Abstract
Objectives
The objective of this study was to determine the fitness of Escherichia coli mutants with reduced susceptibility to tigecycline after exposure to adverse conditions in vitro and in vivo.
Methods
Survival in response to low pH, bile salts, oxidative stress and human serum was examined for E. coli mutants with reduced susceptibility to tigecycline due to single mutations that caused increased efflux (marR, lon) or impaired LPS (rfaC, rfaE, lpcA). An in vitro competition assay was used to determine growth fitness defects. Competitive fitness was assessed using mouse infection models. MICs, exponential growth rates and expression levels of efflux-related genes were measured for genetically reconstructed double and triple mutants.
Results
The LPS mutants were 48–85-fold more susceptible to bile salts compared with the ERN mutants and the WT. As shown by in vitro competitions, the fitness reduction was 0.3%–13% for ERN mutants and ∼24% for LPS mutants. During in vivo survival experiments, LPS mutants were outcompeted by the WT strain in the thigh infection model. Constructed double ERN and LPS mutants showed additive and synergistic increases in tigecycline MICs.
Conclusions
Generally, reduced susceptibility to tigecycline caused a decrease in fitness under stressful in vitro and in vivo conditions with ERN mutants being fitter than LPS mutants. When combined, ERN mutations caused a synergistic increase in the MIC of tigecycline. These findings could explain why clinical resistance to tigecycline in E. coli is mainly associated with up-regulation of the AcrAB efflux system.
Introduction
The spread of antibiotic-resistant bacteria has generated a medical crisis1 and as common antibiotic therapies increasingly fail, last resort drugs such as colistin and tigecycline are used more frequently to treat serious MDR infections. Tigecycline is the only medically approved third-generation tetracycline.2,3 It is a derivative of the second-generation tetracycline minocycline with a 9-t-butylglycylamido group attached to the ring D. This particular side chain leads to stronger binding to the ribosome and evasion of the main tetracycline resistance mechanisms (efflux and ribosomal protection).3 However, resistance to tigecycline has been reported in Gram-negative as well as Gram-positive pathogens.4
We have previously identified two distinct groups of in vitro-selected mutants of Escherichia coli with reduced susceptibility to tigecycline.5 Efflux regulatory network (ERN) mutants contained mutated regulators of the multidrug efflux system AcrAB, which belongs to the resistance–nodulation–division (RND) family of pumps. LPS mutants were impaired in the synthesis and transport of heptose, necessary for the assembly of LPS. To our knowledge, that was the first time defects in LPS were connected to the reduced susceptibility to tigecycline. Both groups of mutations had reduced fitness in in vitro bacterial growth assays.5
In this study, we characterized the reconstructed mutants from both the ERN (lon, marR) and LPS (rfaC, rfaE, lpcA) groups and evaluated their performance under a set of different in vitro conditions. These tests were designed to mimic possible environmental conditions these bacteria might encounter in vivo. Additionally, we determined how these mutants grew during in vitro and in vivo competition assays. We also constructed and characterized double and triple mutants carrying mutations in the ERN and LPS targets. Our findings suggest that the LPS mutations are the more costly single mutations. Thus, single ERN mutants survived the harsh in vitro conditions better and when combined had a synergistic effect on the MIC of tigecycline. These results suggest that clinical resistance to tigecycline in pathogenic E. coli is mainly associated with up-regulation of the AcrAB efflux system because ERN mutants show both a higher fitness and resistance than the LPS mutants.
Materials and methods
Bacterial strains and genetic constructions
Efflux and LPS mutants were reconstructed in the WT E. coli K-12 MG1655 background during our previous study.5 Double and triple mutants with decreased susceptibility to tigecycline were created by moving mutations to the lon (DA25822) or marR (DA25825) mutant background using Lambda Red recombineering.6,7 Both strains contained flippase recognition target scars at different intergenic locations from previous genetic construction experiments.5 The location of the flippase recognition target scar did not affect the MIC and growth rate measurements. The constructions of double and triple mutants were performed by amplifying a neo-sacB cassette (a gift from Diarmaid Hughes) with primers containing 40 bp homology sites to the target genes and inserting it during the first Lambda Red recombination step. The PCR product of the mutated gene was used to exchange the inserted neo-sacB cassette during the second Lambda Red recombination step. All reconstructed mutations were confirmed by PCR and sequencing.
Response to acidic environment
Three independent cultures were grown overnight in LB broth (pH 7.0) at 37°C with shaking (190 rpm). The cells were diluted 100-fold and cultured until OD600 ≈ 0.3. After the cultures reached log phase, the cells were divided into two tubes (1 mL/tube) and pelleted. In one tube the cells were re-suspended in 1 mL of LB broth (pH 4.5) for pre-adaptation to low pH, while the cells in the other tube were re-suspended in 1 mL of LB broth (pH 7.0) to be used as a non-adapted control. After 1 h of incubation at 37°C, the pre-adapted and non-adapted cells were pelleted and re-suspended in 1 mL of LB broth (pH 2.5). The cells were incubated at 37°C for 2 h and the viable count was performed at timepoints 0, 0.5, 1 and 2 h. The data were normalized to the cfu/mL at 0 h.
Sensitivity to bile
Three independent cultures were grown overnight in Mueller–Hinton (MH) broth till saturation at 37°C with shaking (190 rpm). Approximately 5 × 106 cfu of cells were inoculated into MH broth supplemented with different concentrations of bile salts (Sigma–Aldrich). The bacterial suspension was aliquoted in a round-bottom 96-well plate (0.1 mL/well) and incubated at 37°C with shaking (190 rpm). Sixteen hours later OD620 was determined using the Multiskan FC microplate photometer (Thermo Scientific).
Oxidative stress assay
Three independent cultures were grown till saturation in M9 medium supplemented with 0.2% glucose and 10 mg/L thiamine at 37°C with shaking (190 rpm). The cells were diluted 100-fold and cultured till OD600 ≈ 0.3. Exponentially dividing cells were mixed with paraquat (Sigma–Aldrich) at final concentrations of 0.025, 0.1, 0.2, 0.8 and 3.2 mM and incubated at 37°C with shaking (190 rpm) for 4 h. A viable count was performed to estimate the survival of bacteria. The data were normalized to the cfu/mL of untreated cells.
Sensitivity to human serum
Three independent cultures were grown overnight in M9 medium supplemented with 0.2% glucose and 10 mg/L thiamine at 37°C with shaking (190 rpm). The cells were diluted 100-fold and cultured till OD600 ≈ 0.3. A 2-fold dilution series of sterile-filtered human serum (H4522; Sigma–Aldrich) was performed and mixed with exponentially dividing cells. The mixture was incubated at 37°C with shaking (190 rpm) for 2 h and plated for viable count. The data were normalized to the cfu/mL of untreated cells.
In vitro competitions
To evaluate the fitness of reconstructed mutants throughout all growth phases, in vitro competition assays were set up. All competition assays were performed as described previously.8 In short, blue (bfp) and yellow (yfp) fluorescent proteins were inserted in the galK gene of reconstructed mutants using Lambda Red recombineering.6,7 Two independent overnight cultures of reconstructed mutants labelled with one fluorescent dye and their isogenic WTs labelled with the other fluorescent dye were grown in MH broth at 37°C with shaking (190 rpm). As the selection coefficients are independent of initial frequency of resistant mutants,8 the reconstructed mutants harbouring less costly mutations (lon, marR) were mixed 1:1, while mutants with more costly mutations (rfaC, rfaE, lpcA) were mixed 100:1 with their isogenic WTs. This inoculation ratio allowed us to capture the change in competing populations within the 30 generations of growth. The competition mixtures were maintained by 1000-fold dilutions performed every 24 h, for three cycles (∼10 generations of growth per cycle). The ratio of mutant versus WT cells was calculated by counting 105 cells with a fluorescence-activated cell sorter FacsAria (BD Biosciences) and was plotted as the function of generations of growth. The selection coefficients were calculated as described previously.8 The selection coefficients were plotted as a function of different tigecycline concentrations. The concentration of tigecycline at which the cost of the mutation was compensated by the antibiotic enrichment of mutants was defined as minimal selective concentration (MSC).8
In vivo competitions
To determine the competitive ability of the reconstructed mutants in animal models, neutropenic mouse thigh infection and peritonitis models were chosen. Outbred female NMRI mice (26–30 g; Harlan, the Netherlands) were housed at the animal facility at Statens Serum Institut, Copenhagen, Denmark. Four to eight animals per timepoint were used in the experiments.
For the peritonitis model, fresh overnight E. coli colonies from a 5% Horse Blood Agar plate were suspended in sterile saline to ∼108 cfu/mL. One ara+ E. coli strain (mutant) was mixed with the ara− E. coli strain (WT) and diluted to ∼106 cfu/mL in saline with 5% mucin (M-2378; Sigma–Aldrich), used as an adjuvant during inoculation. For each experiment, the size of the inoculum was determined. The ara+ E. coli strains were seen as reddish colonies and the ara− strain as greyish colonies on modified MacConkey (bile salts and Crystal Violet were omitted due to toxicity to LPS mutants)/1% arabinose agar plates (20 g of peptone, 5 g of NaCl, 14 g of bacteriological agar, 0.03 g of neutral red dye and 900 mL of sterile water, autoclaved at 121°C for 15 min and supplemented with a final concentration of 1% l-arabinose). Mice were inoculated intraperitoneally with 0.5 mL of the mixed E. coli suspension. Six hours after infection mice were killed by cervical dislocation and peritoneal washes were performed. Peritoneal fluids were serially diluted (10-fold dilutions) and 20 μL was plated on modified MacConkey/1% arabinose agar plates with subsequent counting of colonies after incubation overnight at 35°C in ambient air.
For the thigh infection model, a suspension of fresh overnight E. coli colonies from a modified MacConkey/1% arabinose agar plate was prepared as described above. The competitor E. coli strains were mixed and diluted to ∼5 × 106 cfu/mL. For each experiment, the size of the inoculum and scoring of competitors were performed as described above. Mice were inoculated intramuscularly with 0.05 mL of the suspensions into the quadriceps femoral muscle close to the femur of the left hind leg. At 24 h mice were killed by cervical dislocation, the thigh muscles were collected, mixed with saline and homogenized using a Dispomix® Drive. Colony count on modified MacConkey/1% arabinose agar plates was made as described above.
The competitive index was calculated by dividing the ratio of the mutant population versus the isogenic WT population at the end of infection by the ratio of the mutant population versus the WT population in the initial inoculum. Values were normalized to the competitive index of arabinose+ and arabinose− WT competition experiments. Approximately 0.5 h before inoculation for both infection models, mice were treated orally with 45 μL of neurofen (20 mg/mL ibuprofen corresponding to ∼30 mg/kg) as pain relief. All animal experiments were approved by the National Committee of Animal Ethics—Animal Experiment Inspectorate under the Danish Ministry of Justice and the animals were cared for in line with national guidance. Ethics approval permit nos 2014-15-0201-00171 (peritonitis) and 2012-15-2934-00619 (thigh infection).
Determination of MIC
MICs of double and triple mutants were measured using Etest strips (bioMérieux). A culture was incubated in a shaking incubator (190 rpm) at 37°C overnight and then diluted 100-fold in saline. The diluted suspension was distributed on a MH agar plate with a cotton swab. An Etest strip was applied and the plate was incubated at 37°C for 16–20 h. MIC was read as the lowest concentration at which no bacterial growth was observed.
Determination of growth rates
The growth rates of double and triple mutants were measured in MH broth at 37°C by taking OD600 measurements every 4 min in a BioscreenC reader (Oy Growth Curves Ab Ltd). Six independent cultures per mutant were grown overnight till saturation. The cultures were diluted to ∼5 × 106 cfu/mL and aliquoted into a BioscreenC plate in quadruplicate (0.25 mL/well). The growth rate was estimated from OD600 interval between 0.01 and 0.07, where the growth was observed to be exponential. Relative growth rates were calculated by dividing the corresponding growth rate of mutant by the average growth rate of an otherwise isogenic WT strain in the same experiment.
Determination of gene expression
Three independent cultures per strain were grown overnight and then diluted 100-fold and cultured till exponential phase. Total RNA was prepared using RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Residual DNA was degraded using the Turbo DNA-free kit (Ambion) and 500 ng of prepared RNA was reverse-transcribed using the High Capacity cDNA Reverse Transcription Kit (Thermo Scientific) following instructions of the manufacturer. Real-time quantitative PCR was performed to measure expression levels of marA, acrA and acrB genes using the Eco Real-Time PCR system from Illumina. A 10 μL reaction contained 1× PerfeCTa SYBR Green FastMix (Quanta BioSciences), 0.5 μM of forward primer (marA_F, ATACATCCGCAGCCGTAAGA; acrA_F, CGGATGACAAAGTGGAAACC; and acrB_F, CTGCTGATGGTCAGATGGTG), 0.5 μM of reverse primer (marA_R, TTTGAAGGTTCGGGTCAGAG; acrA_R, CCTTCTGTCACCAGCCACTT; and acrB_R, GAACCGTACTCCCAACGAGA) and 1 μL of 10−1, 10−2 or 10−3 dilution of cDNA. The PCR was performed with 10 min of polymerase activation at 95°C, followed by 40 cycles of DNA amplification for 10 s at 95°C and 30 s at 60°C. Expression levels were calculated using the ΔCq method with two reference genes (cysG and hcaT) and the following formula: 2[Cq(reference genes) geometric mean−Cq(target gene)]. All expression level values were related to the expression level of the target genes in the isogenic WT strain.
DNA sequencing
Target genes were PCR amplified with primers binding upstream and downstream of the coding region using 2× PCR mix (Thermo Scientific). PCR products were purified with GeneJet Gel Extraction Kit (Thermo Scientific) and premixed with one of the amplification primers. The mixture was sent to Eurofins MWG Operon (Ebersberg, Germany) for DNA sequencing. For the list of primers refer to Table 1 in Linkevicius et al.5
Results
Acid and bile tolerance response in ERN and LPS mutants
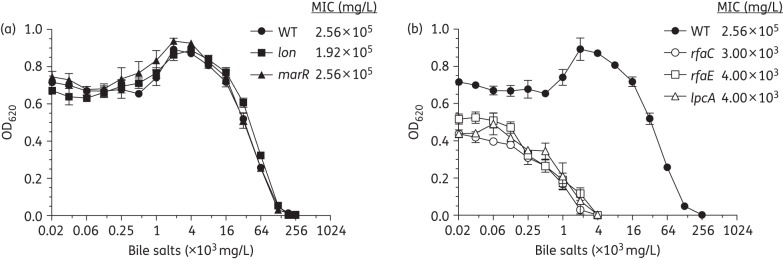
In this study, we evaluated the fitness of reconstructed ERN and LPS mutants by exposing them to a series of adverse conditions, including low pH, bile salts, oxidative stress (paraquat) and human serum, to mimic stress conditions bacteria might encounter in vivo. Viability of the pre-adapted (see the Materials and methods section) WT strain was not affected by a reduction in pH from 7.0 to 2.5, while the non-adapted WT survival was decreased by 4–5 log (Figure S1a, available as Supplementary data at JAC Online). This general trend was also observed for the reconstructed mutants and none of the mutants differed substantially from the WT, except for the lon mutant, which showed slightly higher pH sensitivity. In contrast, a noticeable difference between the ERN and LPS mutant groups was observed in the bile tolerance assay (Figure 1). The ERN mutants behaved like the WT control and could grow at very high concentrations of bile salts with MIC values of 1.92 × 105 mg/mL and 2.56 × 105 mg/mL for the lon and marR mutants, respectively (Figure 1a). However, the LPS mutants were inhibited at much lower bile salt concentrations, with MIC values being as low as 3 × 103 mg/mL for the rfaC mutant and 4 × 103 mg/mL for the rfaE and lpcA mutants, indicating that defects in the outer membrane can significantly contribute to increased sensitivity to bile salts (Figure 1b).
Figure 1.
Bile tolerance. (a) ERN reconstructed mutants. (b) LPS reconstructed mutants. OD measurements were performed after 16 h of growth in MH broth supplemented with bile salts. Values represent the mean of three independent biological replicates. Error bars indicate the standard deviation.
Response to oxidative stress and serum resistance in ERN and LPS mutants
E. coli cells may be exposed to oxidative stress when interacting with phagocytic cells during infection of tissues that are normally not accessible for these bacteria. Similarly, if E. coli cells enter the bloodstream, they get in contact with blood proteins like the complement system. To investigate the effect of such stressful environments on the reconstructed mutants, we performed in vitro oxidative stress and serum resistance assays.
Exposure to the reactive oxygen species generated by paraquat decreased colony counts about 10-fold for all strains exposed to the highest tested concentrations, indicating that all of the reconstructed mutants were as susceptible as the WT control (Figure S2). Similarly, the overall survival in blood serum of reconstructed ERN and LPS mutants was comparable to the WT with a decreasing colony count at increasing serum concentrations (Figure S3). The lon mutant tolerated the inhibitory activity of serum best with viable counts dropping ∼4 log at the highest tested concentration, while the marR mutant behaved similar to the WT with colony counts decreasing about 6 log (Figure S3a). The viability of the LPS mutants was comparable to the WT strain with a decline of 6 log in viable counts at the highest concentration of serum measured (Figure S3b).
In vitro competition assays of ERN and LPS mutants
In our previous study, fitness was measured as the maximum exponential growth rate.5 Here we used in vitro competition assays to evaluate the differences in growth conferred by the resistance mutations during the whole growth cycle and to increase the sensitivity of the assay. Additionally, as shown by our recent work, selection of antibiotic-resistant mutants can occur at antibiotic levels far below the MIC value for the susceptible WT.8 The MSC is defined as the antibiotic concentration above which resistant mutants will be enriched over the susceptible counterpart. As low tigecycline concentrations could be present in human tissues during tigecycline treatment, we also determined the MSCs of tigecycline for both ERN and LPS mutants by performing competition assays in the presence of sub-MIC concentrations of tigecycline (Figure 2).
Figure 2.
In vitro competitions between mutants and WT at different tigecycline concentrations. (a) ERN reconstructed mutants. (b) LPS reconstructed mutants. During the competition experiment, the ratio of mutant versus WT was plotted as a function of growth cycle at different tigecycline concentrations. For each tigecycline concentration a selection coefficient was calculated from the slope. The selection coefficient values were used to generate the curves above. The intercept of the curve on the x-axis represents the MSC and the intercept on the y-axis denotes the fitness cost. Values represent the mean of 4–12 independent biological replicates. Error bars indicate the standard deviation.
In the absence of tigecycline, the ERN mutations (Figure 2a) were generally less costly than the LPS mutations (Figure 2b). The lon and marR mutants showed a 13% and 0.3% reduction in fitness, respectively, whereas all LPS mutants exhibited fitness costs of ∼24%. As the MSC is directly affected by differences in fitness costs, the mutants followed the same pattern where MSC was ∼0.03 mg/L for the LPS mutants, 0.021 mg/L for the lon mutant and 0.003 mg/L for the least costly marR mutant. The MIC of tigecycline for our WT strain was previously determined to be 0.047 mg/L.5 These results suggest that the lon mutation and the LPS mutations confer a substantial cost in in vitro competitions and that they require tigecycline concentrations about half the WT MIC to be selectively enriched, whereas the marR mutant was much less costly and enrichment could be achieved at 16-fold lower tigecycline concentrations than the WT MIC.
MICs for and fitness of mutants with combinations of ERN and LPS mutations
During our in vitro selection of spontaneous tigecycline-resistant E. coli mutants, we found some mutants that contained several ERN mutations or ERN and LPS mutations.5 To evaluate such mutants in terms of resistance and fitness (here only the exponential growth rates were assessed), we reconstructed lon, marR and lpcA mutations in different combinations (lpcA, lon; lpcA, marR; lon, marR; lpcA, lon, marR) and measured MICs, exponential growth rates and relative expression levels of marA, acrA and acrB genes (Table 1).
Table 1.
Characterization of reconstructed single, double and triple mutants
| Strain | Mutated gene(s) | MIC (mg/L) |
Mean ± SD relative growth rate | Mean ± SD relative expressiona |
Reference | |||
|---|---|---|---|---|---|---|---|---|
| TGC | TET | marA | acrA | acrB | ||||
| DA5438 | — | 0.047 | 1 | 1 | 1 | 1 | 1 | 5 |
| DA25822 | lon Q137stop | 0.125 | 1 | 0.96 ± 0.04 | 0.8 ± 0.2 | 1.6 ± 0.3 | 1.6 ± 0.4 | 5 |
| DA25825 | marR K62fs | 0.125 | 1.5 | 0.97 ± 0.03 | 47.0 ± 3.6 | 5.0 ± 0.5 | 3.8 ± 0.5 | 5 |
| DA26800 | lpcA E173G | 0.125 | 1.5 | 0.91 ± 0.01 | 1.7 ± 0.4 | 1.1 ± 0.2 | 1.1 ± 0.3 | 5 |
| DA40498 | lpcA E173G, lon Q137stop | 0.19 | 1 | 0.75 ± 0.02 | 3.3 ± 0.7 | 5.7 ± 1.4 | 4.4 ± 0.6 | this study |
| DA39146 | lpcA E173G, marR K62fs | 0.25 | 2 | 0.79 ± 0.01 | 88.8 ± 6.1 | 6.8 ± 0.2 | 4.7 ± 0.3 | this study |
| DA39150 | lon Q137stop, marR K62fs | 0.75 | 6 | 0.68 ± 0.01 | 100.7 ± 5.4 | 10.8 ± 0.8 | 7.5 ± 0.8 | this study |
| DA39997 | lpcA E173G, lon Q137stop, marR K62fs | 0.25 | 2 | 0.76 ± 0.01 | 2.8 ± 0.3 | 2.6 ± 0.2 | 1.9 ± 0.2 | this study |
TGC, tigecycline; TET, tetracycline; fs, frame shift; stop, stop codon.
aExpression level measurements.
All constructed mutant combinations showed increased MICs of tigecycline and tetracycline and reduced maximum growth rates compared with their isogenic WTs. Additionally, most mutants containing a marR mutation overexpressed the marA and acrAB genes. The lpcA, lon and lpcA, marR double mutants had very similar MICs and growth rates. Tigecycline and tetracycline MICs for the lpcA, lon mutation combination were 0.19 mg/L and 1 mg/L, respectively, with a reduction in growth rate of 25%. The lpcA, marR mutation combination had tigecycline and tetracycline MICs of 0.25 mg/L and 2 mg/L, respectively, and a fitness cost of 21%. The highest increase in MICs (0.75 mg/L for tigecycline and 6 mg/L for tetracycline) and the largest fitness cost (32%) was observed in the lon, marR double mutant. Interestingly, the triple mutant (lpcA, lon, marR) had not only lower MICs (0.25 mg/L for tigecycline and 2 mg/L for tetracycline), but also a lower fitness cost (24%) and lower expression of efflux-related genes compared with the lon, marR double mutant.
In vivo competition assays of ERN and LPS mutants
We set up in vivo competition assays in two well-established model systems, the mouse peritonitis and neutropenic mouse thigh infection models, to assess the fitness of the mutants inside a host. In the latter model the immune system was compromised because of depletion of neutrophils. In these assays, if the competitive index is lower than 1, this indicates that the WT population outcompeted the mutant population and conversely with a competition index higher than 1, the mutant population outcompeted the WT. To distinguish between WT and mutant competitors we tagged the WT strain by inactivating the araB gene, which resulted in red ara+ and white–grey ara− colonies on modified MacConkey agar. The control experiment showed that inactivation of araB was neutral with regard to growth in mice and did not affect the competitive properties of the WT (Figures S4a and S5a).
In the peritonitis infection model (Figure 3a), the lon mutant was as fit as the WT, whereas the marR mutant and all LPS mutants were weakly outcompeted by the WT. Over the course of infection, the bacterial counts dropped for the majority of the mutants and WT as compared with the initial inoculum size (Figure S4), suggesting efficient clearance of the bacteria by the immune system.
Figure 3.

In vivo competitions between mutants and WT. (a) Peritonitis mouse model. (b) Neutropenic mouse thigh infection model. Four to eight animals were used per timepoint. The horizontal line for each timepoint represents the median value. Error bars indicate the range of data points. Relative competitive index values were normalized to the competitive index of arabinose+ and arabinose− WT competition experiments. *Below detection limit.
In the neutropenic mouse thigh infection model, the ERN mutants were as fit as the WT strain, but for the LPS mutants a >3 log reduction in competitive indices was observed (Figure 3b). Contrary to the peritonitis model, in the thigh infection model the immune system was compromised due to the depletion of neutrophils. Thus, in the thigh muscle the drop in bacterial counts observed for the LPS mutants was mainly due to slower growth (Figure S5). Overall, the ERN mutants were able to compete with the WT strain much better within the thigh muscle compared with the LPS mutants.
Discussion
We examined how fitness was affected by mutations that confer reduced susceptibility to tigecycline by subjecting genetically defined mutants to a set of in vitro conditions mimicking environments they might encounter in vivo. First, we tested if the acid response was affected by the accumulation of mutations in efflux regulation and LPS synthesis and assembly targets. Both mutant groups were similar to the WT strain with regard to acid survival (Figure S1), probably because neither up-regulation of the AcrAB efflux nor the deep-rough phenotype is directly involved in acid responses in E. coli. Lon protease, however, has been implicated in acid resistance in E. coli by shutting the response down rather than activating it.9
Bile primarily acts as a detergent that solubilizes fats, and additionally it has antimicrobial properties through its destructive effect on bacterial membranes. E. coli typically has high-level bile tolerance as it normally resides in the gastrointestinal tract of the host where bile is abundant.10 There was a significant decrease in bile tolerance in reconstructed mutants with LPS defects compared with the WT and ERN mutants, in accordance with previous observations.11 As can be seen in Figure 1, the WT strain was able to grow at very high bile salt concentrations. This characteristic of E. coli was previously connected to expression of the AcrAB efflux system.12 It has been demonstrated that bile salts activate the positive transcriptional regulator Rob, which in turn up-regulates the AcrAB pump leading to high bile tolerance.13 Therefore, up-regulation of AcrAB through the MarA regulator, which is directly affected by mutations in lon and marR genes, would not be necessary.
ERN and LPS mutants were exposed to oxidative stress and human serum, environmental conditions that E. coli would encounter during extraintestinal infections. No significant differences in viability were observed between the reconstructed mutants and the WT control during the challenge with paraquat (Figure S2). The Lon protease is involved in turnover of the SoxS regulator,14 which is a principal activator of the superoxide radical response in E. coli, but as discussed for the acid response above, the effect of the Lon protease is not in activation of the response but rather in deactivation.
Gram-negative bacteria are protected from the antibacterial activity of serum by the polysaccharide chains of the O-antigen and K-antigen. However, if LPS is truncated and bacteria express a rough or deep-rough phenotype, they become susceptible to serum.15–18 In a recent study, the production of colanic acid (M-antigen) has been suggested to contribute to serum resistance in E. coli ST131.19 Colanic acid biosynthesis is controlled by an Rcs phosphorelay with the regulator RcsA being a substrate of the Lon protease.20 Over-production of colanic acid, which leads to the mucoid phenotype, has been shown in lon mutants of E. coli where RcsA is not degraded by the Lon protease.21 The lon mutant in our study had a mucoid appearance, which was most likely caused by the over-production of M-antigen. It is possible that the protective effect of M-antigen against human serum could explain the ∼2 log difference in viable counts between the lon mutant and the WT after treatment with the highest concentrations of serum (Figure S3a). Even though up-regulation of colanic acid production has been demonstrated in deep-rough mutants of E. coli,22 the LPS mutants in our study did not behave differently from the WT strain in the serum resistance assay.
In vitro competition experiments were performed to evaluate fitness during the complete growth cycle of the reconstructed ERN and LPS mutants. Moreover, we were interested in determining the MSC values for each mutation (Figure 2). Among the reconstructed mutants the MSC value for the marR mutant was well below the MIC for the WT, indicating that mutants with an up-regulated AcrAB system could be selected by low tigecycline concentrations in the environment or body tissues. Our findings (Table 1) are in accord with the results reported in clinical E. coli isolates with elevated tigecycline MICs, where frameshift mutation in marR led to constitutive overexpression of marA and acrAB.23 The LPS mutants, on the other hand, would be more difficult to enrich at low tigecycline concentrations due to their higher fitness cost.
Combinations of ERN and LPS mutations were evaluated in terms of MICs, exponential growth rates and relative expression of efflux-related genes. As expected, all double mutants had higher MICs and lower growth rates than the corresponding single reconstructed mutants (Table 1). Additionally, the double mutants were also comparable to the second-step selected mutants from our previous study5 with the lon, marR combination being the most costly and conferring the highest resistance and highest expression of acrAB genes.
To evaluate mutant survival in vivo we used the peritonitis and neutropenic mouse thigh infection models. The mutants were competed in vivo against the WT strain inside the peritoneal cavity (Figure 3a) and within the thigh muscle (Figure 3b). In the latter model the levels of neutrophils were reduced. While overall we did not observe any substantial differences in competitions between mutants and the WT within the peritoneal cavity, LPS mutants performed poorly compared with the WT in the thigh infection model. Even though the two animal models in our study are not directly comparable due to different infection sites and duration, an important point is that the neutrophil response was functional in the peritonitis model, but compromised in the thigh muscle. Therefore, we mainly observed killing of bacteria in the peritonitis model with no net growth (Figure S4), while the differences observed between ERN and LPS mutants during the thigh infection were mainly because of different growth rates (Figure S5). These results suggest that a truncated LPS is less deleterious for E. coli in the peritoneal cavity with active neutrophils, but disadvantageous for survival in thigh muscle with a compromised neutrophil response.
In conclusion, our study provides a potential explanation for why increased efflux is the main mechanism observed in the clinic that leads to tigecycline resistance. Thus, the majority of the in vitro and in vivo fitness assays indicate that mutants with reduced susceptibility to tigecycline due to a truncation of LPS are generally less fit than the ERN mutants. Therefore, it is likely that the fitter ERN mutants are more successful in surviving and causing infections in clinical settings.
Funding
This work was supported by a grant from the Swedish Research Council—Medicine and Health (K2013-99X-22208-01-5) to L. S. and grants from the Swedish Research Council—Medicine and Health (K2013-56X-13025-15), the Swedish Research Council—Formas (2013-5476-25194-9) and the European Commission (project 282004 EvoTAR) to D. I. A.
Transparency declarations
None to declare.
Supplementary data
Figures S1 to S5 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
We thank Carina Vingsbo Lundberg and Niels Frimodt Møller for advice on animal experiments.
References
- 1.WHO. Antimicrobial Resistance: Global Report on Surveillance 2014. 2014: 1–256. http://www.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1.
- 2.Petersen PJ, Jacobus NV, Weiss WJ et al. . In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob Agents Chemother 1999; 43: 738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sum PE, Petersen P. Synthesis and structure-activity relationship of novel glycylcycline derivatives leading to the discovery of GAR-936. Bioorg Med Chem Lett 1999; 9: 1459–62. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y, Cai Y, Liu X et al. . The emergence of clinical resistance to tigecycline. Int J Antimicrob Agents 2013; 41: 110–6. [DOI] [PubMed] [Google Scholar]
- 5.Linkevicius M, Sandegren L, Andersson DI. Mechanisms and fitness costs of tigecycline resistance in Escherichia coli. J Antimicrob Chemother 2013; 68: 2809–19. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 2000; 97: 6640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu D, Ellis HM, Lee EC et al. . An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 2000; 97: 5978–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gullberg E, Cao S, Berg OG et al. . Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 2011; 7: e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heuveling J, Possling A, Hengge R. A role for Lon protease in the control of the acid resistance genes of Escherichia coli. Mol Microbiol 2008; 69: 534–47. [DOI] [PubMed] [Google Scholar]
- 10.Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev 2005; 29: 625–51. [DOI] [PubMed] [Google Scholar]
- 11.Picken RN, Beacham IR. Bacteriophage-resistant mutants of Escherichia coli K12. Location of receptors within the lipopolysaccharide. J Gen Microbiol 1977; 102: 305–18. [DOI] [PubMed] [Google Scholar]
- 12.Thanassi DG, Cheng LW, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol 1997; 179: 2512–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg EY, Bertenthal D, Nilles ML et al. . Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol 2003; 48: 1609–19. [DOI] [PubMed] [Google Scholar]
- 14.Griffith KL, Shah IM, Wolf RE. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol Microbiol 2004; 51: 1801–16. [DOI] [PubMed] [Google Scholar]
- 15.Rowley D. Sensitivity of rough gram-negative bacteria to the bactericidal action of serum. J Bacteriol 1968; 95: 1647–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opal S, Cross A, Gemski P. K antigen and serum sensitivity of rough Escherichia coli. Infect Immun 1982; 37: 956–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross AS, Kim KS, Wright DC et al. . Role of lipopolysaccharide and capsule in the serum resistance of bacteremic strains of Escherichia coli. J Infect Dis 1986; 154: 497–503. [DOI] [PubMed] [Google Scholar]
- 18.Burns SM, Hull SI. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect Immun 1998; 66: 4244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phan M-D, Peters KM, Sarkar S et al. . The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet 2013; 9: e1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres-Cabassa AS, Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol 1987; 169: 981–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markovitz A. Regulatory mechanisms for synthesis of capsular polysaccharide in mucoid mutants of Escherichia coli K12. Proc Natl Acad Sci USA 1964; 51: 239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker CT, Kloser AW, Schnaitman CA et al. . Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol 1992; 174: 2525–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeney D, Ruzin A, McAleese F et al. . MarA-mediated overexpression of the AcrAB efflux pump results in decreased susceptibility to tigecycline in Escherichia coli. J Antimicrob Chemother 2008; 61: 46–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.