 11 (3), 14, 15 (3), 17, 22, 35, 147 (2), 250, 252, 258 (41), 321, 336, 491, 507, 512 (9), 661 (5), 678, 709, 732, 791, 833, 896, 1026 (2), 1162 (2), 1163 | 65 | 11 |
| E. cloacae complex | 12 | KPC-2 (4), KPC-4 (8) | 56, 133 (2), 171 (8), 190 | 7 | 3 |
| E. coli | 4 | KPC-2 (4) | 12, 127, 131, 744 | 3 | 2 |
Almost two-thirds (64%; 54/84) of K. pneumoniae isolates belonged to CG258 comprising ST258 (n = 41), ST512 (n = 9), ST11 (n = 3) and ST833 (n = 1). Between 2007 and 2014, ST258 isolates were submitted from 31 laboratories across all UK regions studied (10/11) except Wales; 54% (22/41) carried blaKPC-2 and the remaining 46% (19/41) carried blaKPC-3. One of the earliest ST258 isolates was obtained in 2008 from the urine of a patient previously hospitalized in Israel;17 another ST258 isolate was from a wound swab of a patient hospitalized in Greece in 2011. However, the first ST258 isolate, from 2007, came from a patient who had no known foreign travel history. ST512 isolates were referred from six laboratories between 2008 and 2014 from three UK regions and all carried blaKPC-3. One patient with K. pneumoniae ST512 isolated from sputum in 2012 had been hospitalized in Italy within the previous 6 months. ST11 isolates were obtained between 2009 and 2011 from three patients across two UK regions, one of whom had previously been hospitalized in Curaçao and had undergone urinary catheterization.30 There were four outbreaks in hospitals across three regions caused by members of CG258: one involved ST11 isolates carrying blaKPC-2 in two patients; another involved ST512 isolates carrying blaKPC-3 in three patients; another involved ST258 isolates carrying blaKPC-3 in two patients; and the last involved ST258 isolates carrying blaKPC-2 in three patients. These outbreaks persisted over periods ranging from 1 week to 2 months.
Thirty (36%) characterized K. pneumoniae isolates (of those typed) did not belong to CG258. Of these, five isolates belonged to ST661, were submitted from two laboratories located in different regions between July 2013 and May 2014 and carried blaKPC-2. The remaining 25 isolates, from 16 laboratories, represented 20 different STs and carried either blaKPC-2 (24/25) or blaKPC-3 (1/25) (Figure 2).
Figure 2.
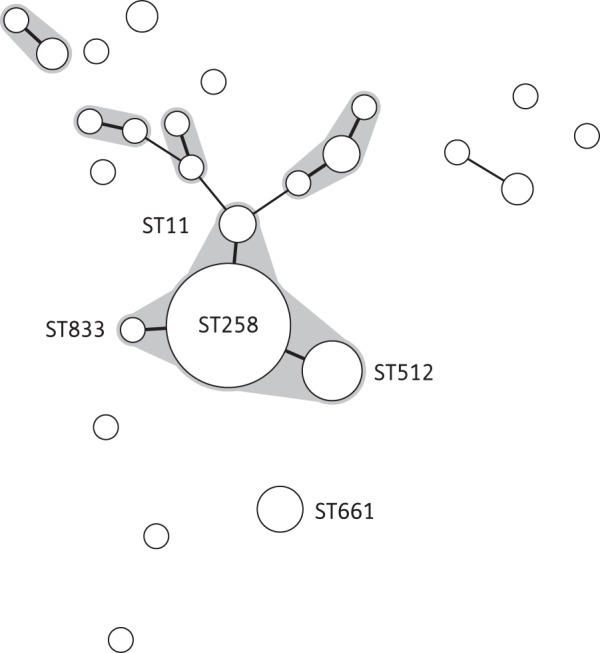
Minimum spanning tree of the MLST profiles of 84 KPC-positive K. pneumoniae isolates, received between September 2007 and July 2014 from 65 submitting laboratories. The shaded areas represent SLVs. Members of the ST258 clonal group are labelled, as are other STs with more than four submissions. The diameter of the circle represents the number of isolates of that particular ST. Thick solid lines represent SLVs, thin solid lines represent double-locus variants and the absence of connecting lines indicates multilocus variants.
Eight of the 12 E. cloacae complex isolates were submitted from five laboratories in Scotland between 2003 and 2013; these 8 were all ST171 and carried blaKPC-4. The four E. coli isolates represented unrelated STs.
Antibiotic susceptibility
MIC distributions for the KPC-positive isolates are shown in Table 3. All isolates were resistant to ertapenem and most were resistant or non-susceptible to imipenem (98%; 202/207) and meropenem (97%; 201/208). All meropenem MICs were above the EUCAST screening concentration of 0.125 mg/L. Sixty-four percent of all isolates were non-susceptible to tobramycin, 46% to amikacin and 35% to gentamicin. Thirty-two of 210 (15%) isolates were resistant to all three aminoglycosides tested, all of which were K. pneumoniae. Seventy-six percent, 39% and 13% of isolates were non-susceptible to ciprofloxacin, tigecycline and colistin, respectively (Table 3). All members of K. pneumoniae CG258 were resistant to tobramycin and most were also non-susceptible or resistant to amikacin (89%; 57/64); however, two-thirds were susceptible to gentamicin (Table S1, available as Supplementary data at JAC Online). By comparison, members of non-CG258 K. pneumoniae STs (n = 36) were more often susceptible to all three aminoglycosides (61%, 86% and 64% were susceptible to tobramycin, amikacin and gentamicin, respectively). Most non-K. pneumoniae isolates also were susceptible to all three aminoglycosides. All members of CG258 were resistant or non-susceptible to ciprofloxacin, compared with 47% (17/36) of other K. pneumoniae STs and 62% (23/37) of all other species. Colistin resistance was observed in 26 isolates, all of which were K. pneumoniae, and 13 of these belonged to CG258 and 3 to unrelated STs. STs of the remaining 10 colistin-resistant isolates were not determined. Colistin MICs for resistant isolates ranged from 4 to 32 mg/L (Table 3) and resistant isolates originated from laboratories/hospitals across six UK regions. Susceptibility to tigecycline was observed in 61% (125/205) of all isolates, but in only 48% (31/64) of CG258 isolates.
Table 3.
MIC distributions for KPC-producing isolates, irrespective of genus and species (n = 210)
| Antibiotic (range tested, mg/L) | EUCAST breakpoints ≤S/>R, mg/L | Number of isolates with MIC (mg/L) |
%S | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ≥128 | NA | |||
| Ertapenem (0.125–16) | ≤0.5/>1 | 1 | 5 | 7 | 11 | 184a | 2 | 0 | ||||||
| Imipenem (0.06–128) | ≤2/>8 | 1 | 1 | 3 | 6 | 24 | 72 | 42 | 31 | 27 | 3 | 2 | ||
| Meropenem (0.06–32) | ≤2/>8 | 1 | 1 | 4 | 1 | 7 | 34 | 44 | 116 | 2 | 3 | |||
| Amikacin (0.5–64) | ≤8/>16 | 5b | 35 | 32 | 22 | 19 | 26 | 69 | 2 | 54 | ||||
| Gentamicin (0.125–32) | ≤2/>4 | 10 | 48 | 44 | 34 | 7 | 13 | 5 | 47 | 2 | 65 | |||
| Tobramycin (0.125–32) | ≤2/>4 | 9 | 44 | 15 | 7 | 2 | 22 | 109 | 2 | 36 | ||||
| Ciprofloxacin (0.125–8) | ≤0.5/>1 | 36 | 5 | 9 | 8 | 3 | 4 | 12 | 131a | 2 | 24 | |||
| Colistin (0.5–32) | ≤2/>2 | 132b | 47 | 2 | 2 | 7 | 1 | 16 | 3 | 87 | ||||
| Tigecycline (0.25–16) | ≤1/>2 | 10b | 37 | 78 | 59 | 12 | 6 | 3 | 5 | 61 | ||||
S, susceptible; R, resistant; NA, not available.
Dark grey cells = resistant, light grey cells = intermediate and white cells = susceptible.
aMIC less than or equal to the indicated value.
bMIC greater than or equal to the indicated value.
Plasmid analysis
Transformants expressing KPC enzymes were obtained for 90/123 (73%) isolates chosen for further analysis. PBRT was performed on all of the transformants and WGS was performed on 61/90 transformants. Results revealed the following replicon types: IncFIB/IncFIIK (n = 49), IncN (n = 17), IncFIIK (n = 8), IncFIB (n = 3), IncR (n = 3), ColE-like (n = 2), IncI2 (n = 2), IncFIA (n = 1), IncP-6 (n = 1) and IncX3 (n = 1) (three plasmids were non-typeable). Most (80%; 39/49) plasmids with the IncFIB/IncFIIk replicon type were isolated from K. pneumoniae CG258. Of the 61 that were sequenced, 34/35 IncFIB/IncFIIK plasmids carried blaTEM-1, blaOXA-9 and either blaKPC-2 or blaKPC-3 and exhibited >99% sequence identity to pKpQIL (GenBank accession no. NC_014016). Twenty-five of these 35 IncFIB/IncFIIK plasmids were found in K. pneumoniae CG258 isolates, 20 ST258 and 5 ST512. The other 10 were found in eight unrelated K. pneumoniae STs: ST15, ST147, ST252, ST678, ST709, ST896, ST1162 and ST1163.
Sequenced KPC plasmids ranged from ∼13 to ∼224 kb, with blaKPC located within Tn4401 isoforms a, b, c or d (Table 4). All plasmids encoded blaKPC-2 or blaKPC-3, except the two ColE-like plasmids found in E. cloacae complex isolates from Scotland, which encoded blaKPC-4. Most plasmid replicon types were recovered from multiple UK regions (Table 4). Some KPC plasmids carried a number of additional antibiotic resistance genes (Table 4).
Table 4.
Features of 61 KPC plasmids sequenced
| Replicon type | Number of plasmids | Approx. size (kb) | Species | KPC variant(s) | Other resistance gene(s) | ST(s)a carrying plasmids | Number of regions | Tn4401 isoform(s) |
|---|---|---|---|---|---|---|---|---|
| IncFIB/IncFIIK | 35 | 106–115 | K. pneumoniae | KPC-2/3 | blaTEM-1, blaOXA-9 | 15/147/252/258/307/321/512/ 678/709/732/896/1162/1163 | 7 | a |
| IncN | 8 | 59–76 | K. pneumoniae/ K. oxytoca | KPC-2 | blaTEM-1, aph(6)-Id, sul1, sul2, dfrA18, dfrA14, qnrB2, tet(A) | 258/336/1838 | 4 | b/c |
| IncFIIK | 4 | 97–224 | K. pneumoniae | KPC-2 | blaTEM-1, blaOXA-9, aph(3′)-la, aadA2, aadA5, dfrA12, dfrA17, catA1, sul1, mph(A), qnrB1 | 258/321/732/1162 | 3 | a |
| IncR | 3 | 48–69 | K. pneumoniae | KPC-2/3 | aac(6′)-Ib, aph(4)-la, aadA1, aadA2, catA1, cmlA1, mef(B), sul3 | 258 | 2 | a/b |
| ColE-like | 2 | 13 | E. cloacae complex | KPC-4 | 171 | 1 | a | |
| IncI2 | 2 | 77 | K. pneumoniae | KPC-3 | blaOXA-9, blaTEM-1, aac(6′)-Ib, aadA1 | 258 | 1 | b |
| IncFIA | 1 | 52 | K. pneumoniae | KPC-2 | blaTEM-1, blaOXA-9 | 15 | 1 | a |
| IncFIB | 1 | 76 | K. pneumoniae | KPC-3 | blaTEM-1, blaOXA-9 | 258 | 1 | a |
| IncP-6 | 1 | 38 | K. pneumoniae | KPC-2 | 11 | 1 | a | |
| IncX3 | 1 | 53 | K. pneumoniae | KPC-3 | blaSHV-11 | 258 | 1 | a |
| Non-typeable | 3 | 62–89 | K. pneumoniae | KPC-2/3 | blaTEM-1, blaOXA-9, aph(6)-Id, aac(6′)-Ib, aadA1, qnrB2, aph(6)-Ib, sul2, | 258/833 | 2 | b/d |
aSTs were determined for K. pneumoniae and E. cloacae complex isolates.
Discussion
This report reviews the first 160 recorded patients infected or colonized by KPC-positive bacteria from across the UK, excluding North-West England (which accounts for most cases), as ascertained from referrals to PHE's AMRHAI Reference Unit. Isolates were submitted over an 11 year period from August 2003 to August 2014.
Isolates with KPC enzymes were submitted from all UK regions. The majority were obtained from clinical specimens (132/210), predominantly urines (70/132). All isolates were multiresistant and exhibited non-susceptibility to at least one carbapenem. The only antibiotics that retained relatively good levels of activity in vitro were colistin (87%), gentamicin (65%) and tigecycline (61%). Use of colistin as monotherapy against KPC producers has limitations due to its nephrotoxicity and neurotoxicity and also the danger of selecting for colistin-resistant mutants.31 The potential for expansion of colistin-resistant variants is evidenced by outbreaks caused by colistin-resistant members of the ST258 clone.14,15 In this study, we found 26 colistin-resistant K. pneumoniae (but no isolates of other species); most were members of CG258 and were found in 10/11 UK regions. Tigecycline usage is limited by its inability to achieve high concentrations in the urine and blood and is licensed for the treatment of complicated skin and skin-structure infections and complicated intra-abdominal infections.32 Several antibiotic combinations have been used for the treatment of infections caused by KPC-producing bacteria, including: colistin with aminoglycosides/carbapenems/fluoroquinolones; tigecycline with aminoglycosides; and several β-lactam and fluoroquinolone/aminoglycoside combinations.31,33 Such combination therapies have been shown to be more effective than monotherapy and are believed to reduce the likelihood of the development of resistant mutants.31,33
Although travel history data was limited, 14 patients reported travel within the previous 6 months to countries (Greece, Italy and Israel) that have previously reported KPC outbreaks in their hospitals.4 Whilst it is not possible to know conclusively where acquisition of the KPC-producing bacteria took place, it is clear that international travel continues to play a significant role in the dissemination of KPC-producing clones and this has been illustrated by the worldwide spread of members of CG258.4 The finding that five patients each had KPC-positive bacteria isolated in two UK regions demonstrates that domestic travel and patient transfers also may play a vital role in the dissemination of these bacteria within the UK. This has the potential to be particularly problematic when one UK region has a long-standing outbreak (North-West England) and may provide a reservoir.
At the time of this study, there were 23 known KPC variants (KPC-2–KPC-24) (www.lahey.org/studies/), but only 3 variants were found here: KPC-2, KPC-3 and KPC-4. KPC-2 and KPC-3 are the most common variants worldwide and their genes are often harboured on pKpQIL and pKpQIL-like plasmids.7,9 We first identified KPC-4 in 2003 and this variant has recently also been found in the USA on IncL/M plasmids in both E. cloacae and Serratia marcescens and encoded by an IncN plasmid in K. pneumoniae.10,11 In this study, all of the isolates with KPC-4 enzymes were E. cloacae complex ST171; they were isolated in five laboratories in Scotland over a 10 year period (from 2003 to 2013). Sequencing of two representative KPC-4 plasmids identified ∼13 kb ColE-like plasmids encoding blaKPC-4 in both isolates. Despite the long-term persistence of this KPC-4-producing clone, it has not caused recognized outbreaks and its plasmid has apparently not spread to other hosts.
Although the worldwide dissemination of bacteria with KPC enzymes is substantially associated with a single CG (K. pneumoniae CG258), these β-lactamases have been found in numerous other K. pneumoniae STs and in other bacterial species.4,10 blaKPC genes have been recorded on several plasmids of different incompatibility groups, including IncF, IncI2, IncN, IncL/M and IncX.7,10–12 Here, we found KPC genes in four bacterial species and in ≥33 different STs, carried by >11 plasmid replicon types, suggesting that both plasmid spread and the mobility between plasmids plays an important role in the dissemination of KPC enzymes in the UK. Even within CG258, we found at least eight different KPC plasmid replicon types, indicative of the success of this CG as a host of KPC plasmids. Nevertheless, most plasmids were of the IncFIB/IncFIIK type and were highly homologous to pKpQIL, indicating that plasmids of this type are the dominant carriers of blaKPC in the UK.
There are numerous reports of outbreaks of bacteria with KPC enzymes from other countries,4,8,12,14 associated particularly with members of CG258. We have shown here that K. pneumoniae ST258 is the dominant host of KPC enzymes in the UK outside of North-West England and that multiple UK hospitals have been challenged by the introduction of this successful clone and its close relatives since 2007. Nevertheless, to date, there have been very few clusters of infections or colonizations caused by K. pneumoniae ST258 in the UK. Whether the lack of CG258 dissemination can be attributed to better screening and/or compliance with infection control practices in the UK is unknown, but this does underline the need for continued surveillance and implementation of rigorous infection prevention and control measures.34
Funding
This work was funded partly internally and by National Institute of Health Strategic Research and Development Fund (project no. 108373).
Transparency declarations
PHE's AMRHAI Reference Unit has received financial support for conference attendance, lectures, research projects or contracted evaluations from numerous sources, including: Achaogen Inc., Allecra Antiinfectives GmbH, Amplex, AstraZeneca UK Ltd, Becton Dickinson Diagnostics, BSAC, Cepheid, Check-Points B.V., Cubist Pharmaceuticals, Department of Health, Enigma Diagnostics, Food Standards Agency, GlaxoSmithKline Services Ltd, Henry Stewart Talks, IHMA Ltd, Merck Sharpe & Dohme Corp., Meiji Seika Kiasya Ltd, Momentum Biosciences Ltd, Nordic Pharma Ltd, Norgine Pharmaceuticals, Rempex Pharmaceuticals Ltd, Rokitan Ltd, Smith & Nephew UK Ltd, Trius Therapeutics, VenatoRx and Wockhardt Ltd. D. M. L.: Advisory Boards or ad hoc consultancy—Achaogen, Adenium, Alere, Allecra, Astellas, AstraZeneca, Basilea, Bayer, BioVersys, Cubist, Curetis, Cycle, Discuva, Forest, GSK, Meiji, Pfizer, Roche, Shionogi, Tetraphase, VenatoRx and Wockhardt; paid lectures—AOP Orphan, Astellas, AstraZeneca, Bruker, Curetis, Merck, Pfizer and Leo; shareholdings in—Dechra, GSK, Merck, PerkinElmer and Pfizer amounting to <10% of portfolio value. All other authors: none to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
References
- 1.Yigit H, Queenan AM, Anderson GJ et al. . Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 2001; 45: 1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villegas MV, Lolans K, Correa A et al. . First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob Agents Chemother 2007; 51: 1553–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robledo IE, Aquino EE, Santé MI et al. . Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob Agents Chemother 2010; 54: 1354–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz-Price LS, Poirel L, Bonomo RA et al. . Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13: 785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breurec S, Guessennd N, Timinouni M et al. . Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin Microbiol Infect 2013; 19: 349–55. [DOI] [PubMed] [Google Scholar]
- 6.Naas T, Cuzon G, Truong H et al. . Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother 2012; 56: 4751–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leavitt A, Carmeli Y, Chmelnitsky I et al. . Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob Agents Chemother 2010; 54: 3002–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hrabák J, Papagiannitsis CC, Študentová V et al. . Carbapenemase-producing Klebsiella pneumoniae in the Czech Republic in 2011. Euro Surveill 2013; 18: pii=20626. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Chavda KD, Melano RG et al. . Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob Agents Chemother 2014; 58: 2871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant KA, Van Schooneveld TC, Thapa I et al. . KPC-4 is encoded within a truncated Tn4401 in an IncL/M plasmid, pNE1280, isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob Agents Chemother 2013; 57: 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Chavda KD, Fraimow HS et al. . Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 2013; 57: 269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Chavda KD, Al Laham N et al. . Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother 2013; 57: 5019–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Martinez L, Gonzalez-Lopez JJ. Carbapenemases in Enterobacteriaceae: types and molecular epidemiology. Enferm Infecc Microbiol Clin 2014; 32 Suppl 4: 4–9. [DOI] [PubMed] [Google Scholar]
- 14.Toth A, Damjanova I, Puskas E et al. . Emergence of a colistin-resistant KPC-2-producing Klebsiella pneumoniae ST258 clone in Hungary. Eur J Clin Microbiol Infect Dis 2010; 29: 765–9. [DOI] [PubMed] [Google Scholar]
- 15.Mezzatesta ML, Gona F, Caio C et al. . Outbreak of KPC-3-producing, and colistin-resistant, Klebsiella pneumoniae infections in two Sicilian hospitals. Clin Microbiol Infect 2011; 17: 1444–7. [DOI] [PubMed] [Google Scholar]
- 16.Palepou MF, Woodford N, Hope R et al. . Novel class A carbapenemase, KPC-4, in an Enterobacter isolate from Scotland. In: Abstracts of the Fifteenth European Congress of Clinical Microbiology and Infectious Diseases, Copenhagen, 2005. Abstract 1134_01_20. European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland. [Google Scholar]
- 17.Woodford N, Zhang J, Warner M et al. . Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J Antimicrob Chemother 2008; 62: 1261–4. [DOI] [PubMed] [Google Scholar]
- 18.Central Manchester University Hospitals NHS Foundation Trust. Infection Prevention and Control Annual Report 2013/2014. https://www.cmft.nhs.uk/media/892227/agenda%20item%2084%20annual%20infection%20control%20report.pdf.
- 19.Haraoui LP, Lévesque S, Lefebvre B et al. . Polyclonal outbreak of KPC-3-producing Enterobacter cloacae at a single hospital in Montreal, Quebec, Canada. J Clin Microbiol 2013; 51: 2406–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz-Garbajosa P, Curiao T, Tato M et al. . Multiclonal dispersal of KPC genes following the emergence of non-ST258 KPC-producing Klebsiella pneumoniae clones in Madrid, Spain. J Antimicrob Chemother 2013; 68: 2487–92. [DOI] [PubMed] [Google Scholar]
- 21.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 2001; 48 Suppl 1: 5–16. [DOI] [PubMed] [Google Scholar]
- 22.EUCAST. Clinical Breakpoint Table, Version 4.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf.
- 23.Woodford N, Warner M, Pike R et al. . Evaluation of a commercial microarray to detect carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 2011; 66: 2887–8. [DOI] [PubMed] [Google Scholar]
- 24.Doumith M, Day M, Ciesielczuk H et al. . Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J Clin Microbiol 2015; 53: 160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diancourt L, Passet V, Verhoef J et al. . Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 2005; 43: 4178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N et al. . Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 2013; 8: e66358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wirth T, Falush D, Lan R et al. . Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 2006; 60: 1136–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carattoli A, Bertini A, Villa L et al. . Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 2005; 63: 219–28. [DOI] [PubMed] [Google Scholar]
- 29.Johnson TJ, Bielak EM, Fortini D et al. . Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 2012; 68: 43–50. [DOI] [PubMed] [Google Scholar]
- 30.Erkens-Hulshof S, Virginia-Cova L, van Dijk W et al. . The occurrence of Enterobacteriaceae producing KPC carbapenemases in a general hospital in Curaçao. Antimicrob Resist Infect Control 2014; 3: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qureshi ZA, Paterson DL, Potoski BA et al. . Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother 2012; 56: 2108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noskin GA. Tigecycline: a new glycylcycline for treatment of serious infections. Clin Infect Dis 2005; 41 Suppl 5: S303–14. [DOI] [PubMed] [Google Scholar]
- 33.Tumbarello M, Viale P, Viscoli C et al. . Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis 2012; 55: 943–50. [DOI] [PubMed] [Google Scholar]
- 34.PHE. Carbapenemase-Producing Enterobacteriaceae: Early Detection, Management and Control Toolkit for Acute Trusts. 19 June 2014 https://www.gov.uk/government/publications/carbapenemase-producing-enterobacteriaceae-early-detection-management-and-control-toolkit-for-acute-trusts.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.