Abstract
Mutations in the human telomerase RNA (hTR), the telomerase RNP component dyskerin (DKC1), and the poly(A) ribonuclease (PARN) can lead to reduced levels of hTR and dyskeratosis congenita (DC). However, the enzymes and mechanisms responsible for hTR degradation are unknown. We demonstrate that defects in dyskerin binding lead to hTR degradation by PAPD5-mediated oligoadenylation promoting 3’ to 5’ degradation by EXOSC10, as well as decapping and 5’ to 3’ decay by the cytoplasmic DCP2 and XRN1 enzymes. PARN increases hTR levels by deadenylating hTR, thereby limiting its degradation by EXOSC10. Telomerase activity and proper hTR localization in dyskerin- or PARN-deficient cells can be rescued by knockdown of DCP2 and/or EXOSC10. Prevention of hTR RNA decay also leads to a rescue of localization of DC-associated hTR mutants. These results suggest that inhibition of RNA decay pathways might be a useful therapy for some telomere pathologies.
Telomere elongation by telomerase is required for continuous proliferation of human stem cells1. Insufficient telomerase levels are responsible for the inherited disorder dyskeratosis congenita and contribute to more common diseases such as aplastic anemia and idiopathic pulmonary fibrosis2. Telomerase is a ribonucleoprotein (RNP) enzyme, and disease-causing mutations have been identified in its RNA component (hTR), its catalytic protein subunit (hTERT), and the small nucleolar ribonucleoprotein dyskerin, which stabilizes hTR in the nucleus even in the absence of hTERT3–6. More recently, mutations in the poly(A) ribonuclease (PARN) have been identified in patients suffering from a severe form of dyskeratosis congenita, as well as idiopathic pulmonary fibrosis7–9. An unaddressed issue is how hTR is degraded and the interplay between those RNA degradation systems and mutations in PARN and dyskerin. Our goal was to investigate the quality control pathways of hTR when dyskerin or PARN is depleted from cells, and whether inhibition of RNA quality control pathways can rescue hTR levels and function under these conditions.
Results
Loss of dyskerin binding leads to hTR degradation
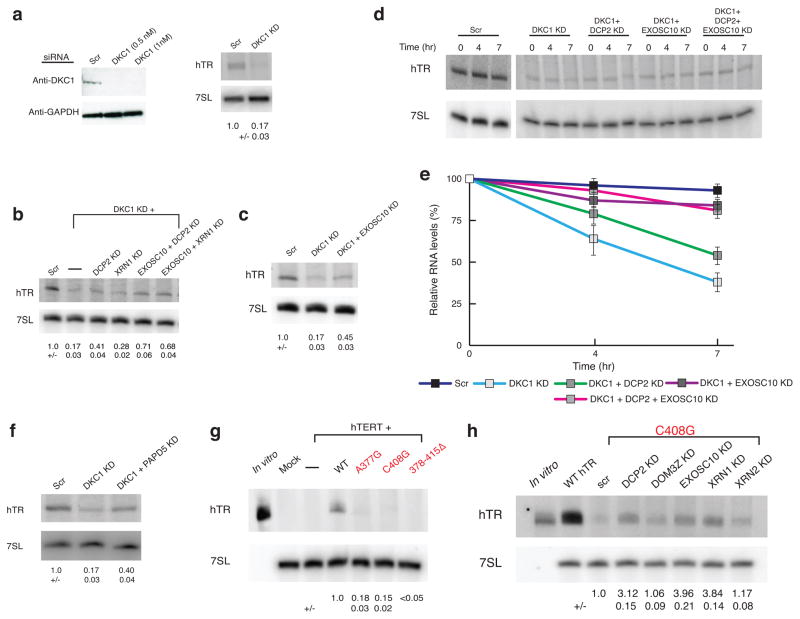
To determine how hTR is degraded when dyskerin levels are reduced, we examined how knockdowns of RNA decay enzymes affected endogenous hTR levels when dyskerin was depleted in HeLa cells. We observed that knockdown of dyskerin reduced hTR to 17% of wild-type levels (Fig. 1a), but could be partially rescued by knockdown of DCP2 (~2.5X, from 17% to 41% of control) or of XRN1 (~1.6X, from ~17% to 28% of control) (Fig. 1b). DCP2 and XRN1 are components of a cytoplasmic decapping and 5’ to 3’ decay pathway. We also observed that hTR levels upon DKC1 knockdown can be rescued by knockdown of the nuclear 3’ to 5’ exonuclease EXOSC10 (~2.6X from 17% to 45% of control) (Fig. 1b, c). Moreover, double knockdown of EXOSC10 and DCP2 or XRN1 restored hTR levels in the dyskerin-depleted cells to ~70% of the wild-type levels.
Figure 1.
Lack of dyskerin binding reduces hTR levels by two different RNA decay mechanisms. a, Western blot for DKC1 knockdown and northern blot for hTR in HeLa cells (mean +/−s.d., n=5 independent experiments). Scr, scrambled negative control siRNA. b, c, Representative northern blot for hTR upon DKC1 knockdown and rescue in HeLa cells (mean+/−s.d., n=4 independent experiments. d,e, Representative northern blot for hTR upon Actinomycin D shutoff in dyskerin knockdown and rescue in HeLa cells. Measurement of hTR decay rate in dyskerin knockdown HeLa cells. Error bars, s.d. (n=3 independent experiments). f, Northern blot showing hTR upon DKC1 knockdown with or without PAPD5 knockdown (mean+/−s.d., n=3 independent experiments). g, Northern blot for wild type (WT) disease-causing hTR mutants in U2OS cells (mean+/−s.d., n=3 independent experiments). h, Northern blot for C408G hTR RNA upon DCP2, EXOSC10 or XRN1 knockdown (mean+/−s.d., n=4 independent experiments).
To determine if these differences in RNA levels were due to changes in hTR stability, we measured the decay of hTR following inhibition of transcription with actinomycin D. As expected, we observed that in dyskerin knockdown cells, hTR was degraded faster than in control cells (Fig. 1d). More importantly, knockdown of DCP2, EXOSC10, or both led to a decrease in hTR decay in the dyskerin knockdown cells (Fig. 1d, e). These results indicate that in the absence of dyskerin, hTR can be degraded by DCP2-mediated decapping leading to 5’ to 3’ degradation by XRN1, as well as by nuclear 3’ to 5’ degradation by EXOSC10.
The exonucleolytic activity of EXOSC10 is enhanced by the poly(A) polymerase PAPD5, which adds 3’ oligo(A) tails to promote substrate recognition by EXOSC1010–14. To determine if PAPD5 affected hTR degradation, we examined if PAPD5 knockdown could rescue the reduced hTR levels in dyskerin-deficient cells. We observed that PAPD5 knockdown increased hTR levels 2.4X (from 17% to 40% of control) in the dyskerin knockdown (Fig. 1f). Consistent with PAPD5 adding oligo(A) tails to hTR, a subset of endogenous hTR population exists in an oligoadenylated state15.
Pathogenic point mutations in hTR that disrupt either the dyskerin binding site (A377G, 378–415Δ) or the BIO box (C408G) also reduce hTR levels6,16,17. To determine if these reduced levels could also be rescued by inhibition of RNA decay enzymes, we transfected wild-type or mutant hTR along with hTERT into U2OS cells, which do not express detectable endogenous hTR by northern blotting, and examined hTR levels by northern blots with or without knockdown of nucleases (Supplementary Fig. 1a). Compared to the wild-type hTR, the A377G, C408G, and 378–415Δ mutants have <20% steady-state hTR levels in U2OS cells (Fig. 1g). The reduced levels of the A377G and C408G hTR RNAs could be partially rescued by knockdown of DCP2 (3x), XRN1 (4X), EXOSC10 (4X) or PAPD5 (2X) (Fig. 1h and Supplementary Fig. 1b, c and d). Knockdowns of several other RNA decay enzymes did not rescue hTR levels (Fig. 1h and Supplementary Fig. 1e).
To test whether the low levels of mutant hTR might be due to reduced transcription rather than RNA stability, we examined the association of RNA polymerase II with the hTR coding gene by ChIP in U2OS cells transfected with plasmids encoding either wild-type or C408G RNA. We observed that both genes showed a similar level of RNA polymerase pol II occupancy, with the C408G plasmid-borne copy showing slightly higher occupancy (Supplementary Fig. 1f). This suggests that these mutations in hTR do not reduce hTR transcription, but rather RNA quality control pathways lead to low levels of mutant hTR under these conditions. Thus, when either dyskerin is deficient or its binding site in hTR is mutated, hTR molecules are degraded by PAPD5-mediated adenylation coupled to EXOSC10-mediated 3' to 5' degradation, as well as degradation by DCP2 and XRN1.
EXOSC10 competes with PARN for processing of mature hTR
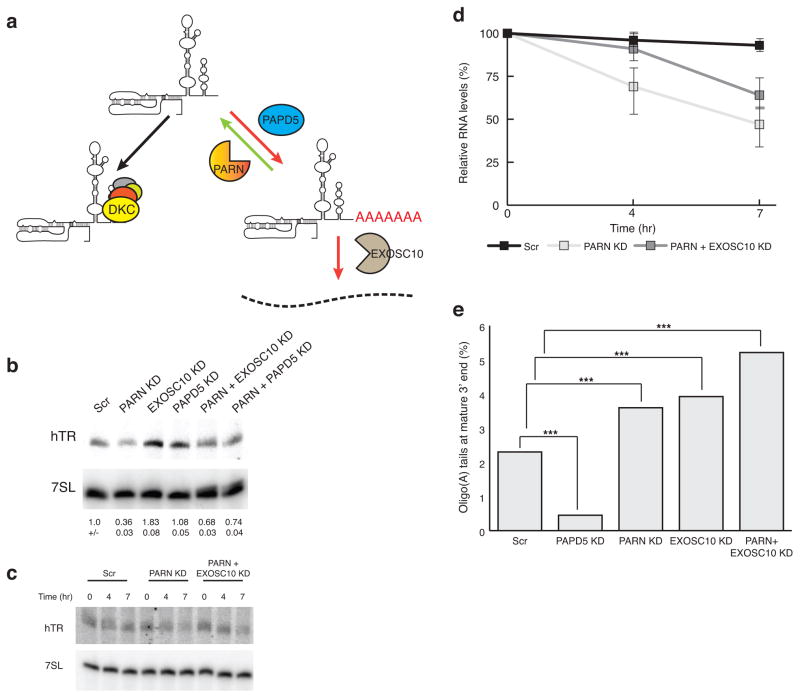
The degradation of hTR by PAPD5-promoted EXOSC10 nucleolytic activity led us to hypothesize that PARN would contribute to hTR stability by removing oligo(A) tails added by PAPD5, thereby limiting the ability of EXOSC10 to degrade hTR (Fig. 2a). Consistent with this model, PARN knockdown in HeLa cells led to reduced levels of hTR, and those were partially restored by knockdown of PAPD5 or EXOSC10 (Fig. 2b). Moreover, measurement of hTR stability following inhibition of transcription with actinomycin D showed that PARN knockdown leads to reduction in the stability of hTR in HeLa cells, which could be partially rescued by knockdown of EXOSC10 (Fig. 2 c, d). We interpret this observation to argue that PARN and EXOSC10 are competing for hTR 3’ end processing (Fig. 2a).
Figure 2.
PARN knockdown reduces hTR levels due to competing activity of EXOSC10. a, Model for competition between PARN and EXOSC10 for access to adenylated hTR 3’ end mediated by PAPD5. b, Northern blot for hTR upon PARN knockdown and rescue by EXOSC10 or PAPD5 co-knockdown (mean+/−s.d., n=6 independent experiments). c, d, Representative northern blot for Actinomycin D shutoff in PARN knockdown and rescue in HeLa cells. Measurement of hTR decay rate in PARN knockdown HeLa cells. Error bars, s.d. (n=4 independent experiments). e, Relative abundance of oligoadenylated reads at the mature hTR 3’ end with different components knocked down. Reads were normalized to the total number of mature end-containing reads under each condition.P<0.001 by two tailed Student’s t test for total number of reads in each condition(Supplementary Table 1).
To examine how 3’ adenylation of hTR was modulated by these enzymes, we sequenced the 3’ end of hTR in control cells and in cells with knockdowns of PARN, EXOSC10, PAPD5, or a double knockdown of PARN and EXOSC10. We observed a fraction of hTR with oligo(A) tails, consistent with earlier results, and this oligoadenylated fraction decreased upon PAPD5 knockdown and increased upon PARN, EXOSC10, or PARN & EXOSC10 knockdowns (Fig. 2e) (See Supplementary Table 1). The approximately fivefold reduction in the percentage of adenylated hTR molecules in the PAPD5 knockdown argues that PAPD5 adenylates hTR. Moreover, the increase in adenylated hTR molecules in PARN, EXOSC10, or the PARN and EXOSC10 knockdowns provides evidence that PARN and EXOSC10 can deadenylate and/or degrade adenylated hTR molecules (Fig. 2a). We also observed that the length distribution of residual oligo(A) tail lengths on the mature 3’ end of hTR slightly decreases with PAPD5 knockdown and increases in PARN or PARN and EXOSC10 knockdowns (Supplementary Fig. 2). Oligo(A) tails at the mature 3’ end were slightly shorter in the EXOSC10 knockdown, which is consistent with increased PARN activity in the absence of EXOSC10-mediated degradation (Supplementary Fig. 2). Thus, PARN competes with PAPD5 to limit the length of oligo(A) tails on hTR and thereby prevent EXOSC10-mediated degradation of hTR.
EXOSC10 & PARN compete for PAPD5-mediated 3’ end processing
The sequencing of the 3' ends of hTR and their adenylation status also provides several observations arguing that PAPD5, PARN and EXOSC10 also affect 3’ end processing and/or the degradation of 3’ extended hTR precursors. Compared to the mature hTR 3' end, where 2.3% of the total reads were oligoadenylated, those hTR molecules extended up to 10 nucleotides past the mature 3’ end had a much greater proportion of oligoadenylation (74.5% of total reads; see Supplementary Tables 1, 2). The percentage of these adenylated 3’ extended molecules decreased upon PAPD5 knockdown (down to 60%, Supplementary Table 2), and increased upon knockdown of PARN (up to 90.1%, Supplementary Table 2) or EXOSC10 (87.4%, Supplementary Table 2).
We also observed that PARN knockdown led to an increase in the total amount of 3' extended hTR molecules detected, from 7.2% in the control to 15.7 % upon PARN knockdown (Supplementary Table 2). We interpret this result to suggest that PARN promotes the efficient 3' end processing of at least a subset of hTR molecules, which could occur if 3' end maturation requires deadenylation prior to trimming to the mature end.
Rescue of telomerase activity in DKC1 or PARN depleted cells
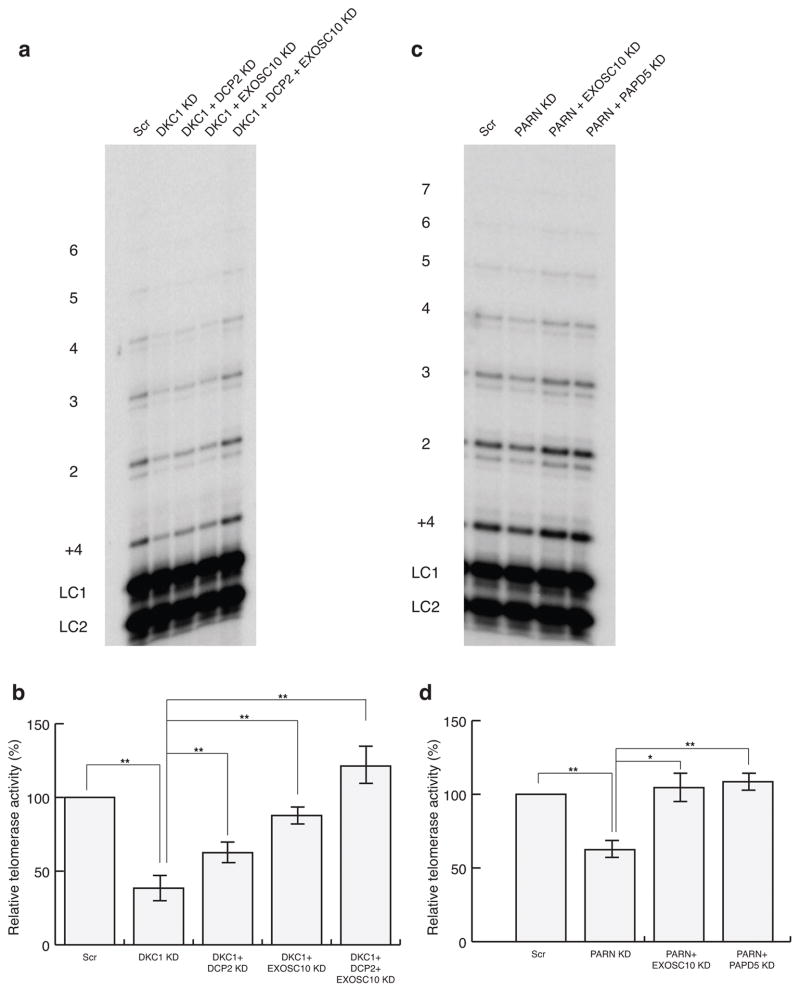
Our results indicate that knocking down RNA decay enzymes restores hTR levels in dyskerin- or PARN-deficient cells. To determine if the stabilized hTR molecules were functional, we examined the endogenous telomerase activity in dyskerin- or PARN-deficient HeLa cells, with or without rescue of hTR levels by knockdown of various nucleases. Immunoprecipitation with an anti-TERT antibody allowed a direct assay of the low endogenous telomerase activity18.
We observed that knockdown of dyskerin in HeLa cells reduced telomerase activity (Fig. 3a), but activity could be partially restored by knockdowns of DCP2 or EXOSC10 (Fig. 3a, b). A co-knockdown of EXOSC10 and DCP2 restored telomerase activity to essentially wild-type levels, in correlation with an increase in hTR levels to ~70% of WT levels under this condition (Fig. 3b, 1b). Similarly, PARN-deficient cells showed reduced telomerase activity, and this decrease was rescued by a co-knockdown of EXOSC10 or PAPD5, similar to what was observed for the hTR levels in the cells under this condition (compare Fig. 3c, d with Fig. 2b). These results indicate that the defect in telomerase activity in dyskerin- or PARN-deficient cells is due to the reduced levels of hTR, and restoration of the RNA by inhibiting its decay is sufficient to restore telomerase activity.
Figure 3.
Rescue of telomerase activity in dyskerin- or PARN-depleted HeLa cells by co-knockdown of competing nucleases. a, Autoradiograph for telomerase activity assay in HeLa DKC1 knockdown and rescue cells. LC1 and LC2, oligonucleotide loading controls.1–6, telomeric repeats added to the telomeric primer. b, Telomerase activity relative to the Scr control sample. Error bars, s.d. (n=5 independent experiments). ** P<0.01 by one-tailed unpaired Student’s t test. c, d, Autoradiograph for telomerase activity assay in HeLa PARN knockdown and rescue cells. Labeling of blot as in a. Error bars, s.d. (n=5 independent experiments), * P<0.05, ** P<0.01 by one-tailed unpaired Student’s t test.
Rescue of hTR subcellular localization by decay inhibition
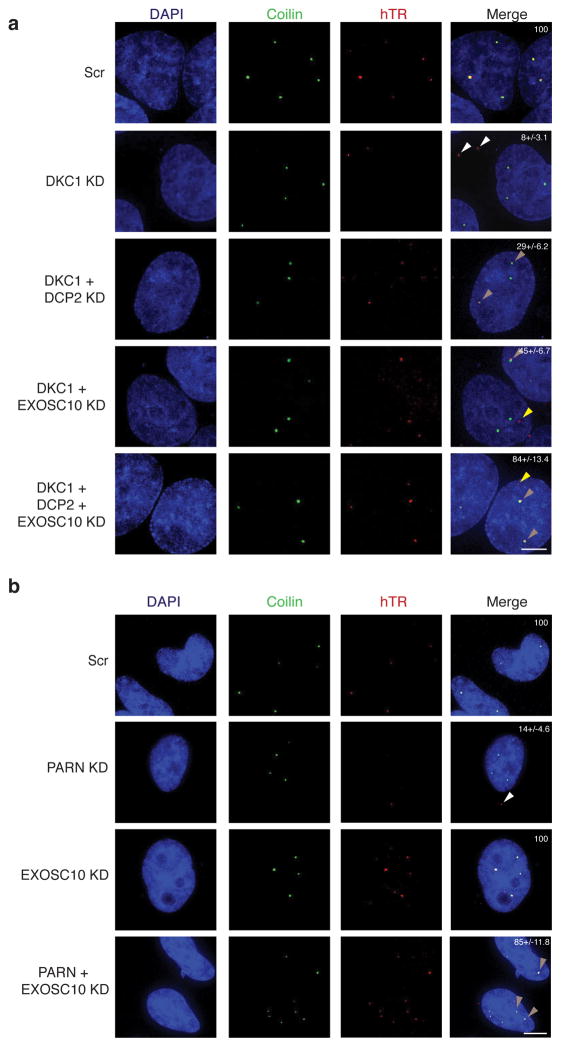
Normally, hTR is predominantly localized to Cajal bodies and to telomeres (depending on the cell cycle stage) in telomerase-positive human cells19–21. Since knockdown of nucleases increased hTR levels in cells that were deficient in dyskerin or PARN, we determined if these hTR RNPs could localize to Cajal bodies by FISH.
Similar to previous results19,21, we observed that endogenous hTR was localized to Cajal bodies in essentially all HeLa cells (Fig. 4a). However, in cells with knockdown of dyskerin or PARN, we observed a reduction in FISH signal, consistent with the reduced hTR levels; only 8% or 14% of the cells maintained hTR in Cajal bodies upon DKC1 or PARN knockdown, respectively (Fig. 4a, b). Surprisingly, in a fraction of the dyskerin (23%) or PARN (13%) knockdown cells, we observed the residual FISH signal for hTR in cytoplasmic puncta that have not been previously described (Fig. 4a, white arrowheads). We refer to these puncta as cytoplasmic TER (cyTER) -bodies.
Figure 4.
Mislocalization of hTR in dyskerin or PARN knockdown HeLa cells can be corrected by knockdown of competing nuclease. a, Subcellular localization of hTR by FISH in HeLa cells upon dyskerin knockdown (white arrowheads), and the localization to Cajal bodies upon rescue by DCP2 or DCP2 and EXOSC10 co-knockdown (light brown arrowheads) (Scale bar 5 μm). White numbers in Merge, % of cells showing hTR localization to CBs (mean+/−s.d., n=3 independent experiments). b, Subcellular localization of hTR by FISH to the cytoplasm or to CBs upon PARN knockdown and rescue, respectively (light brown arrowheads) (Scale bar 5 μm). White numbers in Merge, % of cells showing hTR localization to CBs (mean+/−s.d., n=4 independent experiments).
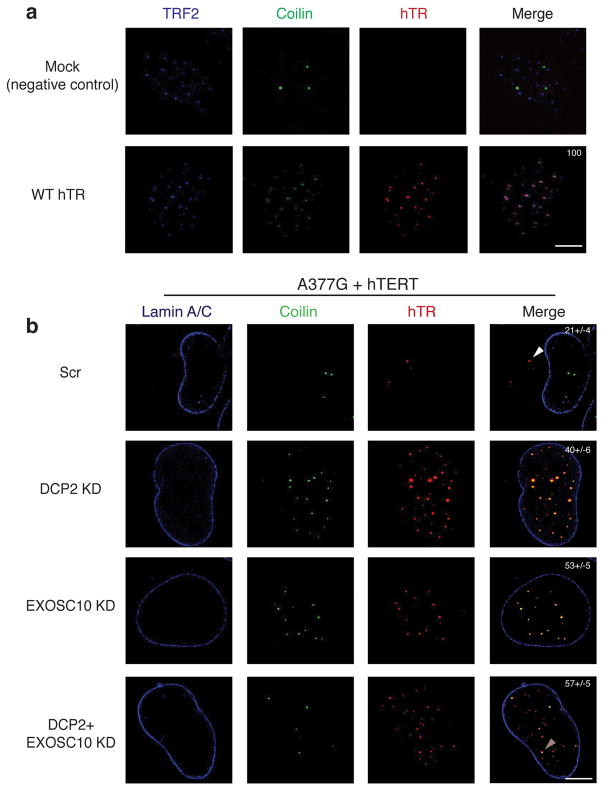
Similar aberrant localization of hTR was observed when we analyzed hTR point mutations. In U2OS cells transfected with hTERT and wild-type hTR, we observed that hTR localized to telomeres along with Cajal bodies, which have been defined as neo-Cajal bodies (Fig. 5a) 22. In contrast, cells expressing hTERT and the A377G or C408G mutant hTR showed reduced FISH signal for hTR and loss of hTR association with Cajal bodies, and only 21% and 18% of cells respectively still showed hTR in Cajal bodies (Supplementary Fig. 3a). The residual hTR signal was observed in cyTER-bodies (82% of cells for C408G hTR) (Supplementary Fig. 3a, b, white arrowheads).
Figure 5.
Mutant hTR localization to Cajal bodies can be rescued by knockdown of RNA decay pathways. a, WT hTR localization to neo-Cajal bodies at telomeres (labeled with TRF2) by FISH in cells overexpressing hTERT (Scale bar 5μm). White numbers in Merge, % of cells showing hTR localization to CBs (mean+/−s.d., n=4 independent experiments). b, Nuclear localization of A377G mutant hTR to Cajal bodies by FISH (light brown arrowheads) in hTERT co-transfected U2OS cells upon knockdown of nucleases (Scale bar 5μm). White numbers in Merge, % of cells showing hTR localization to CBs (mean+/−s.d., n=3 independent experiments).
To test if cyTER-bodies might represent sites of promiscuous hybridization of the hTR probes, we performed a FISH experiment with a combination of differently labeled hTR probes. The two different probes clearly co-localize in Cajal bodies for wild-type hTR. For the C408G mutant, the probes co-localize in cyTER-bodies but not with Cajal bodies (Supplementary Fig. 4a, white arrowheads). cyTER-bodies also do not co-localize with DCP1 or SMN, indicating these assemblies are not P-bodies, U-bodies or GEMS, respectively (Supplementary Fig. 4b). Thus, defects in PARN or dyskerin binding reduce hTR levels, disrupt proper hTR localization to Cajal bodies and telomeres, and lead to the identification of a novel cytoplasmic pool of hTR.
An important result was that knockdown of DCP2 and/or EXOSC10 could restore hTR levels and proper localization of defective hTR RNPs by FISH in three cases. First, in dyskerin knockdown cells, EXOSC10, DCP2 or a double DCP2 and EXOSC10 knockdown led to restoration of hTR localization to Cajal bodies (29%, 45% and 84% of cells respectively) (Fig. 4a, light brown arrowheads). For EXOSC10 knockdown, the RNA sometimes localized to sites other than Cajal bodies in the nucleus (yellow arrowheads), which could possibly be telomeres. Second, we observed that knockdown of EXOSC10 in PARN-deficient cells restored localization of hTR to Cajal bodies (85% of cells) (Fig. 4b, light brown arrowheads). Finally, we observed that knockdowns of DCP2, EXOSC10, or DCP2/EXOSC10 could restore the localization of A377G hTR to Cajal bodies (Fig. 5b). Thus, stabilization of hTR in either dyskerin or PARN-deficient cells can restore both telomerase activity (Fig. 3) and localization to Cajal bodies (Fig. 4).
Discussion
The absence of dyskerin protein or mutations in hTR that alter RNP assembly lead to decreased hTR levels, insufficient telomerase, and failure to maintain human stem cells. Here we identify decapping followed by XRN1-mediated 5’ to 3’ degradation and 3’ to 5’ degradation by the nuclear exosome as two independent quality control pathways for hTR in response to defects in dyskerin protein or mutations in hTR. Importantly, we show that these reduced levels of hTR and of telomerase activity can be rescued by knockdown of DCP2, XRN1, or EXOSC10 (Fig. 1 and Supplementary Fig. 1).
Evidence that decapping/XRN1 and the nuclear exosome comprise independent pathways is that double knockdowns of DCP2 or XRN1 with EXOSC10 have an additive effect on hTR levels (Fig. 1). Additional evidence that the nuclear exosome affects hTR degradation comes from the observation that knockdowns of the nuclear exosome lead to the accumulation of 3’ extended hTR molecules, as well as an increase in the levels of mature hTR23,24.
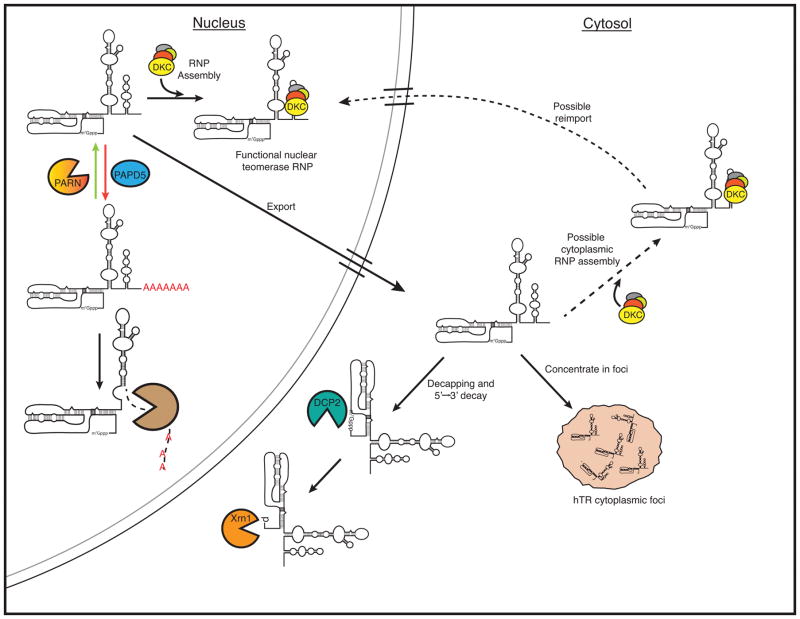
These observations support a model for hTR biogenesis that involves competition between dyskerin binding and hTR decay in the cell (Fig. 6). Under normal conditions, dyskerin binding allows hTR accumulation, either through co-transcriptional association or through cytoplasmic binding followed by nuclear re-import. When dyskerin binding to hTR is compromised, hTR can be degraded in the nucleus by EXOSC10 as well as exported to the cytoplasm and degraded by DCP2–XRN1.
Figure 6.
Model for hTR biogenesis, depicting competition between hTR assembly with H/ACA snoRNP proteins, 3’ end processing by PAPD5 and PARN and degradation by EXOSC10, and cytoplasmic export and degradation by DCP2 and XRN1.
Several observations provide a molecular explanation for the requirement of PARN for hTR stability, wherein it specifically removes oligo(A) tails on hTR added by PAPD5, which would otherwise recruit EXOSC10 (the nuclear exosome) to degrade hTR. First, knockdown of PARN leads to reduced levels of hTR due to faster hTR degradation (Fig. 2) 25, which can be rescued by knockdown of EXOSC10 or PAPD5 (Fig. 2). Second, we observe that the oligo(A) tails on hTR are reduced when PAPD5 is depleted, and increase upon PARN or EXOSC10 depletion (Fig. 2). Since PARN is specific for adenosine residues, deadenylation by PARN would limit the recruitment of EXOSC10 to hTR, thereby increasing it stability.
Our observations suggest that PARN is not primarily required for hTR biogenesis, since when PARN is limiting, knockdowns of EXOSC10 or PAPD5 can restore hTR levels (Fig. 2), telomerase activity (Fig. 3) and localization of hTR to Cajal bodies (Fig. 4). Instead, we suggest that PARN continually functions to remove oligo(A) tails added to hTR by PAPD5. This is likely to be a general phenomenon wherein stable non-coding nuclear RNAs with accessible 3’ ends will be constant substrates for PAPD5, and therefore their equilibrium concentrations would be maintained by PARN removing such oligo(A) tails to limit their degradation by the nuclear exosome.
There are two important implications of our observation that defects in hTR levels, telomerase activity, and proper subcellular location of hTR in cells deficient in PARN or dyskerin could all be rescued by knockdowns of EXOSC10, PAPD5 and/or DCP2. First, this demonstrates that neither PARN nor dyskerin is required for the biogenesis, activity, or in certain cases, localization of hTR. Second, this restoration of function suggests that inhibition of hTR degradation could be a viable therapeutic strategy for telomere pathologies. As such, dyskeratosis congenita is now seen to fall in the broader category of a ‘RNP hypo-assembly disease’, where a competing RNA quality control pathway degrades the RNA and limits RNP assembly due to reduced binding by the RNA’s protein partner(s)26.
A surprising observation was that in cells deficient in PARN or dyskerin, or those expressing mutant hTR molecules, hTR accumulated in discrete cytoplasmic foci, in a fraction of cells, referred to as cyTER-bodies. This provides evidence that hTR can be exported from the nucleus, as described previously for the budding yeast telomerase RNA27,28 and reveals a completely new aspect of human telomerase RNA cell biology. These foci do not co-localize with markers of P-bodies or GEMs, and thus are of unknown composition. Future work should reveal if these puncta are sites of hTR storage, or play a role in normal hTR biogenesis, possibly licensing the hTR RNP for nuclear import.
Online Methods
Cell culture
HeLa cells were purchased from ATCC, and the U2OS cells were a kind gift from Dr. Sabrina Spencer (University of Colorado Boulder), and were tested for mycoplasma contamination. All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS), L-Glutamax (Thermo Fisher), Penicillin/Streptomycin (Thermo Fisher) and Normocin (Invivogen), at 37° C and 5% CO2.
Plasmid transfection and siRNA knockdown
The 3xFLAG-hTERT and WT U1-hTR plasmid were gifts of Joachim Lingner (ISREC, Lausanne, CH). The disease-causing mutations were introduced in the WT hTR plasmid using site-directed mutagenesis, and the mutations were verified using Sanger sequencing. Plasmid transfection was performed on U2OS cells using JetPrime (Polyplus transfection) according to manufacturer’s protocol. Total plasmid concentration was limited to 2 μg for 6-well plates and 10 μg for 10 cm dishes. Plasmid transfection was allowed to take place between 24 and 48 hours before harvesting cells. siRNAs were purchased from Qiagen (Scr, DKC1, EXOSC10 and DCPS) and Dharmacon (PARN, XRN1, DCP2, XRN2, DOM3Z, PAPD5 and NUDT16). siRNA transfection was performed using Interferin (Polyplus transfection) according to manufacturer’s protocol, and final siRNA concentration was limited to 5 nM except for DKC1 siRNA, where final concentration of the siRNA was kept at 0.5 nM. Knockdown was allowed to take place for at least 48 hours and was verified using western blot analysis using antibodies against XRN1 (Bethyl #A300-443A) , EXOSC10 (Thermo #PA5-28672), DCP2 (Bethyl #A302-597A), DPCS (PA5-30532) and NUDT16 (LSbio #LC-C117055) described previously 26, while the other antibodies are as follows: anti-Dyskerin (Bethyl #A302-591A), anti-XRN2 (Cell Signaling #13760S), anti-DOM3Z (Pierce #PA5-29505), anti-PARN (Abcam #ab27778) and anti-PAPD5 (Atlas #HPA042968). Anti-GAPDH (Cell Signaling #2118S) was used as loading control. Antibody validation is provided on the manufacturer’s website. Uncropped blot image is available in Supplementary Data Set 1.
Northern blot analysis of hTR
RNA extraction was carried out using Quick RNA miniprep kit (Zymo Research). RNA was separated on a 5% polyacrylamide 7M urea gel at 20W, and transferred to a Nylon membrane (Whatman). hTR and 7SL northern probes have been previously described 26,29. In vitro transcribed hTR was used as a marker and has been previously described 29. For hTR decay rate measurement, cells were treated with Actinomycin D (Sigma) to final concentration of 5 μg/ml and harvested at 0, 4 and 7 hours post-treatment. All blots were imaged on a Typhoon phosphorimager, and hTR levels were quantified using Image Quant 5.2 and normalized to the 7SL levels under each condition. Uncropped blot images are available in Supplementary Data Set 1.
Immunofluorescence and FISH in U2OS and HeLa cells
For imaging, HeLa or U2OS cells were grown directly on coverslips in six well plates, and siRNA knockdown/plasmid transfection was performed on these cells before fixing them with 4% formaldehyde. Immunofluorescence was performed with Mouse anti-TRF2 (Novus Biologicals # NB100-56506), Rabbit anti-coilin (Santa Cruz #sc-32860), Mouse anti-lamin A/C (Cell Signaling #4777S), Rabbit anti-DCP1B (Cell Signaling #13233S), Rabbit anti-pan cadherin (Abcam #ab16505). Secondary antibodies used were Anti-rabbit Alexa Fluor 405 (Life Technologies #A-31556), Anti-mouse Alexa Fluor 405 (Life Technologies #A-31553), Anti-rabbit Alexa Fluor 594 (Life Technologies #A-11037), Anti-mouse Alexa Fluor 594 (Life Technologies #A-11005) and Anti-mouse Alexa Fluor 647 (Abcam #ab150119). Antibody validation is provided on the manufacturer’s website. After IF protocol, the antibodies were again fixed using 4% formaldehyde, and FISH for hTR was performed using Alexa 488 or Alexa 647 5’-labeled DNA oligos as described previously 30. The coverslips were sealed using ProLong Diamond Antifade Mountant (Thermo) with or without DAPI (Thermo Fisher). For FISH combination experiments in U2OS cells, transfected cells grown on coverslips were hybridized by equal amounts of each individual probe such that the total amount of hTR probe used was the same as that for single probe FISH experiment. In all U2OS imaging experiment, hTERT was co-transfected along with the indicated hTR RNA. Cells were imaged on a Deltavision deconvolution microscope using either a 60x or a 100x zoom, and all images were deconvolved and quick projected using the SoftWoRx software. All images taken for a particular experiment were normalized to the same threshold. Representative images are shown under each condition. For quantification, at least 50 cells were quantified for three biological replicates. Cells showing hTR localization to at least one Cajal body were scored and depicted as a percentage of total cells counted.
Telomerase activity assay for HeLa and U2OS cells
For direct assays of endogenous telomerase activity, a similar number of HeLa cells for each experiment was trypsinized, pelleted and lysed using lysis buffer described previously 18. hTERT IP was performed using an anti-hTERT antibody (generous gift from Dr. Scott B. Cohen, Children’s Medical Research Inst., Sydney AU). The activity assay conditions 18,31 and the primer 31 used have been described previously. Reactions were quenched using a stop buffer containing 3.6 M ammonium acetate, 20mg/ml glycogen and two 32P-labeled telomeric template oligos used as loading controls. The reaction products were separated on a 10% polyacrylamide 7 M urea sequencing gel at 90W, and imaged on a Typhoon phosphorimager. Intensity of the bands was quantified using Image Quant 5.2 software and normalized to one of the two loading controls (labeled LC1 and 2). Uncropped blot image is available in Supplementary Data Set 1.
hTR 3’ end sequencing
hTR sequencing was performed from total RNA isolated under various knockdown conditions. The RNA was DNase-treated, followed by RiboZero (Illumina) treatment as per manufacturer’s protocol. The resultant RNA was treated with rAPid Alkaline phosphatase (Sigma Aldrich) as per manufacturer’s protocol. ~125 ng of the resultant RNA was used for library preparation as previously described, except that a different hTR barcoded forward primer downstream of the original primer was used for 3’ RACE15. The amplified libraries were quantified on Bioanalyzer and Qubit, and 3 pM pooled libraries containing 30% phiX control were sequenced on the Illumina MiSeq as per manufacturer’s instructions. Data analysis was performed as previously described15. More than 160,000 reads containing both the RNA appendix and the hTR search primer were generated for each condition. For analysis of oligo(A) distribution at the mature 3’ end, total number of reads containing the mature 3’ end (UGC-451), whether adenylated or non-adenylated, was calculated, and reads that contained the first A (UGCA-452) were assumed to be non-adenylated, since the mechanism of generating those reads (whether genomically encoded or adenylation of the mature end) is ambiguous. Reads that contained two or more A’s were identified as oligoadenylated, and these were pooled to calculate the relative percentage of oligoadenylated mature 3’ ends under each transfection condition. To calculate the frequency distribution of the A tails, the relative percentage of each tail length was calculated relative to the total number of reads containing two or more A’s. For PAPD5 knockdown, the total number of mature oligoadenylated reads vs non-adenylated reads was very small (2388 reads against 529628 reads), and the hTR oligo(A) tail length distribution at the mature end shown in Supplementary Fig. 2 overestimates the abundance of such reads under this condition.
hTR Pol II chromatin immunoprecipitation (ChIP)
U2OS cells were grown and transfected in 15 cm dishes with TERT plasmid alone (mock control), or TERT with wild-type or C408G hTR plasmid. Cells were harvested ~48 hours post-transfection and cross-linked, and ChIP was performed as previously described32. The signal obtained from the mock control was ~ 25-fold lower than from either the wild-type or C408G hTR transfected cells, validating the absence of any detectable hTR in U2OS cells by northern blotting or FISH. hTR enrichment in Pol II immunoprecipitate over IgG control was calculated using two different hTR gene primer sets and qRT-PCR as previously described32.
Supplementary Material
Acknowledgments
We would like to thank A. Zaug for his assistance with the telomerase direct activity assays. We would like to thank A. Webb for her help with the illustrations and figures in this manuscript. We would like to thank members of the Parker and Cech labs for their comments and suggestions. J.C.S is supported as a Merck fellow of the Damon Runyon Cancer Research Foundation (DRG-2169-13). This work is supported by US National Institutes of Health R01 GM45443 (R.P.) and GM099705 (T.R.C.). T.R.C. and R.P. are investigators of the Howard Hughes Medical Institute.
Footnotes
The other authors declare no competing financial interest.
Supplementary Information is linked to the online version of the paper at www.nature.com/nsmb.
Author contributions
S.S., J.C.S., T.R.C. and R.P. conceptualized and designed the experiments. S.S. performed the experiments. J.C.S. performed the Pol II ChIP for hTR. K.C.G. and S.S. analyzed the hTR 3’ end sequencing reads. S.S., J.C.S., T.R.C. and R.P. analyzed and interpreted data. S.S., J.C.S., T.R.C. and R.P. wrote the manuscript.
References
- 1.Blackburn E, Collins K. Telomerase: An RNP Enzyme Synthesizes DNA. Cold Spring Harb Perspect Biol. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armanios M, Blackburn E. The telomere syndromes. Nat Rev Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heiss NS, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–8. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 4.Vulliamy T, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–5. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–5. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 6.Vulliamy & Dokal. Dyskeratosis congenita: The diverse clinical presentation of mutations in the telomerase complex. Biochimie. 2008;90:122–130. doi: 10.1016/j.biochi.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Stuart BD, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet. 2015;47:512–7. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhanraj S, et al. Bone marrow failure and developmental delay caused by mutations in poly(A)-specific ribonuclease (PARN) J Med Genet. 2015;52:738–48. doi: 10.1136/jmedgenet-2015-103292. [DOI] [PubMed] [Google Scholar]
- 9.Tummala H, et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J Clin Invest. 2015;125:2151–60. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rammelt C, Bilen B, Zavolan M, Keller W. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA. 2011;17:1737–1746. doi: 10.1261/rna.2787011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanácová S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–37. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 13.Kadaba S, et al. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–40. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–24. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 15.Goldfarb K, Cech T. 3′ terminal diversity of MRP RNA and other human noncoding RNAs revealed by deep sequencing. BMC Mol Biol. 2013;14:23. doi: 10.1186/1471-2199-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu D, Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell. 2003;11:1361–72. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- 17.Ueda Y, et al. A mutation in the H/ACA box of telomerase RNA component gene (TERC) in a young patient with myelodysplastic syndrome. BMC Medical Genetics. 2014;15:68. doi: 10.1186/1471-2350-15-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen S, Reddel R. A sensitive direct human telomerase activity assay. Nat Meth. 2008;5:355–360. doi: 10.1038/nmeth.f.209. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell. 2006;17:955–65. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venteicher AS, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–8. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong F, et al. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–94. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng CKK, et al. Human Telomerase RNA Processing and Quality Control. Cell Rep. 2015;13:2232–43. doi: 10.1016/j.celrep.2015.10.075. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen D, et al. A Polyadenylation-Dependent 3′ End Maturation Pathway Is Required for the Synthesis of the Human Telomerase RNA. Cell Reports. 2015;13:2244–2257. doi: 10.1016/j.celrep.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Moon D, et al. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat Genet. 2015;47:1482–1488. doi: 10.1038/ng.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla S, Parker R. Quality control of assembly-defective U1 snRNAs by decapping and 5’-to-3' exonucleolytic digestion. Proc Natl Acad Sci USA. 2014;111:E3277–86. doi: 10.1073/pnas.1412614111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teixeira MT, Forstemann K, Gasser SM, Lingner J. Intracellular trafficking of yeast telomerase components. EMBO Rep. 2002;3:652–9. doi: 10.1093/embo-reports/kvf133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 2008;27:748–57. doi: 10.1038/emboj.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xi L, Cech T. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res. 2014;42:8565–8577. doi: 10.1093/nar/gku560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abreu E, Terns R, Terns M. Visualization of human telomerase localization by fluorescence microscopy techniques. Methods Mol Biol. 2011;735:125–37. doi: 10.1007/978-1-61779-092-8_12. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt J, Dalby A, Cech T. Identification of human TERT elements necessary for telomerase recruitment to telomeres. eLife. 2014 doi: 10.7554/eLife.03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N, Cech TR. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 2015;29:2219–24. doi: 10.1101/gad.269498.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.