Abstract
Objectives
The Gram-negative bacillus Stenotrophomonas maltophilia (SM) is an emerging MDR opportunistic pathogen. Recent studies identify a potentially relevant activity of azithromycin against Gram-negative bacteria overlooked in standard bacteriological testing. We investigated azithromycin activity against SM in testing conditions incorporating mammalian tissue culture medium and host defence factors.
Methods
MIC testing, chequerboard assays, time–kill assays and fluorescence microscopy were performed for azithromycin, the cationic peptide antibiotic colistin and the human defence peptide cathelicidin LL-37 alone or in combination in cation-adjusted Mueller–Hinton broth or mammalian tissue culture media. Azithromycin sensitization of SM to host immune clearance was tested in a human neutrophil killing assay and a murine pneumonia model.
Results
We observed potent bactericidal activity of azithromycin against SM in mammalian tissue culture medium absent in bacteriological medium. Colistin and LL-37 strongly potentiated azithromycin killing of SM by increasing drug entry. Additionally, azithromycin sensitized SM to neutrophil killing and increased SM clearance in the murine pneumonia model.
Conclusions
Despite lack of activity in standard MIC testing, azithromycin synergizes with cationic peptide antibiotics to kill SM in medium mimicking tissue fluid conditions. Azithromycin, alone or in combination with colistin, merits further exploration in therapy of drug-resistant SM infections.
Introduction
Stenotrophomonas maltophilia (SM) is a ubiquitous MDR Gram-negative bacillus and opportunistic pathogen. Risk factors for infection include malignancy, neutropenia, HIV, cystic fibrosis, mechanical ventilation, ICU admission, long-term central venous catheter use, recent surgery, trauma and prior broad-spectrum antibiotic administration. Common SM infections are pneumonia and bacteraemia, with reported mortality of 18%–69%.1 The incidence of SM infections ranges from 7.1 to 14.1 per 10 000 inpatients and is increasing, with intrinsic or acquired antibiotic resistance presenting great therapeutic challenges in severe infection.2,3 Trimethoprim/sulfamethoxazole and ticarcillin/clavulanic acid (no longer manufactured in the USA) are first- and second-line treatments for serious SM infections, but recent studies have found in vitro resistance to these agents to be as high as 30% and 41%, respectively.4,5
Clinical data regarding optimal therapy for SM infection are limited. Moreover, in vitro antimicrobial susceptibility testing results may not correctly predict SM clinical treatment response with reported discrepancies among testing methods (e.g. Etest, disc diffusion versus agar dilution) and differing guidelines (e.g. EUCAST versus CLSI). Consequently, SM MIC results vary according to the conditions and methods implemented.3 There is a great need to identify innovative treatment strategies for SM infection and a consistent antimicrobial susceptibility scheme to guide such therapy.
Macrolides are never recommended for SM therapy based on high MIC values in standard assays. However, recent studies have indicated that conventional antimicrobial susceptibility testing conditions may overlook macrolide activity and antibiotic synergies against other MDR Gram-negative bacteria including Pseudomonas aeruginosa, Klebsiella pneumoniae and Acinetobacter baumannii.6,7 Here, we examine the potential of the commonly prescribed macrolide azithromycin to combat SM under more physiological medium conditions, in synergy with colistin or endogenous host defence factors (cathelicidin LL-37 and neutrophils) in vitro, and in vivo using a murine pneumonia model.
Materials and methods
Bacterial strains, media and antibiotics
MDR SM isolate K279a (ATCC BAA-2423) was utilized in all experiments. Additional MDR SM clinical isolates were obtained from the University of California, San Diego (UCSD) Center for Advanced Laboratory Medicine (three strains) and the Laboratory of Bacterial Drug Resistance, Gunma University Graduate School of Medicine (five strains). Isolates were stored in LB broth + 50% glycerol at −80°C until use. Azithromycin and colistin were purchased from Sigma–Aldrich and cathelicidin LL-37 from American Peptide. Bacteriological medium Mueller–Hinton broth (Spectrum Chemicals) was supplemented with 20–25 mg/L Ca2+ and 10–12.5 mg/L Mg2+ (CA-MHB). Tissue culture medium Roswell Park Memorial Institute 1640 (RPMI) (ThermoFisher Scientific) was supplemented with 10% LB broth (Hardy Diagnostics) (RPMI + 10%LB).
MIC, chequerboard and time–kill assays
Broth microdilution antimicrobial susceptibility testing was performed in CA-MHB according to CLSI guidelines and in RPMI + 10%LB. Chequerboard panels were likewise performed in both media.8,9 Synergy, indifference and antagonism were defined by the fractional inhibitory concentration index (FICI): FICI ≤ 0.5 = synergy, FICI > 0.5 to ≤4 = indifference and FICI > 4 = antagonism. Time–kill assays were performed in CA-MHB and RPMI + 10%LB as previously described, with bactericidal activity defined as a reduction in viable bacteria by ≥3 log10 cfu/mL; synergy, ≥2 log10 cfu/mL reduction; indifference, <2 log10 cfu/mL reduction; and antagonism, >2 log10 cfu/mL increase.10
Fluorescence microscopy
Fluorescence microscopy was performed as previously described with the following modifications.11 For ready visualization, SM was used at a 2 log10 cfu/mL higher concentration than in MIC testing, along with higher, but pharmacologically achievable, concentrations of antibiotics [NBD-tagged azithromycin (20× MIC), colistin (8× 1/4 MIC) and LL-37 (10× 1/4 MIC)].12 Bacteria were labelled with 1 μg/mL FM4-64 (membrane) and 2 μg/mL DAPI (nucleic acid) (Molecular Probes) prior to transfer onto a 1.2% agarose pad containing 20% LB broth for microscopy and image analysis using ImageJ software v1.48f and CellProfiler 2.0.
Neutrophil killing assays
Human neutrophils were isolated from healthy donors using the PolymorphPrep system (Axis-Shield) under protocols approved by the UCSD Human Subjects Institutional Review Board for use in established bacterial killing assays with minor modifications.13 Neutrophils were resuspended in RPMI to 2 × 106 cells/mL, stimulated with 25 nM phorbol-12-myristate-13-acetate for 2 h at 37°C, washed and used to seed a 96-well plate (2 × 105 cells/well). Cells were infected at an moi equal to 50 with SM that were untreated or exposed overnight to a sub-bacteriostatic concentration (0.03 mg/L) of azithromycin.
Murine pneumonia model
Adapting a published pneumonia model, A/J mice (8–10 weeks old, Jackson Laboratory) were infected intratracheally with 2 × 106 cfu (low inoculum) or 2 × 107 cfu (high inoculum) of SM (K279a) and treated with PBS control or 50 mg/kg azithromycin subcutaneously (sc) every 24 h for one dose (low inoculum) or two doses (high inoculum).14 Animals were euthanized and lungs harvested, weighed, homogenized, serially diluted and plated onto LB agar to enumerate cfu after 24 h of incubation at 37°C. In a subset of mice infected at the high inoculum, bronchoalveolar lavage fluid (BALF) was collected at 48 h by exposing the trachea, injecting 0.8 mL of cold PBS and retrieving BALF using an 18 gauge needle connected to a 1 mL syringe. BALF was centrifuged at 500 g for 10 min and supernatants frozen at −80°C for later cytokine analysis using commercial ELISA kits for IL-1β (R&D Systems) and macrophage inflammatory protein 2 (MIP-2) (BD Biosciences). Pelleted BALF cells were resuspended in 0.25 mL cold PBS and enumerated utilizing a haemocytometer, light microscopy and Wright–Giemsa staining to determine the differential leucocyte count. Excised lungs from four mice in each group were fixed using 4% formalin, embedded in paraffin, sectioned and stained with haematoxylin–eosin for blinded analysis by a pathologist. A graded scale of 0 (absent) to 3 (extensive) was used to assess overall airspace involvement, perivascular/peribronchial inflammation, evidence of inflammation and oedema/debris or macrophages in the intra-alveolar space. All procedures were performed by a protocol approved by the UCSD Institutional Animal Care and Use Committee.
Statistics
Statistical analyses were performed using GraphPad Prism 6.0f (GraphPad Software). One-way ANOVA, two-way ANOVA or unpaired Student's t-test were utilized where appropriate. P values <0.05 were regarded to be statistically significant.
Supplementary methods
Supplementary methods (growth curve analysis, electron microscopy, neutrophil extracellular trap and oxidative burst studies) are available as Supplementary data at JAC Online.
Results
Markedly increased activity of azithromycin against SM in tissue culture medium versus bacteriological medium
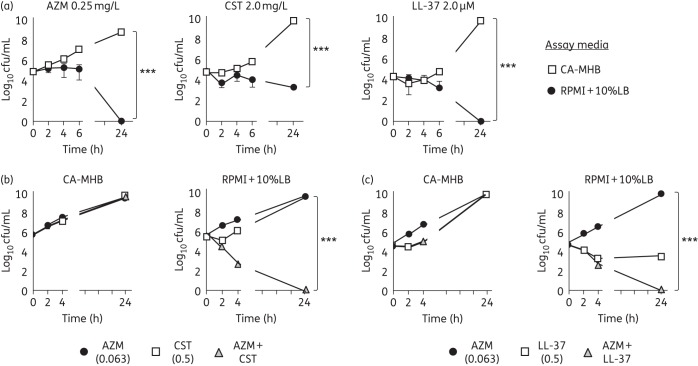
Azithromycin MICs for several MDR SM strains were determined by CLSI broth microdilution methodology using either the recommended bacteriological medium, CA-MHB, or the common mammalian tissue culture medium RPMI supplemented with 10% LB broth to support equivalent bacterial growth (Figure S1). A >500-fold reduction in MIC (≤0.25 versus ≥128 mg/L) was observed for nearly all isolates tested with RPMI + 10%LB compared with CA-MHB (Table 1). Transmission electron microscopy of SM K279a cells treated with azithromycin, a ribosomal protein synthesis inhibitor, in RPMI + 10%LB revealed ribosomal clustering consistent with its known target of action (Figure S2). In a kinetic killing assay, 0.25 mg/L azithromycin had no effect on SM K279a growth in CA-MHB, but reduced bacterial cfu below detectable levels within 24 h in RPMI + 10%LB (Figure 1a).
Table 1.
MICs of azithromycin for nine different SM strains
| SM strain | MIC (mg/L) |
|
|---|---|---|
| CA-MHB | RPMI + 10%LB | |
| K279a (ATCC BAA-2423) | 256 | 0.25 |
| SM 5 | 32 | 0.06 |
| SM 12 | 256 | 0.03 |
| SM 26 | 128 | 0.03 |
| SM 57 | 128 | 0.03 |
| SM 65 | 256 | 0.06 |
| ATCC 51331 | 256 | 0.06 |
| SM BWR | 256 | 0.03 |
| SM NP | 256 | 0.25 |
Comparative MIC testing performed in standard bacteriological (CA-MHB) and supplemented mammalian tissue culture (RPMI + 10%LB) media.
Figure 1.
Azithromycin bactericidal activity against SM observed in media alone and in synergy with cationic antimicrobial peptides. (a) Time–kill curve demonstrating azithromycin, colistin and LL-37 activity against SM K279a in bacteriological (CA-MHB) versus tissue culture-based (RPMI + 10%LB) media. (b and c) Bactericidal synergy, defined as a ≥2 log10 decrease in cfu/mL for time–kill assays, was observed for both azithromycin + colistin and azithromycin + LL-37 at 1/4 MIC of all agents (FICI ≤ 0.5), but only in RPMI + 10%LB. Data are plotted as mean ± SEM and represent the combination of three experiments performed in triplicate. ***P < 0.001 by two-way ANOVA. AZM, azithromycin; CST, colistin.
Cationic peptides potentiate the bactericidal activity of azithromycin against SM by increasing drug entry
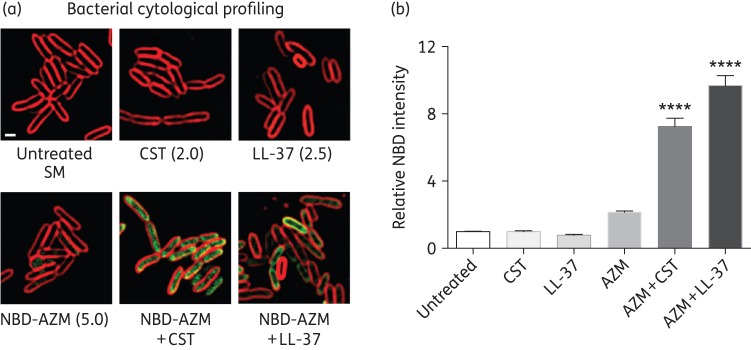
We recently observed synergy of azithromycin with cationic antimicrobial peptides against other Gram-negative bacilli.7 The cationic peptide antibiotic colistin, a drug of last resort for certain MDR pathogens, is bacteriostatic against SM in RPMI + 10%LB, but not in CA-MHB (Figure 1a). At 1/4 MIC of both drugs, azithromycin showed potent bactericidal synergy with colistin in RPMI + 10%LB, but not CA-MHB (Figure 1b). The human cathelicidin LL-37 is a cationic endogenous host defence peptide abundantly produced by neutrophils and epithelial cells. Once again, at 1/4 MIC of each agent, azithromycin showed bactericidal synergy with LL-37 in RPMI + 10%LB, but not CA-MHB (Figure 1c). Using fluorescence microscopy-based bacterial cytological profiling, both colistin and LL-37, which are known to form pores and disrupt bacterial membranes, markedly enhanced entry of fluorescently labelled (NBD-tagged) azithromycin into bacterial cells (Figure 2a and b), facilitating azithromycin access to the 50S ribosomal subunit.
Figure 2.
Bacterial cytological profiling showing that cationic peptides facilitate azithromycin entry into SM cells. (a) Fluorescence microscopy performed using log-phase SM K279a treated for 1 or 2 h with NBD-tagged azithromycin (5 mg/L) (green), colistin (2 mg/L) or LL-37 (2.5 μM) alone or in combination. (b) Bar graphs generated from software analysis of multiple random fluorescent microscopy fields of cells treated with NBD-tagged azithromycin, colistin, LL-37, NBD-tagged azithromycin + colistin and NBD-tagged azithromycin + LL-37 (with >500 cells counted per condition). Azithromycin + colistin- and azithromycin + LL-37-treated bacteria had 10-fold higher NBD intensity than azithromycin, colistin or LL-37 alone. Data are plotted as mean ± SEM and represent the combination of three experiments performed in triplicate. ****P < 0.001 by one-way ANOVA. AZM, azithromycin; CST, colistin.
Azithromycin sensitizes SM to neutrophil killing and increases SM clearance in vivo
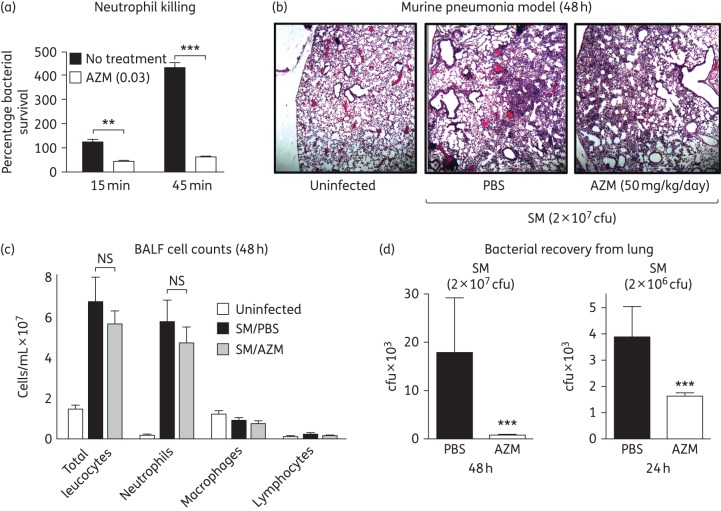
Azithromycin synergizes with LL-37 against SM, suggesting it could sensitize the opportunistic pathogen to killing by neutrophils, which produce the defence peptide as part of their antimicrobial arsenal. SM induced oxidative burst and the formation of neutrophil extracellular traps (in which LL-37 is deployed) from freshly isolated human neutrophils (Figure S3). Overnight pretreatment of SM with a sub-bacteriostatic concentration of azithromycin sensitized the bacterium to human neutrophil killing (Figure 3a). We next assessed the efficacy of azithromycin monotherapy in an adapted murine model of SM pneumonia, using immune susceptible A/J mice that have functional C5 complement deficiency. Under the experimental conditions, azithromycin treatment did not affect the acute histopathological features of inflammatory pneumonia (Figure 3b and Figure S4A), the amount or composition of BALF leucocyte infiltration (Figure 3c) nor BALF levels of pro-inflammatory cytokines MIP-2 or IL-1β (Figure S4B) at 48 h post-infection. However, the recovered cfu of SM from the lungs was markedly reduced by 58% in azithromycin-treated mice (1629 ± 123 cfu/g) versus PBS controls (3898 ± 1154 cfu/g) at 24 h in a lower inoculum challenge and reduced by 95% in azithromycin-treated mice (824 ± 184 cfu/g) versus PBS controls (17 915 ± 11 186 cfu/g) at 48 h in a higher inoculum challenge (Figure 3d).
Figure 3.
Azithromycin sensitizes SM to neutrophil killing and increases SM clearance in vivo. (a) Percentage survival of untreated versus azithromycin-pretreated SM K279a bacteria in a human neutrophil killing assay. (b) Representative lung histopathology of A/J mice infected intratracheally with high-inoculum SM K279a or PBS control and treatment with daily azithromycin or PBS control. (c) Total enumeration of leucocytes in BALF of mice including neutrophils, macrophages and lymphocytes. (d) Bacterial recovery (cfu/g) from lung homogenates of A/J mice intratracheally infected with different inocula of SM K279a and treated with azithromycin (50 mg/kg sc daily) for 24 or 48 h. n = 20 per group (24 h) and 11 per group (48 h). For all panels (a–d), data represent the mean ± SEM from the combination of three experiments performed in triplicate. **P < 0.01 and ***P < 0.001 by two-way ANOVA (a) or unpaired Student's t-test (d). NS, no statistical significance by unpaired Student's t-test (c). AZM, azithromycin.
Discussion
Using SM K279a, a strain with several genes conferring resistance to heavy metals and antimicrobials (including nine resistance–nodulation–division-type efflux pump genes), and several contemporary clinical SM isolates, this study demonstrated a striking difference of azithromycin potency in standard bacteriological testing medium (negligible activity, MIC as high as 256 mg/L) compared with mammalian tissue culture medium (MIC ≤0.25 mg/L) supplemented with 10% LB broth.15 While there is no current clinical MIC breakpoint of azithromycin for SM, the EUCAST breakpoint used for the Gram-negative bacillus Campylobacter jejuni is 4 mg/L. Azithromycin also showed potent synergy with colistin, an antibiotic of last resort for MDR Gram-negative bacilli infections, and LL-37, an endogenous cationic antimicrobial peptide. As disruptors of bacterial membranes, colistin and LL-37 potentiate azithromycin entry into bacterial cells and enhance azithromycin access to its ribosomal target.
Recent studies suggest different antibiotics may work in concert or at odds with our innate immune system in the setting of infection.13,16 However, our understanding of the interactions between common antibiotics prescribed in clinical practice and endogenous antimicrobials such as LL-37 and the cells that produce them (e.g. neutrophils) remains limited. Whereas SM was impervious to human neutrophil killing in our ex vivo assay, it was sensitized to neutrophil killing by azithromycin pretreatment. Moreover, azithromycin showed the ability to significantly reduce SM bacterial counts in an in vivo murine pneumonia challenge model in which a predominantly neutrophilic inflammatory infiltrate was elicited in the lungs.
Traditional SM antimicrobial susceptibility testing can be problematic given inconsistencies between the results of different methods, the lack of a gold standard or universal reference method and the inability of testing results to reliably and appropriately translate to clinical efficacy. Conventional in vitro studies also inadequately account for the dynamic interactions between conventional antibiotics and the host immune response. Our study and others suggest that current standardized susceptibility testing in bacteriological medium may fail to adequately detect the bactericidal activity of azithromycin and its interactions with the innate immune system against various MDR Gram-negative bacilli, including SM.6,7
Limitations of our study include the modest number of SM isolates tested and the reliance on a murine pneumonia model for in vivo azithromycin activity against SM without human clinical data. Nevertheless, this preliminary investigation highlights a potential utility of azithromycin, a safe and established antibiotic, used alone or in combination with colistin, in SM infection. The work also emphasizes the importance of accounting for the dynamic interaction between conventional antibiotics and host immunity in the setting of infection. Future randomized clinical trials in humans with azithromycin will be required to determine the true utility of this finding.
Funding
This work was supported by National Institutes of Health grants T32-AI007036 (to M. K.), HD071600 (to G. S. and V. N.), AI052453 and AR052728 (to V. N.) and AI113295 (to J. P.).
Transparency declarations
None to declare.
Supplementary data
Acknowledgements
We thank Dr Sharon Reed (University of California, San Diego), Dr Nicholas P. Cianciotto (Northwestern University Medical School) and Dr Koichi Tanimoto (Gunma University Graduate Medical School) for providing bacterial isolates. We also thank Ying Jones, Timo Meerloo and Dr Marilyn G. Farquhar of the UCSD Electron Microscopy Core Facility for their assistance with electron microscopy studies.
References
- 1.Paez JI, Costa SF. Risk factors associated with mortality of infections caused by Stenotrophomonas maltophilia: a systematic review. J Hosp Infect 2008; 70: 101–8. [DOI] [PubMed] [Google Scholar]
- 2.Morrison AJ Jr, Hoffmann KK, Wenzel RP. Associated mortality and clinical characteristics of nosocomial Pseudomonas maltophilia in a university hospital. J Clin Microbiol 1986; 24: 52–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 2012; 25: 2–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang CH, Lin JC, Lin HA et al. . Comparisons between patients with trimethoprim-sulfamethoxazole-susceptible and trimethoprim-sulfamethoxazole-resistant Stenotrophomonas maltophilia monomicrobial bacteremia: a 10-year retrospective study. J Microbiol Immunol Infect 2014; doi:10.1016/j.jmii.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Nicodemo AC, Araujo MR, Ruiz AS et al. . In vitro susceptibility of Stenotrophomonas maltophilia isolates: comparison of disc diffusion, Etest and agar dilution methods. J Antimicrob Chemother 2004; 53: 604–8. [DOI] [PubMed] [Google Scholar]
- 6.Buyck JM, Plesiat P, Traore H et al. . Increased susceptibility of Pseudomonas aeruginosa to macrolides and ketolides in eukaryotic cell culture media and biological fluids due to decreased expression of oprM and increased outer-membrane permeability. Clin Infect Dis 2012; 55: 534–42. [DOI] [PubMed] [Google Scholar]
- 7.Lin L, Nonejuie P, Munguia J et al. . Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant Gram-negative bacterial pathogens. EBioMedicine 2015; 2: 690–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fifth Informational Supplement M100-S25. CLSI, Wayne, PA, USA, 2015. [Google Scholar]
- 9.Sopirala MM, Mangino JE, Gebreyes WA et al. . Synergy testing by Etest, microdilution checkerboard, and time–kill methods for pan-drug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 2010; 54: 4678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haste NM, Hughes CC, Tran DN et al. . Pharmacological properties of the marine natural product marinopyrrole A against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2011; 55: 3305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nonejuie P, Burkart M, Pogliano K et al. . Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc Natl Acad Sci USA 2013; 110: 16169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matijasic M, Munic Kos V, Nujic K et al. . Fluorescently labeled macrolides as a tool for monitoring cellular and tissue distribution of azithromycin. Pharmacol Res 2012; 66: 332–42. [DOI] [PubMed] [Google Scholar]
- 13.Sakoulas G, Okumura CY, Thienphrapa W et al. . Nafcillin enhances innate immune-mediated killing of methicillin-resistant Staphylococcus aureus. J Mol Med 2014; 92: 139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouf R, Karaba SM, Dao J et al. . Stenotrophomonas maltophilia strains replicate and persist in the murine lung, but to significantly different degrees. Microbiology 2011; 157: 2133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crossman LC, Gould VC, Dow JM et al. . The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 2008; 9: R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kristian SA, Timmer AM, Liu GY et al. . Impairment of innate immune killing mechanisms by bacteriostatic antibiotics. FASEB J 2007; 21: 1107–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.