Abstract
Protein aggregates are the hallmark of stressed and ageing cells, and characterize several pathophysiological states1,2. Healthy metazoan cells effectively eliminate intracellular protein aggregates3,4, indicating that efficient disaggregation and/or degradation mechanisms exist. However, metazoans lack the key heat-shock protein disaggregase HSP100 of non-metazoan HSP70-dependent protein disaggregation systems5,6, and the human HSP70 system alone, even with the crucial HSP110 nucleotide exchange factor, has poor disaggregation activity in vitro4,7. This unresolved conundrum is central to protein quality control biology. Here we show that synergic cooperation between complexed J-protein co-chaperones of classes A and B unleashes highly efficient protein disaggregation activity in human and nematode HSP70 systems. Metazoan mixed-class J-protein complexes are transient, involve complementary charged regions conserved in the J-domains and carboxy-terminal domains of each J-protein class, and are flexible with respect to subunit composition. Complex formation allows J-proteins to initiate transient higher order chaperone structures involving HSP70 and interacting nucleotide exchange factors. A network of cooperative class A and B J-protein interactions therefore provides the metazoan HSP70 machinery with powerful, flexible, and finely regulatable disaggregase activity and a further level of regulation crucial for cellular protein quality control.
To investigate the possibility of a potent protein disaggregation activity in metazoans, we focused on the HSP70 chaperone system, which displays some in vitro capacity to disentangle and refold aggregated polypeptides when powered by an HSP110 co-chaperone4,7. The HSP70–J-protein–HSP110 functional cycle described in Extended Data Fig. 1a, by generally accepted extrapolation, occurs on protein aggregate surfaces. Homodimeric J-proteins are essential components of this cycle8,9. Three classes of J-proteins (A, B and C) with >50 members in humans, determine HSP70 substrate selection, with some functional redundancy among members9. For example, class A and B J-proteins (Fig. 1a) implicated in protein quality control have common functions, but independent and differing efficacies9–11. The basis for the evolutionary maintenance of these two classes of J-proteins (despite appreciable internal diversity12,13), and the relation of class to function and principles governing substrate selection, remain unknown.
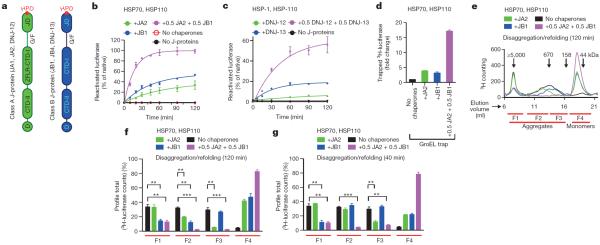
Figure 1. Simultaneous presence of class A and B J-proteins unleashes protein disaggregation activity and broadens target aggregate range of the HSP70 machinery.
a, Two distinct classes (A and B) display highly conserved domain organization involving the HSP70-intertacting HPD motif (red) containing amino-terminal J-domain (JD), Gly/Phe-rich flexible region (G/F), C-terminal β-sandwich domains (CTD-I and II), with class A J-proteins distinguished mainly by a zinc-finger-like region (ZFLR) that inserts into the CTD-I subdomain and a dimerization domain (D)9,23. CTD together with ZFLR provide substrate specificity24,25. b, Disaggregation and reactivation of preformed luciferase aggregates using human HSP70–HSP110 with human J-proteins JA2 (green), JB1 (blue), JA2+JB1 (magenta) or with no J-proteins (black) (n=3). c, Reactivation of heat-aggregated luciferase by nematode HSP70 machinery containing HSP-1, HSP-110 and either alone or in combination with the nematode J-proteins DNJ-12 (A) and DNJ-13 (B) (n=2). d, Fold change in trapped luciferase; control, GroELD87K without other chaperones (black). Values normalized to total 3H counts in each reaction (n=2). e, SEC profile after disaggregation/refolding (120 min) with either J-protein alone or combined. Elution fractions labelled F1–F4 (red lines); F4, disaggregated monomers (~63 kDa). f, Aggregate quantification for fractions F1–F4 from the SEC profile in e. Disappearance of 3H-luciferase from aggregates (F1–F3) occurs with concomitant accumulation of disaggregated monomer (F4). g, Aggregate quantification, after 40-min disaggregation. Values normalized to total counts in each reaction. Two-tailed t-test, **P<0.01, ***P<0.001 (n=3). Data are mean±s.e.m. Precise concentrations are shown in Extended Data Table 1.
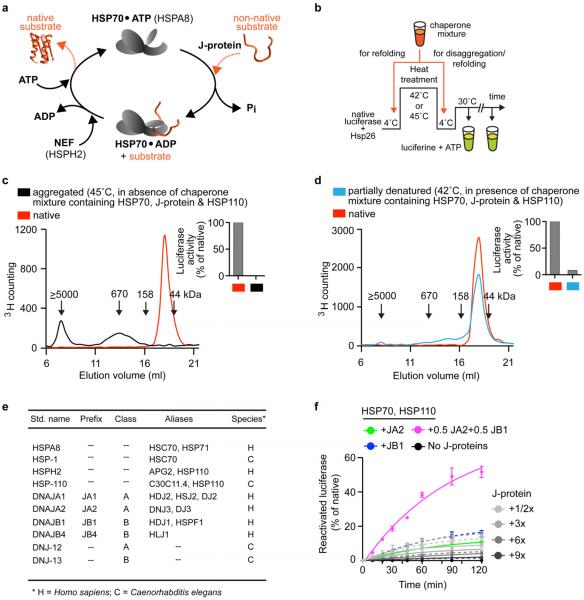
Here we explore the full potential of the metazoan HSP70–J-protein–HSP110 system in protein disaggregation, by examining the functional relationship between class A and B J-proteins. Using thermally denatured luciferase from Photinus pyralis as model substrate4, we investigate the in vitro protein disaggregation/refolding versus protein refolding-only (Extended Data Fig. 1b–d) capacities of the human and Caenorhabditis elegans HSP70–HSP110 systems (also known as HSPA8–HSPH2 in humans, and HSP-1–HSP-110 in C. elegans) in conjunction with class A and B J-proteins (Fig. 1, Extended Data Fig. 1e and Extended Data Table 1).
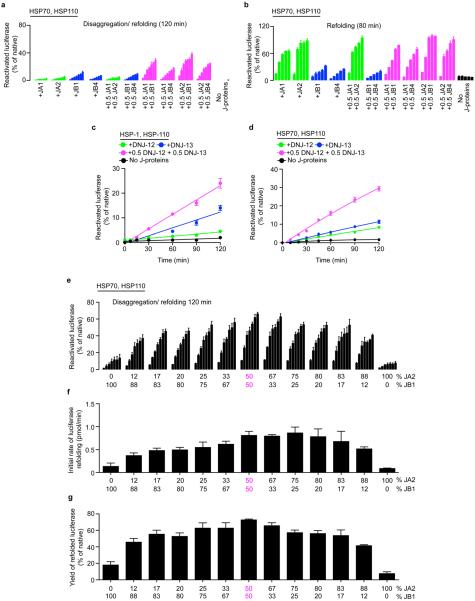
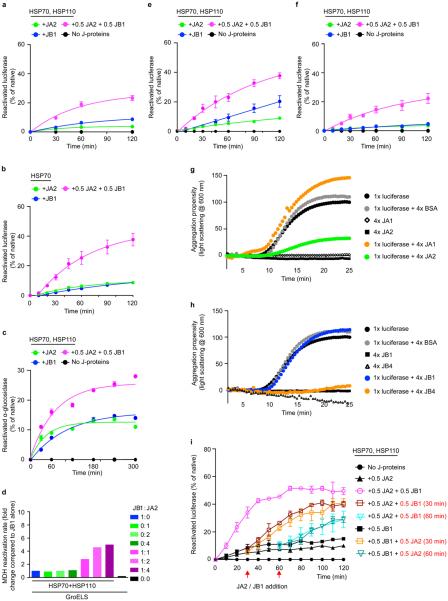
In disaggregation/refolding reactions with a class A (JA2) and B (JB1) J-protein present together (Fig. 1b, magenta), rather than either class J-protein alone (Fig. 1b, green or blue), we observed unprecedented reactivation of pre-formed heat-aggregated luciferase, indicating synergistically accelerated protein disaggregation. This was also seen under limiting chaperone concentrations (maintained in all further experiments) with multiple class A (JA1 and JA2) and class B (JB1 and JB4) human J-proteins (Extended Data Figs 1f and 2a). Disaggregation reactions with the corresponding nematode HSP-1–HSP-110 system and J-proteins DNJ-12 (class A) and DNJ-13 (class B) show similar synergic acceleration (Fig. 1c and Extended Data Fig. 2c, d). In reactions containing only one J-protein class (Extended Data Fig. 1f, JA2, solid lines; or JB1, dashed lines), with increased J-protein levels of threefold or more relative to the mixed-class J-protein reaction (Extended Data Fig. 1f, magenta), protein disaggregation/refolding slows and is inhibited. We infer that the presence of class A and B J-proteins together, rather than J-protein amount, determines reaction efficiency. Both the disaggregation/refolding rate (Extended Data Fig. 2e, f) and yield (Extended Data Fig. 2g) of renatured luciferase peak with equal proportions of class A to B. A broad range of flanking reciprocal A to B J-protein stoichiometries also show appreciable activity, suggesting that efficient disaggregation/refolding is supported by minimal amounts of preferentially interacting A and B J-proteins. Increased initial rates at higher stoichiometries of JA2 (Extended Data Fig. 2f) reflect intrinsically higher refolding capacity of class A J-proteins with HSP70 (Extended Data Fig. 2b, green)14.
Disaggregation synergy in mixed J-protein class reactions occurs with and without small HSP (Saccharomyces cerevisiae Hsp26) incorporation into aggregates for both human (Extended Data Figs 1f and 3a) and nematode J-protein containing systems (Fig. 1c and Extended Data Fig. 2d). Synergy is independent of nucleotide exchange factors (NEFs) (Extended Data Fig. 3b), protein substrate (Fig. 1b and Extended Data Fig. 3c, d) and substrate concentration variations affecting density, size4 and therefore the architectural nature of the aggregate generated (Extended Data Fig. 3e). Synergy also occurs at lower chaperone to substrate ratios (Fig. 1b and Extended Data Figs 1f and 3f), and at different and characteristic ranges of substrate to J-protein ratio for malate dehydrogenase (MDH) versus luciferase or α-glucosidase disaggregation (Fig. 1b and Extended Data Fig. 3c, d). MDH aggregates resolve considerably with non-limiting concentrations of JB1 alone (not shown), but with limiting JB1 concentrations in the presence of JA2, synergic MDH disaggregation occurs (Extended Data Fig. 3d). Synergy in disaggregation therefore appears generic, operating over a range of ratios and concentrations, with room for substrate-linked variation. By contrast, refolding-only reactions show no synergism (Extended Data Fig. 2b). We conclude that efficient protein disaggregation, but not refolding, requires cooperation between class A and B J-proteins.
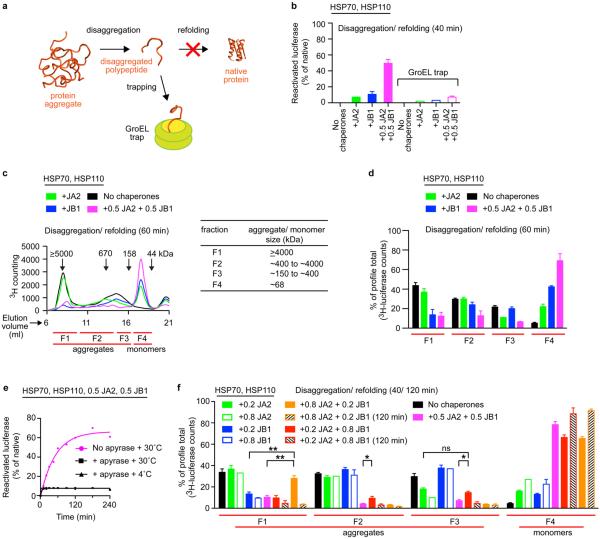
Three non-exclusive mechanisms could explain the synergistic action of class A and B J-proteins. In a mechanism involving sequential action, one J-protein class interacts with HSP70–HSP110 to extract polypeptides from aggregates. The other J-protein class then prevents re-aggregation of extracted polypeptide (holdase function) and/or in combination with HSP70–HSP110 promotes substrate refolding. Of the four J-proteins tested for holdase function, only JA2 and JB4 prevent luciferase aggregation at 42 °C (Extended Data Fig. 3g, h). However, disaggregation synergy is indistinguishable for J-protein combinations with (JA2 or JB4) and without (JA1 or JB1) holdase function (Extended Data Fig. 2a). Furthermore, disaggregation/refolding rates are unaffected by the order of JA2 and JB1 addition during the reaction (Extended Data Fig. 3i), indicating that J-proteins act in no strict order. For direct validation, we quantified tritium-labelled luciferase extracted from aggregates using a mutant GroEL protein (GroELD87K) as a trap15 for extracted luciferase molecules, preventing refolding. Decreased luciferase activity in disaggregation/refolding reactions in the presence of GroELD87K reflects trapping of labelled disaggregated polypeptides (Extended Data Fig. 4a, b), counted by measuring tritium scintillation (Fig. 1d). Disaggregation/refolding reactions containing only one class of J-protein show similar amounts of trapped 3H-labelled luciferase polypeptides. With class A and B J-proteins present together, however, we see synergistically accelerated accumulation of disaggregated 3H-labelled luciferase trapped in GroEL (Fig. 1d). Together, these results exclude a strictly sequential function of J-protein classes in disaggregation/refolding, corroborating the inference that synergy occurs at the protein disaggregation step.
A second model stipulates that each J-protein class acts specifically, in parallel, distinguishing protein aggregates by size and/or compactness during the disaggregation step. We tested this by adding different J-protein–HSP70–HSP110 mixtures to preformed 3H-labelled luciferase aggregates, which display a range of sizes, and probably variations in molecular architecture. We analysed the disaggregation of aggregate populations by size-exclusion chromatography (SEC; Fig. 1e–g and Extended Data Fig. 4c, d). Reactions were run in parallel, stopped by depleting ATP with apyrase, and held on ice until SEC (Extended Data Fig. 4e). Eluted fractions (F1–F4, Fig. 1e–g) reveal JA2-containing chaperone mixes preferentially solubilize smaller aggregates (F3; ~200–700 kilodaltons (kDa)). Conversely, JB1-containing mixes preferentially solubilize larger aggregates (F1, ≥5,000 kDa; F2, ~700–4,000 kDa), but solubilize small aggregates less efficiently. These results are consistent with distinct, parallel class activity. JA2 plus JB1 combinations, however, in much shorter reactions (40 min instead of 120 min), solubilize both larger and smaller aggregates far more efficiently than the added efficiencies of separate JA2 and JB1 reactions allow (Fig. 1g). Similar results obtain throughout for α-glucosidase aggregate solubilization (Extended Data Fig. 4d). This suggests concerted action on the same target.
This prompts a third model, in which synergy results from the formation of mixed-class J-protein complexes exerting concerted activity to facilitate disaggregation. A range of approaches rigorously tests this model.
To visualize individual versus complexed J-protein function, we biased disaggregation/refolding reactions by combining JA2:JB1 in 5:1 to 1:5 ratios, then analysed aggregate resolution by SEC (Extended Data Fig. 4f). The 1:1 ratios dissolve all aggregates (F1–F3, magenta). In contrast, limiting JB1 concentration and excess JA2 in shorter reactions (40 min, orange solid) barely resolves the largest aggregates (F1), whereas the smaller aggregates (F2–F3) disappear completely; F1 aggregates resolve only in longer reactions (120 min, orange hash). Limiting JB1 concentrations alone, however, readily resolve large F1 aggregates (blue solid). We infer that scarce JB1 molecules preferentially sequester with excess JA2 into complexes that efficiently process all sizes of aggregates; the smaller F2 and F3 aggregates accordingly disappear first. Reciprocal titration with scarce JA2 and excess JB1 concentration shows less disaggregation of the smaller F2 and F3 aggregates (magenta versus red solid, Extended Data Fig. 4f), which fully resolve with a longer reaction time (120 min, red hash). Specific J-protein stoichiometries evidently modulate HSP70 targeting and disaggregation efficacy. We infer that J-proteins preferentially form efficient mixed-class complexes, supporting a model for concerted action.
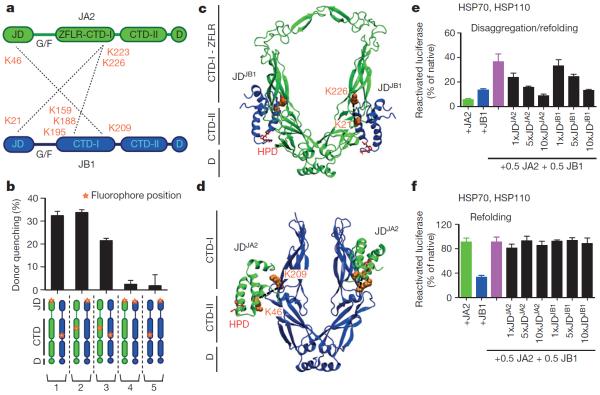
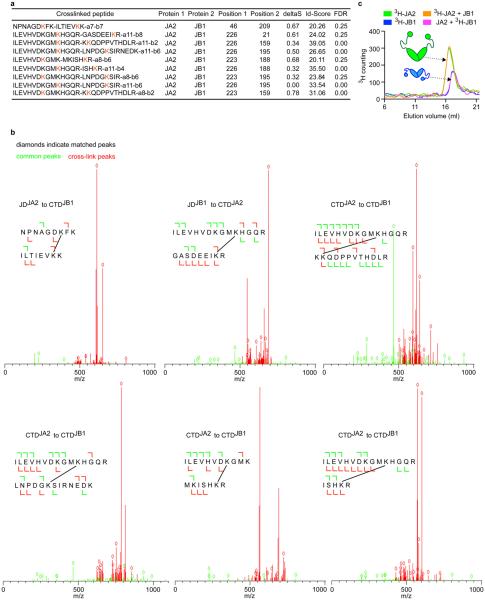
Independent tests for physical interactions between class A and B J-proteins consistently reveal intermolecular J-domain–C-terminal-domain (JD–CTD) and CTD–CTD contacts. Approaches include chemical cross-linking coupled to mass spectrometry (Fig. 2a), Förster resonance energy transfer (FRET; Fig. 2b), docking simulations (Fig. 2c, d) and competition assays (Fig. 2e).
Figure 2. Intermolecular JD–CTD interaction is required for mixed-class J-protein complex formation.
a, Intermolecular cross-links (dashed lines) between Lys residues (orange) on JA2 (green) and JB1 (blue). b, JA2 and JB1 interactions analysed by FRET. Bars show donor quenching efficiency of JD–CTD interactions; cartoons below show fluorophore positions in J-protein protomer pairs 1–5. N-termini of JDJA2 and JDJB1 are labelled with acceptor fluorophore ReAsH. CTDJA2 and CTDJB1 are labelled with donor fluorophores FlAsH and Alexa Fluor 488 at residues 241 and 278, respectively (n=3). c, d, Ribbon diagrams showing representative positions of JDs on CTD dimers from docking simulations; cross-linked Lys residues (space filling, orange, connected with black dashed lines) established in a; HPD motif (stick representation, red). c, JDJB1 (blue) and CTDJA2 (green). d, JDJA2 (green) and CTDJB1 (blue). e, f, Competition of excess isolated JD fragments for classes A and B J-protein complex formation and effect on luciferase disaggregation. e, f, Protein disaggregation/refolding (e) and refolding-only (f) (n=3). Data are mean±s.e.m. Precise concentrations are shown in Extended Data Table 1.
Mass spectrometry of JA2 and JB1 combinations treated with lysine-specific cross-linker (disuccinimidyl suberate) identifies three intermolecular cross-linked regions between JDJA2–CTDJB1, JDJB1–CTDJA2 and CTDJA2–CTDJB1 (Fig. 2a and Extended Data Fig. 5a, b). FRET measured by donor quenching indicates JD–CTD and CTD–CTD interactions between JA2 and JB1 in solution (Fig. 2b, J-protein pairs 1, 2 and 3; Extended Data Fig. 6a). This corroborates our cross-linking data and favours biological relevance. We detect neither JD–JD interactions between classes (J-protein pair 4), nor intermolecular same-class JD–CTD interactions (J-protein pair 5). However, in agreement with structures from small-angle X-ray scattering of class B J-proteins16,17, we detect JDJB1–CTDJB1 cross-links (not shown). Presumably these reflect intramolecular interactions, preventing intermolecular JDJB1–CTDJB1 but not JDJA2–CTDJB1 interactions, as indicated by FRET (Fig. 2b).
We further defined the interface of the JA2–JB1 complex using unbiased docking simulations between J-domain and CTD dimers of JA1, JA2, JB1 and JB4 (Fig. 2c, d and Extended Data Fig. 7a, b). Simulations show a preferred binding arrangement of JDJB1 on CTDJA2 and conversely JDJA2 on CTDJB1 (Fig. 2c, d), again corroborating cross-linking data (Fig. 2a).
Furthermore, in competition experiments, the addition of moderate excess of isolated J-domain fragments inhibits JA2–JB1–HSP70–HSP110-dependent disaggregation/refolding of heat-aggregated luciferase (Fig. 2e), although not refolding alone (Fig. 2f). J-domain fragments carrying the HPD motif mutated to QPN, which abolishes the JD–HSP70 interaction and ATP hydrolysis stimulation on HSP70 (refs 18, 19), have the same effect (Extended Data Fig. 6e), confirming that inhibition of disaggregation is not due to HSP70 being titrated out by J-domain fragment binding. Unlabelled full-length J-proteins and isolated J-domains compete with mixed-class JD–CTD interactions, indicated by decreased FRET efficiency between JA2 and JB1 (Extended Data Fig. 6f, g), explaining the inhibitory effects. However, JD–CTD interaction sites do not overlap CTD binding sites for substrate, since JA2 holdase activity remains unaffected with isolated J-domains present (Extended Data Fig. 7c, d). Molecular docking modelling supports this also (Extended Data Fig. 8). J-protein complexing involving mixed-class J-domains and CTDs is therefore crucial for efficient protein disaggregation, but not for refolding.
Non-ionic detergent affects neither disaggregation activity (Extended Data Fig. 6b) nor FRET efficiency between class A and B molecules (Extended Data Fig. 6a). Increasing salt concentrations, however, weaken both (Extended Data Fig. 6c, d), suggesting ionic interactions. Independent methodologies therefore confirm specific JD–CTD interactions of a predominantly electrostatic nature directly implicated in disaggregation efficiency.
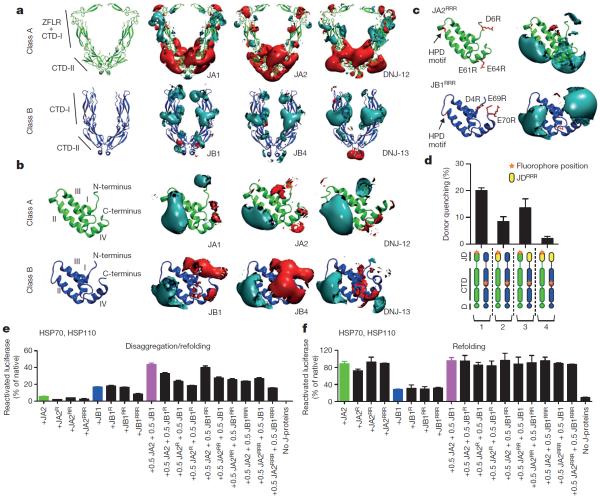
J-domain and CTD regions display highly conserved, class-specific electrostatic potentials (Fig. 3a, b). Class A J-proteins show distinct polarity in the CTDs, with negatively charged regions (red) in the CTD-II and dimerization subdomains, and positively charged regions (cyan) along the zinc-finger-like region and CTD-I hook (Fig. 3a). Conversely, class B CTDs are relatively non-polar, with positively charged regions in the CTD where JDJA2 cross-linking occurs. J-domains in both classes are markedly bipolar, although class A J-domains have smaller negatively charged regions (Fig. 3b). In all J-domains, positive charge (near the HPD motif and helix-II) is implicated in binding to HSP70 (refs 18, 19). We deduce conserved negatively charged regions exposed in the J-domains interact with positively charged CTD regions in opposite class J-proteins.
Figure 3. Conserved electrostatic potential distributions in A and B J-protein classes are complementary and direct mixed-class J-protein interactions for complex formation.
a, Electrostatic isopotential maps of CTD dimers comparing human (JA1, JA2, JB1 and JB4) and nematode (DNJ-12 and DNJ-13) class A and B J-proteins. Electrostatic potential around proteins is contoured at +1 (positive, cyan) and −1 (negative, red), kcal mol−1 e−1. Protein structures are represented by ribbon diagrams. b, Conserved α-helices and electrostatic isopotential maps contoured as in a of human and nematode J-domains. I–IV (from N-terminus) denote conserved α-helices. c, The J-domains of charge-reversal triple mutants (JA2RRR and JB1RRR); and their electrostatic isopotential maps compare with wild-types in b. RRR denotes triple amino acid substitutions D6R, E61R and E64R in JA2, and D4R, E69R and E70R in JB1. d, FRET determination of JA2 and JB1 triple charge-reversa; mutants (n=3). Bars show donor quenching efficiency of JD–CTD interactions; cartoons below show fluorophore positions in J-protein protomer pairs 1–4. Triple charge mutants are yellow. e, Luciferase disaggregation/refolding at 120 min with J-domain charge-reversal mutants (JA2: D6R (R); E61R+E64R (RR); D6R+E61R+E64R (RRR). JB1: D4R (R); E69R+E70R (RR); D4R+E69R+E70R (RRR)) (n=3). f, As in e, refolding-only at 80 min (n=3). Data are mean±s.e.m. Precise concentrations are shown in Extended Data Table 1.
We therefore generated triple charge-reversal variants of the J-domain (JA2RRR or JB1RRR), replacing negatively charged Asp or Glu residues with positively charged Arg residues in and around helices-I and -IV (Fig. 3c). FRET interactions between the JDJA2 and CTDJB1 regions diminish with charge-reversal mutations in either JDJA2 or JDJB1 (Fig. 3d, J-protein pairs 2 and 3), and are abrogated with charge reversals in both interacting J-domains (Fig. 3d, J-protein pair 4). Partial FRET reduction with triple charge reversals in only one interacting JD–CTD domain pair suggests some degree of intermolecular tethering by the other pair, although insufficient for full J-protein cooperation and disaggregation efficiency (Fig. 3e). In refolding-only reactions, recovered luciferase activity remains unaffected by J-domain charge reversals (Fig. 3f). Physically complexed and cooperating mixed-class J-proteins are therefore essential for efficient HSP70-dependent disaggregase activity, and are thought (but not directly shown) to act on the surface of aggregates. A paradoxical sequence dispensability of these highly conserved helices-I and -IV observed in early data, which assayed exclusively for HSP70 interaction and protein folding18,20, is also now explained. These data together strongly support a mixed-class J-protein interaction with vital function conserved in evolution.
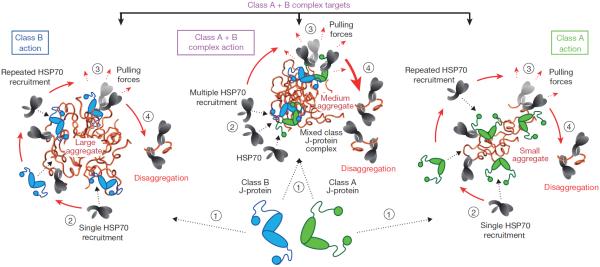
Size separation of tritiated JB1 mixed with unlabelled, larger JA2, or the reciprocal labelling, reveals only JB1 (blue) or JA2 (green) homodimers (Extended Data Fig. 5c), indicating that J-protein complexes are transient. Transient interactions would support an HSP70 disaggregation machinery with a flexible range of tailored activities. Single-class J-protein function shows HSP70–HSP110-mediated disaggregation activity limited to aggregates of specific size ranges (Fig. 4, large or small aggregates). Mixed-class J-protein complexes efficiently disaggregate a wide range of aggregate sizes (Fig. 4, large, medium and small). On the basis of our results, we reason a minimum complex consists of one class A J-protein homodimer binding to one class B homodimer in a 1:1 ratio, indicating that there are four J-domains per complex. Assuming two J-domains engage in interactions sufficient to complex the J-proteins, one J-domain per homodimer remains free to interact with one HSP70, allowing for recruitment of two interacting HSP70 molecules per complex without steric hindrance (Fig. 4, medium aggregates, Extended Data Fig. 8). We conclude that each mixed class J-protein complex recruits at least two HSP70 molecules per targeting event, possibly seeding dynamic, higher order chaperone assemblies on aggregate surfaces.
Figure 4. Model of individual versus complexed class A and class B J-protein function in protein disaggregation.
Size-specific aggregate targeting: large aggregates are targeted by J-proteinclass B–HSP70–HSP110 (blue); small aggregates are targeted by J-proteinclass A–HSP70–HSP110 (green); all aggregates sizes are targeted by J-protein-mixed-class-complex–HSP70–HSP110 (magenta).HSP70 molecules are in grey. Sequential reaction steps (encircled numbers): 1, J-protein targets aggregate; 2, J-protein recruits HSP70; 3, surface-bound chaperones generate pulling forces (dashed red arrows); and 4, polypeptide extraction leading to protein disaggregation. Chaperone recruitment denoted by dashed black arrows.
Our computational models of the structures of mixed class J-protein complexes (Extended Data Fig. 8) incorporate the constraints defined by all our cross-linking, FRET and docking data. In each model, space in the J-protein complex allows for substrate binding via several interfaces, HSP70 interaction with J-domains, and HSP110 interaction with each HSP70 protein. These models accommodate the concept of entropic pulling, in which HSP70 binding to entangled polypeptides decreases entropy, generating reciprocal forces that pull polypeptides from aggregates21. Such higher order chaperone complexes would be expected to increase pulling forces and stabilize disaggregating polypeptides by providing increased substrate binding surface, thereby accelerating protein disaggregation (Fig. 4; class A+B complex). Although also likely, direct verification of mixed-class J-protein–HSP70 complexes interspersed with single-class J-protein–HSP70 complexes on aggregate surfaces is currently experimentally intractable.
In summary, we demonstrate potent protein disaggregation activity in metazoans, mediated by the central HSP70–J-protein–HSP110 chaperone network. Disaggregation efficacy comparable to that of non-metazoan HSP100–HSP70 bi-chaperone systems, over a broad aggregate size range, requires transient physical interaction between class A and B J-proteins. The assembly of higher order chaperone complexes on protein aggregate surfaces is expected to increase coordinated pulling forces on multiple trapped polypeptides, providing a plausible mechanistic basis for increased disaggregation efficacy. Mixed-class J-protein complexes form preferentially and interact with HSP70–HSP110 to resolve a broad range of aggregates efficiently, whereas single-class J-protein–HSP70–HSP110 interaction targets specific aggregate sizes. This suggests intracellular J-protein stoichiometry will differentially regulate HSP70-dependent protein disaggregation efficiency. The transitory nature of J-protein complexes would, in this context, facilitate flexible response according to need. As in nematodes, human cytosol contains several members of J-protein classes: four class A and nine class B J-proteins9. A wide range of complexed J-protein combinations is therefore available in humans and other metazoa, providing flexible target selectivity. This opens the further possibility of physiological function in assembly/disassembly of other macromolecular cell structures. These findings may also impinge on the amorphous, oligomeric, most toxic prefibrilar phase of amyloidic fibre formation characterizing neurodegenerative diseases22. Overall, our work identifies a physically interacting J-protein network that adds another level of functional flexibility to cellular protein quality control. The underlying functional basis for hitherto unexplained evolutionary maintenance of distinct J-protein classes now also becomes clear. In essence, we reveal a J-protein gearbox regulating efficacy of protein disaggregation and consequently, refolding reactions, with fundamental effect on the cellular physiology, and therefore health, of metazoan organisms.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
METHODS
Plasmids and protein purification
Clones of human J-proteins (DNAJA1, DNAJA2, DNAJB1 and DNAJB4) were obtained either directly from Addgene or as gifts from H. Kampinga in pcDNA5/FRT/TO plasmids. C. elegans dnj-12, dnj-13, C30C11.4 (hsp-110) and hsp-1 genes were amplified by PCR using complementary DNA preparations from heat-shocked three-day-old animals as a template. The above-mentioned genes were then recloned into protein expression vector pCA528 or pSUMO with a 6×His–Smt3 tag as previously described26. Mutants of J-proteins were generated by standard PCR mutagenesis techniques and verified by sequencing. JA2 and JB1 variants for N-terminal FlAsH and ReAsH labelling were generated by incorporating the Cys-Cys-Pro-Gly-Cys-Cys tag (5′-TGTTGTCCAGGGTGCTGC-3′) after N-terminal methionine. The same labelling motif was generated in JA2 CTD by inserting two Cys residues before Pro241 and two Cys residues before Val243. The JB1 CTD labelling mutant was generated by mutating Gly278 to Cys. To obtain isolated J-domains, the J-domains of JA2 (1–77 amino acids) and JB1 (1—76 amino acids) were PCR-amplified with a C-terminal TAG site and cloned into pCA528. HPD motifs were mutated to QPN by changing H36Q+D38N and H32Q+D34N in isolated J-domains of JA2 and JB1, respectively. Purification of J-proteins and their variants was performed by affinity (Ni-IDA, Macherey-Nagel; Ni-NTA, Pierce), size-exclusion and ion-exchange chromatographic methods. In brief, BL21(D3E)/pRARE Escherichia coli strains carrying the corresponding expression vectors were induced for protein expression with 0.5 mM isopropyl-1-thio-d-galactopyranoside (IPTG, Sigma-Aldrich) for 3 h at 30 °C. All C. elegans chaperones were expressed at 20 °C with 1 mM IPTG overnight. Cells were lysed either in 50 mM HEPES-KOH, pH 7.5, 750 mM KCl, 5 mM MgCl2, protease inhibitor cocktail (Roche), 2 mM phenylmethylsulphonyl fluoride and 10% glycerol (for human J-protein purifications), or in 30 mM HEPES-KOH, pH 7.4, 500 mM K-acetate, 5 mM MgCl2, 1 mM β-mercaptoethanol, 2 mM phenylmethylsulphonyl fluoride, protease inhibitor cocktail (Roche) and 10% glycerol (for nematode chaperone purifications). After centrifugation at 30,000g (30 min, 4 °C) the resulting supernatants were applied to a Ni-NTA/Ni-IDA matrix and incubated for 60 min at 4 °C. Subsequent washing steps were performed with high-salt buffers (50 mM HEPES-KOH, pH 7.5, 750 and 500 mM KCl, 5 mM MgCl2 and 10% glycerol) for human J-protein purifications. Worm chaperones were first washed in high-salt buffer (30 mM HEPES-KOH, pH 7.4, 1 M K-acetate, 5 mM MgCl2, 1 mM β-mercaptoethanol, 2 mM phenylmethylsulphonyl fluoride and 10% glycerol), followed by a low-salt wash (identical to the high-salt buffer with 50 mM instead of 1 M K-acetate). Protein elution was performed with 300 mM imidazole in the corresponding low-salt buffers. Dialysis was performed overnight at 4 °C in the presence of 4 μg His-tagged Ulp1 per mg substrate protein for proteolytic cleavage of the 6×His–Smt3 tag. The 6×His–Smt3 tag and His–Ulp1 were removed by incubating the dialysed proteins in Ni-NTA/Ni-IDA matrix for 60 min at 4 °C. The targeted proteins were further purified using Superdex 200 (human J-proteins and their variants), ion exchange using the Resource Q (anion exchange for DNJ-12, HSP-1, HSP-110 and isolated human J-domain fragments) or Resource S (cation exchange for DNJ-13) columns (GE Healthcare). Firefly luciferase and human HSPA8 and HSPH2 were purified as previously described4. Pyruvate kinase and α-glucosidase were purchased from Sigma-Aldrich. Pig heart muscle MDH was purchased from Roche.
Luciferase refolding and disaggregation/refolding assays
For refolding-only assays, 20 nM luciferase plus 750 nM HSPA8, 40 nM HSPH2, 380 nM J-protein and 100 nM Hsp26 was incubated at 42 °C for 10 min in HKM buffer (50 mM HEPES-KOH, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol (DTT), 2 mM ATP, pH 7.0 and 10 μM BSA) to generate thermally denatured luciferase. Luciferase refolding was initiated by adding an ATP regenerating system (3 mM phosphoenolpyruvate and 20 ng μl−1 pyruvate kinase) and by shifting the reaction to 30 °C. Luciferase activity was measured at the indicated time points with a Lumat LB 9507 luminometer (Berthold Technologies) by transferring 1 μl of sample to 100 μl of assay buffer (25 mM glycylglycin, pH 7.4, 5 mM ATP, pH 7, 100 mM KCl and 15 mM MgCl2) mixed with 100 μl of 0.25 mM luciferin. Luciferase aggregates for disaggregation/refolding reactions were generated as previously described4. Either 25 nM or 2 μM luciferase with fivefold excess of Hsp26 was aggregated at 45 °C for 15 min. Protein disaggregation/refolding was initiated by adding the indicated chaperone mixtures to preformed luciferase aggregates and shifting the reaction temperature to 30 °C. Luciferase disaggregation/refolding assays with C. elegans HSP70 chaperone system were performed at either 20 °C or 22 °C (in Fig. 1c and Extended Data Fig. 2c, d).
In cells from bacteria to human, aggregates that form under thermal stress contain small HSPs (sHSPs) such as S. cerevisiae Hsp26. sHSP incorporation facilitates aggregate resolution through inherent chaperone holdase activity, possibly changing the density, size and architecture of aggregates, making these more accessible and manageable for disaggregation machineries4,27. The in vitro assay system incorporating Hsp26 therefore more closely approximates the situation in the cell. We used S. cerevisiae Hsp26 because it is the only sHSP induced and activated in yeast cells after heat shock, and has been extensively characterized in vivo and in vitro4,27,28. Hsp26 is therefore the generic heat-induced sHSP of yeast, which justifies its use for our study. By contrast, there are 10 human sHSPs, each with different substrate binding specificities and affinities29,30, and there is no clear basis for choosing one above another. Also, some of these, including those recognizing luciferase as substrate, interact to form hetero-oligomers, which display yet further different properties31 resulting in complicated assay analysis. Furthermore, the activation mechanism of at least one of these is subject to controversy (phosphorylation and dephosphorylation have both been reported32; P. Goloubinoff, personal communication).
α-glucosidase disaggregation assay
α-glucosidase aggregation was achieved by incubating 50 nM of substrate with 500 nM Hsp26 in HKM buffer without DTT at 50 °C for 15 min. Disaggregation/refolding of aggregates was initiated by adding indicated chaperone mixtures supplemented with an ATP regenerating system and by shifting the reaction to 30 °C. Reactivation of α-glucosidase was measured with an α-glucosidase assay kit from Abnova using a FLUOstar Omega plate-reader (BMG LABTECH).
MDH disaggregation assay
Pig heart muscle MDH (Roche) aggregation was achieved by incubating 150 nM of substrate with 750 nM Hsp26 in HKM buffer at 47 °C for 30 min. Disaggregation/refolding of aggregates was initiated by adding indicated chaperone mixtures supplemented with an ATP regenerating system and by shifting the reaction to 30 °C. MDH activity was measured using a potassium phosphate (150 mM, pH 7.6) buffer containing 1 mM oxaloacetate, 2 mM DTT and 0.56 mM NADH. Activity measurements were taken using a FLUOstar Omega plate-reader. Refolding rates were calculated from the linear increase of substrate activities.
SEC and aggregate profiling
Tritium (3H) labelling of firefly luciferase, α-glucosidase, JA2 and JB1 was performed with N-succinimidyl-[2,3-3H]propionate (Hartmann Analytic) according to manufacturer's guidelines. Unincorporated N-succinimidyl-[2,3-3H]propionate was removed using dialysis with HKM buffer and either 150 mM (for luciferase and α-glucosidase) or 500 mM (for J-proteins) plus KCl at 4 °C, overnight. 3H-labelled luciferase and α-glucosidase were aggregated as described in the disaggregation/refolding assays. Luciferase and α-glucosidase reactivation were performed by adding specified chaperone cocktails and incubating at 30 °C. Reactions were quenched with apyrase (0.8 μg μl−1) at 40 or 120 min and placed on ice. Aggregated luciferase/α-glucosidase complexes were separated using an ÄKTA purifier system with a Superose 6 Tricorn column (10/300 GL, GE Healthcare Life Sciences). Samples were centrifuged at 9,000g for 5 min at 4 °C before loading. Running buffer contained 50 mM HEPES-KOH, pH 7.5, 150 mM KCl, 5 mM MgCl2, 2 mM DTT (0.2 mM DTT for α-glucosidase) and 10% glycerol. A similar approach was used to separate 3H-labelled J-protein dimers, with the exception of using a buffer with 50 mM KCl. The 3H signal in each fraction (500 μl) was quantified by scintillation counting (Beckman LS6000 IC). The amount of 3H-luciferase trapped in GroELD87K was calculated by subtracting the total counts between elution volumes 11 and 16 in reactions without the trap from that of the reaction containing the trap (10 μM). The 3H signal in each elution fraction was normalized to the total counts of the corresponding SEC run after background subtraction, and presented as a percentage of the total counts (F1–F4). A SEC standard (BIO-RAD) was used to determine the size of the elution peaks. Void volume contains any complexes ≥5,000 kDa. Notably, ~40–50% of the input material was lost during SEC as a result of nonspecific binding to filters and column matrix.
Luciferase aggregation prevention assay
In the aggregation prevention assay, 200 nM luciferase was mixed with indicated concentrations of chaperones or BSA (control) in a buffer containing 50 mM HEPES-KOH, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT and 2 mM ATP, pH 7.0. Aggregation of luciferase was initiated by increasing the temperature to 42 °C. The extent of luciferase aggregation was monitored by light scattering at 600 nm (Hitachi Fluorometer F4500, λex/em=600 nm, slit widths of 5.0 nm) for 25 or 30 min.
Chemical crosslinking coupled to mass spectrometry
For chemical cross-linking, 100 μl of sample containing 2 μM JA2 and 2 μM JB1 was directly cross-linked with 1 mM disuccinimidyl suberate d0/d12 (DSS, Creativemolecules Inc.), and subsequently enzymatically digested with trypsin and enriched for cross-linked peptides, essentially as previously described33. Liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis was carried out on an Orbitrap Elite mass spectrometer (Thermo Electron). Data were searched using xQuest34 in iontag mode with a precursor mass tolerance of 10 p.p.m. For matching of fragment ions, tolerances of 0.2 Da for common ions and 0.3 Da for cross-link ions were used. False discovery rates of cross-linked peptides were assigned using xProphet35. Cross-linked peptides were identified with a delta score <0.95 and a linear discriminant score >20, and additionally analysed by visual inspection to ensure good matches of ion series on both cross-linked peptide chains for the most abundant peaks.
FlAsH and ReAsH labelling of J-proteins
J-protein variants with introduced tetracysteine motif (CCPGCC, for FlAsH and ReAsH labelling) or with single cysteine residue (for JB1 Alexa Fluor 488 labelling) were reduced with 50-fold molar excess of TCEP for 30 min at room temperature, and incubated with FlAsH or ReAsH (gift from A. Krezel) at a 1:1.5 protein/label ratio for 4 h at 4 °C, or 20-fold excess of Alexa-Fluor-488-maleimide for 2 h at room temperature. Progress of labelling reaction for the biarsenical dyes was monitored in a spectrophotometer, using A280 nm (JA2 ε280 nm=24,000 M−1 cm−1, JB1 ε280 nm=19,035 M−1 cm−1) for protein concentration and absorption at A510 nm for FlAsH (ε510 nm=41,000 M−1 cm−1), A590 nm for ReAsH (ε590 nm=68,000 M−1 cm−1) and A494 nm for Alexa Fluor 488 (ε494 nm=71,000 M−1 cm−1). The excess of unbound dye was removed on a self-packed Sephadex G-25 column (GE Healthcare), and the activity of labelled proteins was confirmed by luciferase refolding assay.
FRET measurements
FRET was used to validate distances between labelled J-domains and CTDs of class A and B J-proteins. Emission spectra were recorded on a Jasco FP6500 spectrofluorimeter between 510 and 650 nm, at excitation wavelength of 508 nm for FlAsH and 488 nm for Alexa Fluor 488 (donor fluorophores). Quenching of donor fluorescence (at 519 nm for Alexa Fluor 488 and 533 nm for FlAsH) and an increase in acceptor emission (at 608 nm for ReAsH) were quantified. Acceptor fluorescence measurement was refined by subtracting the fluorescence from donor-labelled J-protein to minimize background. The Förster radius of the FlAsH–ReAsH FRET pair was calculated to 39 Å (refs 36, 37) and Alexa-Fluor-488–ReAsH to 62 Å (ref. 38). For FRET experiments, J-proteins labelled with donor and acceptor fluorophores were mixed at 0.1 μM (donor) and 1 μM (acceptor) in a buffer containing 25 mM HEPES, pH 7.5, 50 mM KCl and 5 mM MgCl2, and allowed to equilibrate for 15 min at 30 °C before measuring the steady-state fluorescence. For competition experiments, 1-, 5- and 10-fold excess (relative to acceptor concentration) of unlabelled full-length proteins or isolated J-domains were added and allowed to equilibrate for 15 min at 30 °C. All samples were measured at least in duplicate. For Figs 2b and 3d, FRET efficiencies were calculated based on the donor fluorescence quenching, and presented as a percentage of donor fluorescence in the absence of acceptor.
Protein structure preparation
Protein structures used in simulations were either crystal or NMR structures from the RCSB Protein Data Bank (PDB; http://www.rcsb.org) or comparative models that were either present in the SWISS-MODEL database and found using the Protein Model Portal (PMP) or were modelled with SWISS-MODEL (SM)39–42. The structure of the CTDJB1 dimer was taken from the crystal structure (PDB code 3AGZ, resolution: 2.51 Å)43 and that of JDJB1 from the first entry of the NMR structure (PDB code 1HDJ)44. Since the N-terminus of chain B in the CTDJB1 dimer was missing three residues compared to chain A, the N-terminal nine residues from chain A were superimposed on the N-terminus of chain B to obtain coordinates for the missing three residues. Comparative models of the CTDJB4 dimer and JDJB4 were both found in the PMP and were based on the template structures 3AGZ and 1HDJ, respectively. To add the missing three residues at the N-terminus of chain B, the same procedure as for CTDJB1 was used. Comparative models of CTD monomers of JA1 and JA2 were both taken from the PMP. Both structures were modelled with SM based on the template crystal structure, 1NLT (resolution 2.70 Å)24. The structure of JDJA1 was the first entry of the NMR structure, 2LO1 in the RCSB PDB. A comparative model of JDJA2 was taken from the PMP and was based on the template structure, 2LO1. Structures of the CTD dimer were generated for JA1 and JA2 as follows: the dimerization site was modelled with SM based on the template crystal structure, 1XAO (resolution 2.07 Å). Then, the structures of the CTD monomers were superimposed on the corresponding dimerization site model and only C-terminal missing residues of the dimerization site were added to the CTD domains. The structure of the J-domain of DNJ-12 was taken from the crystal structure PDB code 2OCH (resolution 1.86 Å). The CTD dimer of DNJ-12 was modelled based on the crystal structure, 1NLT, using SM. The J-domain of DNJ-13 was modelled based on 1HDJ and the dimer structure of the CTD of DNJ-13 was based on 3AGZ A/B. Further editing of the following structures was performed to generate a set of comparable structures of the J-proteins. N-terminal Gly was deleted in JDJA1, because it was not part of the UniProt entry P31689. The last seven residues of JDJB1 were deleted to have a comparable C-terminal end to the J-domains of JA1 and JA2. Similarly, the last four C-terminal residues in JDJB4 were deleted to obtain comparable C-terminal ends to the J-domains of JA1 and JA2.
Protein–protein docking
Protein–protein docking was performed with a rigid-body treatment of the protein structures using the Simulation of Diffusional Association (SDA) program (version 7, http://mcm.h-its.org/sda7)45,46. SDA uses Brownian dynamics (BD) simulation to perform the sampling of protein configurations subject to inter-protein forces and torques due to electrostatic and non-polar interactions. The docking protocol required the following steps:
Structure preparation: polar hydrogen atoms were added to the protein structures with WHATIF5 (ref. 47) assuming pH 7.2.
Calculation of protein electrostatic potentials: the electrostatic potential of each protein structure was calculated by numerically solving the linearized Poisson–Boltzmann equation with UHBD (ref. 48). Electrostatic potential grids with 2503 grid points with a 1 Å spacing were used for all proteins. The dielectric constants of the solvent and the protein were set to 78.0 and 4.0, respectively, and the dielectric boundary was defined by the protein's van der Waals surface. The ionic strength was set to 50 mM at a temperature of 300K, with an ion exclusion radius (Stern layer) of 1.5 Å. The protein atoms were assigned OPLS atomic partial charges and radii49.
Calculation of effective charges: these were derived with ECM50. The effective charges for each protein were fit to reproduce the electrostatic potential in a 3-Å-thick layer extending outwards from the protein's solvent-accessible surface computed as defined by a probe of radius 4 Å. The effective charges for proteins were placed on the carboxylate oxygen atoms of Asp and Glu amino acid residues and the C-terminus, and the amine nitrogen atoms of Lys and Arg amino acid residues and the N-terminus. For the Zn21 ion, an effective charge site with a formal charge of −2e was placed on the ion, corresponding to the summed charge of the ion and its four coordinating cysteine side-chains.
Calculation of polar desolvation grids: the desolvation penalty of each effective charge was computed as the sum of desolvation penalties due to the low dielectric cavity of each atom of the other protein51, which was precomputed on a grid. The grid dimensions were set to 1503 grid points with a spacing of 1 Å. Ionic strength and dielectric constants were assigned as for the electrostatic potential calculations. The ion radius was assigned as 1.5 Å.
Calculation of non-polar desolvation grids: the non-polar desolvation forces were computed using precomputed grids52. The distance parameters (a) and (b) were assigned values of 3.10 Å and 4.35 Å, respectively. The parameter (c) was assigned as 1.0 and the conversion factor to β=−0.0065 kcal mol−1 Å−2. The grid dimensions were set to 1503 grid points with a spacing of 1 Å.
Calculation of excluded volume grids: protein shape was described by a grid with a 0.25 Å spacing. A probe of radius of 1.77 Å was used to determine the protein shape. The radius of the solvent probe to determine surface atoms was set to 1.4 Å.
Docking simulations: for each protein pair docked, 10,000 trajectories were generated with SDA. Trajectories were started with the proteins at a separation distance of 100 Å and a random relative orientation. A trajectory was terminated if the protein separation exceeded 300 Å or a simulation time of 500 ns was reached. The protein–protein separation was calculated as the distance between their centres of geometry (CoG). Up to 3,000 configurations sampled during the BD trajectories with a separation of less than 105 Å were saved. During the BD simulations, if a new docking pose was considered similar to a previously saved pose, that is, had an approximate root mean squared deviation (r.m.s.d.) less than 2 Å, then the configuration with the lower intermolecular energy was saved and the counter of this docking pose, the occupation, was incremented. The relative translational diffusion coefficient was set to 0.027 Å2 ps−1. The rotational diffusion coefficient for both proteins was set to 3.92×104 radian2 ps−1. The time step was 1 ps at separations less than 120 Å and increased linearly beyond this threshold with slope of 2 ps Å−1.
Clustering: for each protein pair docked, the configurations saved were clustered with a hierarchical clustering algorithm. The backbone r.m.s.d. between each docked protein configuration was calculated to produce an inter-configuration distance matrix. Initially, each docked structure was assigned to a separate cluster. The closest clusters were found and merged; the distance matrix was updated. This process was repeated until all docked protein structures were in one cluster. The distance between clusters was defined as the average backbone r.m.s.d. between docked protein structures in one cluster relative to structures in another cluster. The representative of a cluster is the protein configuration with the smallest r.m.s.d. to every other member of the cluster. In each clustering cycle, the mean and s.d. of the r.m.s.d. of all members of each cluster to the corresponding cluster representative were calculated. The number of configurations in each cluster in each clustering cycle was determined, taking account of cluster occupation during the BD simulations, and the clusters were ranked by size. The number of generated clusters was chosen using the following criteria. Starting with the largest cluster, the minimum number of clusters accounting for 90% of the total number of configurations docked and satisfying the criterion that the mean r.m.s.d. plus s.d. of the clusters is less than 10Å, was determined. This threshold results in configurations with similar CoG of the J-domains but orientations differing by about 90° being assigned to different clusters.
Of note, class B CTD dimers mostly showed a higher number of selected and total clusters with less favourable interaction energies than class A CTD dimers. This indicates a higher diversity in the docking poses of J-domains to class B CTD dimers (see clustering at N-termini of class B CTD dimers, Extended Data Fig. 7). However, in a full-length protein, this clustering region would be occupied by glycine-phenylalanine-rich linkers and J-domains as in the crystal structure of class B DnaJ2 from Thermus thermophilus (PDB code 4J80)53. Consistently, a docking simulation performed with full-length DnaJ2 showed a much more specific interaction of the DnaJ2 J-domain centering between CTD-I and CTD-II regions (data not shown). To take this into account, we also analyzed the docking clusters for JDJA2 and CTDJB1 by requiring docking positions to satisfy distance requirements from cross-linking data (see Fig. 2d and Extended Data Fig. 7).
Modelling of JA2 and JB1 complexes
Using PyMOL (http://www.pymol.org) software, two putative arrangements (compact and open) of the JA2 and JB1 CTD homodimers were generated to satisfy the maximum number of observed cross-links and FRET constraints possible. The J-domains of JA2 and JB1 were added ensuring consistency with docking, cross-linking and FRET results and a distance to the corresponding CTD that would be allowed by the missing residues that connect J-domain and CTD. The positions of the J-domains of JB1 and JA2 in the two models are expected to provide a FRET efficiency of 8 and 3% (within experimental error), correlating with the lack of experimental observation of FRET between these domains. Note that the structures were treated as rigid bodies and flexibility of the CTD dimers parallel and perpendicular to the dimer plane is very likely and would allow other configurations of these complexes.
Extended Data
Extended data Figure 1. Characterization of protein disaggregation/refolding and refolding-only reactions.
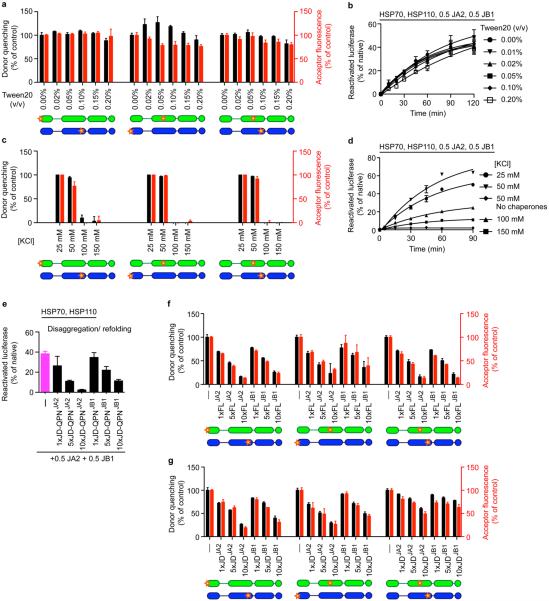
a, HSP70–J-protein–HSP110 (HSPA8–J-protein–HSPH2) functional cycle. Concomitant interaction of HSP70 with a J-protein and substrate results in allosteric stimulation of ATP hydrolysis; this traps the substrate in HSP70 (ref. 8). Subsequent NEF (for example, HSP110) promoted ADP dissociation from HSP70, then allows ATP rebinding, which triggers substrate release to complete the cycle54,55. b, Scheme for in vitro disaggregation/refolding and refolding-only reactions. The aggregates used in disaggregation/refolding assays are preformed by heating luciferase with yeast small heat-shock protein (sHSP) Hsp26 (ref. 4), which is known to co-aggregate with misfolded proteins in vivo27,28 (see Methods for detailed description). If HSP70, J-protein and HSP110 are instead heated together with substrate and Hsp26, luciferase is denatured into a more easily refoldable, inactive and largely monomeric substrate form used in refolding-only assays. c, SEC profiles of aggregated 3H-labelled luciferase (black; size range 200 kDa to ≥5,000 kDa representing ~2 to >50 aggregated luciferase molecules) and monomeric native luciferase (red; size ~63 kDa). Arrows indicate elution size (kDa). Inset, activity of loaded material. d, SEC profile of partially denatured and largely monomeric luciferase (starting material for refolding-only reactions). Inset, activity of loaded material. e, Chaperone nomenclature. f, Disaggregation and reactivation of preformed luciferase aggregates using human HSP70–HSP110 with human J-proteins JA2, green; JB1, blue; JA2+JB1, magenta or no J-protein, black. Under limiting chaperone (HSP70/HSP110) and increasing J-protein concentrations (A, solid or B, dashed) (n = 3). Data are mean ± s.e.m. Precise concentrations are shown in Extended Data Table 1.
Extended data Figure 2. Effects of mixed-class J-proteins on disaggregation/refolding and refolding-only activity of the HSP70 system.
a, Disaggregation/refolding of aggregated luciferase compared for human class A (JA1 and JA2) and class B (JB1 and JB4) J-proteins (n = 3). b, Luciferase refolding-only compared for JA1, JA2, JB1 and JB4 (n = 3). c, Reactivation of heat-aggregated luciferase with nematode HSP70 machinery, using reduced substrate:HSP70 ratio of 1:20, containing DNJ-12 (A), DNJ-13 (B) or DNJ-12+DNJ-13 (A+B) (n = 2). d, Disaggregation/refolding of luciferase using human HSP70 and HSP110 combined with nematode J-proteins (n = 3). e, Reactivation of luciferase showing optimal JA2:JB1 ratio for disaggregation/refolding (n = 2). f, Initial disaggregation/refolding rates for e. g, Final yields of refolded luciferase (120 min) for e. Data are mean ± s.e.m. Precise concentrations are shown in Extended Data Table 1.
Extended data Figure 3. Disaggregation synergy is independent of sHSP incorporation, NEF, substrate and aggregate character, and is not explained by sequential J-protein class activity.
a, Disaggregation/refolding reaction for luciferase aggregates without incorporating sHSP Hsp26 (n = 3). b, Reactivation without NEF (HSPH2) (n = 3). c, Reactivation of α-glucosidase aggregates (n = 3). d, Reactivation of preformed MDH aggregates in the presence of GroEL plus the GroES protein foldase system (GroELS) (n = 2). GroELS is required for efficient MDH refolding56. GroELS alone is in black. JB1:JA2 denotes the stoichiomety of each reaction. e, Disaggregation/refolding of stringent aggregates (≥5,000 kDa) formed using 2 μM luciferase (n = 3). f, Disaggregation/refolding of aggregated luciferase at reduced substrate:HSP70 ratio (luciferase:HSP70:J-protein:HSP110 = 1:7.5:3.8:0.4) (n = 3). The aggregated luciferase concentration is 100 nM. g, h, Holdase function of J-proteins (class A (g) and class B (h)) during luciferase aggregation at 42 °C, shown by decreased light scattering. Concentrations: 1× luciferase; 4× J-protein; 4× BSA (control) (n = 2). i, Reactivation with sequential JA2 and JB1 addition. J-protein added at t = 0 min (black graph legends); J-protein added after 30 or 60 min (red graph legends and arrows) (n = 2). Data are mean ± s.e.m. Precise concentrations are shown in Extended Data Table 1.
Extended data Figure 4. Stoichiometry of class A and B J-proteins determines the range of aggregate sizes resolved.
a, The GroEL trap (GroELD87K) facilitates the capture of 3H-luciferase monomers liberated by protein disaggregation before the refolding step. b, Refolding of disaggregated 3H-luciferase monomers (40 min) in the absence (solid bars) and presence of GroEL trap (open bars). c, SEC profile after disaggregation/refolding of aggregated tritiated α-glucosidase (60 min) with either J-protein class alone (green (A) or blue (B)) or J-proteins combined (magenta). Control reaction without chaperones (black). Elution fractions F1–F4 (red lines). Table shows size distribution of aggregates in each fraction; F1 luciferase aggregates ≥4,000 kDa; F2, aggregates ~400–4,000 kDa; F3, aggregates ~150–400 kDa, F4 disaggregated monomers (~68 kDa). d, Quantification of SEC profile measuring disaggregation of tritiated α-glucosidase from aggregates (F1–F3) from c, also showing concomitant accumulation of disaggregated monomer (F4) from c (n = 3). e, ATP depletion by apyrase abrogates disaggregation. f, Quantification of SEC profile measuring disaggregation of tritiated luciferase from aggregates (F1–F3) with concomitant accumulation of disaggregated monomer (F4), using the HSP70–HSP110 system with JA2 or JB1 alone, or with JA2 plus JB1. Stoichiometry range used for JA2:JB1, 1:1 to 4:1 to 1:4. Specifically, 0.2 JB1:0.8 JA2 (orange); 0.2 JA2:0.8 JB1 (red). Solid colours denote 40-min reaction time; hash denotes 120 min. Control reaction without chaperones (black). Two-tailed t-test, *P<0.05, **P<0.01 (n = 3). Data are mean ± s.e.m. Precise concentrations are shown in Extended Data Table 1.
Extended data Figure 5. JA2 and JB1 form homodimers and interact transiently.
a, Identified JA2 and JB1 inter-molecular cross-links; `Id', amino acid sequence of peptides showing cross-linked lysines (K, orange). Protein 1 and 2 denote source proteins for cross-linked peptides; position 1 and 2 denote positions of cross-linked lysines within proteins; deltaS is the delta score for each crosslink; cut-off = 0.9. ld-Score is the linear discriminant score. b, Representative mass spectrometry spectra for inter-molecular JA2 and JB1 cross-links. Common peaks, green; cross-linked, red; matched peaks, diamonds (no peaks above 1,100 m/z detected). c, SEC profiles of 3H-labelled JA2 dimer (green cartoon) and 3H-labelled JB1 dimer (blue cartoon) mixed with unlabelled J-protein from the other class. Precise concentrations are shown in Extended Data Table 1.
Extended data Figure 6. Electrostatic interactions between J-domain and CTD predominate in JA2 and JB1 complexes.
a, FRET efficiencies for JD–CTD and CTD–CTD interactions with 0–0.2% Tween20 titration. Percentage efficiency is relative to untreated (0% Tween20) samples. Donor quenching (black); acceptor fluorescence (red); below, fluorophore positions in J-protein protomers (JA2, green; JB1, blue). N-termini of JDJA2 and JDJB1 labelled with acceptor fluorophore ReAsH. CTDJA2 and CTDJB1 labelled with donor fluorophores FlAsH and Alexa Fluor 488 at residues 241 and 278, respectively. b, Disaggregation/refolding of preformed luciferase by JA2 and/or JB1 with increasing amounts of Tween20 (n = 2). c, FRET efficiencies for JA2 and JB1 interactions at increasing salt concentrations. d, Disaggregation/refolding of preformed luciferase aggregates by JA2 and JB1 with increasing salt concentrations; control, 50 mM salt, no chaperones (n = 2). e, Luciferase disaggregation/refolding in the presence of excess J-domain fragments carrying JD-QPNJA2/JB1 mutation of the HPD motif (n = 3). f, g, FRET between class A and B J-proteins. f, Competition with unlabelled full-length wild-type J-protein (FL); unlabelled competitor is 1–10× acceptor; (–), no competitor. g, Competition with unlabelled isolated JDJA2 and JDJB1. Data are mean ± s.e.m., average of at least two experiments for FRET experiments. Precise concentrations are shown in Extended Data Table 1.
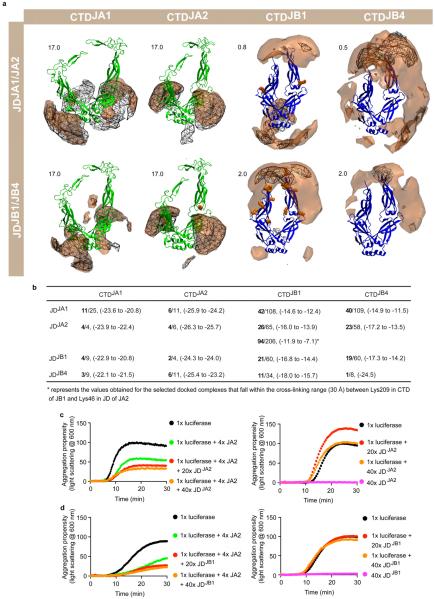
Extended data Figure 7. In silico prediction of JD–CTD interactions between class A and B J-proteins and in vitro evidence that physical interactions between J-proteins do not overlap J-protein substrate binding sites.
a, Preferred positions of the centres of geometry (CoG) of J-domains (y axis, JA1, JA2, JB1 and JB4) around CTD dimers (x axis, class A, green, class B, blue) obtained from molecular docking simulations. JDJA1/JB1, wireframe meshes; JDJA2/JB4, brown contours, each contoured at the isovalue given in the top left of each image. The higher scores for class A CTDs indicate greater specificity of the complexes formed with J-domains; the lower scores for class B CTDs indicate much less specific interactions. Lysines in inter- and intra-J-protein JA2–JB1 cross-links, orange spheres. b, Properties of the docking arrangements obtained after clustering. Total number of clusters per simulation, denominator; number of selected clusters (corresponding to 90% of all docked complexes), numerator, bold. In parentheses, the range of average energy values (in units of kT) for the selected clusters. Lower energy values indicate more favourable binding; fewer clusters indicate a more defined binding mode (see Methods). JDJA2 docking to CTDJB1 is much weaker and less specific than JDJB1 docking to CTDJA1, but docking arrangements compatible with cross-linking results still obtain (Fig. 2d). c, Competition of isolated JDJA2 fragments against JA2 holdase function in luciferase aggregation at 42 °C (n = 2). d, Competition of isolated JDJB1 fragments against JA2 holdase function (n = 2). Luciferase, 1×; JA2, 4×; isolated J-domain fragments, 20× (red; 5-fold excess over JA2), or 40× (orange; 10-fold excess over JA2). Light scattering measured at 600 nm. Precise concentrations are shown in Extended Data Table 1.
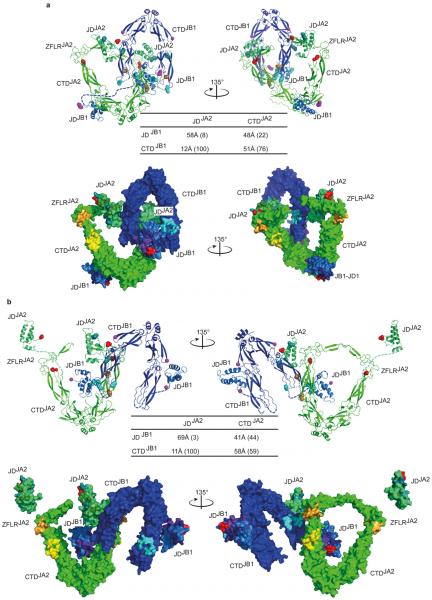
Extended data Figure 8. Possible configurations of the JA2–JB1 mixed-class complex.
a, Compact configuration. b, Open configuration. Configurations were derived from computational docking, using constraints from experimental FRET and cross-linking data (Fig. 2a–d and Extended Data Fig. 5a). Each configuration is shown from two views (left and right) rotated by 135 degrees with respect to each other and in ribbon (top) and molecular surface (bottom) representations. In both cases J-domains of JA2 dock onto the CTD dimer of JB1, and similarly J-domains of JB1 dock to the CTD dimer of JA2. Both CTDJB1 protomers are within cross-linking distance of CTDJA2. Unstructured glycine/phenylalanine (G/F)-rich flexible regions connecting J-domains and CTDs shown by dark blue (JB1) or green (JA2) dashed lines. Residues at FRET fluorophore sites are shown in space-filling representation (red on JA2, magenta on JB1). Inter-molecular crosslinking lysine pairs (gold and cyan, space-filling) are connected by dotted lines. Bottom left within a: molecular surface representation of compact configuration of the JA2–JB1 complex, showing substrate binding sites from crystallographic24 (yellow) and biochemical57 (orange, cyan) data. HPD motif, red. Residues implicated in JD–HSP70 interactions19,58,59 (dark teal and dark green on JDJA2; purple and dark blue on JDJB1). Bottom right within a: rotated image. Table shows fluorophore separation distances; calculated percentage FRET efficiencies in parentheses. a, Both CTDJB1 protomers are within cross-linking distance of CTDJA2. b, As in a, but with only a single CTDJB1 protomer within cross-linking distance to CTDJA2; one JDJA2 docks onto CTDJB1, the other JDJA2 is free. Similarly, one JDJB1 docks onto CTDJA2, the other JDJB1 docks onto its own CTD, consistent with SAXS-determination of class B J-proteins16,17. Model of JB1 (blue) based on the crystal structure of CTD and NMR structure of J-domain. Homology model of JA2 (green) based on the crystal structure of Ydj1 (see Methods).
Extended Data Table 1.
Tabulated reaction conditions
| Panel | Heat treatment time/ Temp | Luciferase/ Hsp26 rxn concentrations (aggregating or denaturing conditions) | Chaperone mixture | Reaction time/ Temp |
|---|---|---|---|---|
| Fig.1b | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [2 μM], NEF [0.1 μM], J-protein [1 μM], 0.5 J-protein [0.5 μM] | 120 min/ 30°C |
| Fig.1c | 15 min/ 45°C | 20 nM (25 nM) | HSP-1 [8 μM], NEF [4 μM], J-protein [4 μM], 0.5 J-protein [2 μM] | 120 min/ 20°C |
| Fig.1d | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], GroELD87K [10 μM], J-protein [380 nM], 0.5 J-protein [190 nM] | 40 min/ 30°C |
| Fig.1e,f | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 120 min/ 30°C |
| Fig.1g | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 40 min/ 30°C |
|
| ||||
| Fig.2e | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM], 1x JD [380 nM], 5x JD [1.9 μM], 10x JD [3.8 μM] | 120 min/ 30°C |
| Fig.2f | 10 min/ 42°C | 20 nM/ 100 nM (20 nM/ 100 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM], 1x JD [380 nM], 5x JD [1.9 μM], 10x JD [3.8 μM] | 80 min/ 30°C |
|
| ||||
| Fig.3e | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 120 min/ 30°C |
| Fig.3f | 10 min/ 42°C | 20 nM/ 100 nM (20 nM/ 100 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 80 min/ 30°C |
| Panel | Heat treatment time/ Temp | Luciferase/ Hsp26 rxn concentrations (aggregating or denaturing conditions) | Chaperone mixture | Reaction time/ Temp |
|---|---|---|---|---|
| EDFig.1c | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | NA | 0 min |
| EDFig.1d | 10 min/ 42°C | 20 nM/ 100 nM (20 nM/ 100 nM) | HSP70 [750 nM], NEF [40 nM], 0.5 JA2 [190 nM], 0.5 JB1 [190 nM] | 0 min |
| EDFig.1f | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 1/2x J-protein [190 nM], 3x J-protein [1140 nM], 6x J-protein [2280 nM], 9x J-protein [3420 nM] | 120 min/ 30°C |
|
| ||||
| EDFig.2a | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 120 min/ 30°C |
| EDFig.2b | 10 min/ 42°C | 20 nM/ 100 nM (20 nM/ 100 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 80 min/ 30°C |
| EDFig.2c | 15 min/ 45°C | 20 nM (25 nM) | HSP-1 [400 nM], NEF [20 nM], J-protein [200 nM], 0.5 J-protein [100 nM] | 120 min/ 20°C |
| EDFig.2d | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [3 μM], NEF [0.1 μM], J-protein [1 μM], 0.5 J-protein [0.5 μM] | 120 min/ 22°C |
| EDFig.2e,f,g | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], 100% J-protein [380 nM] | 120 min/ 30°C |
|
| ||||
| EDFig.3a | 15 min/ 45°C | 20 nM (25 nM) | HSP70 [750 nM], NEF [40nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 120 min/ 30°C |
| EDFig.3b | 15 min/ 45°C | 20 nM/100 nM (25 nM/ 125 nM) | HSP70 [750 nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 120 min/ 30°C |
| EDFig.3e | 15 min/ 45°C | 20 nM/ 100 nM (2 μM/ 10 μM) | HSP70 [2 μM], NEF [0.1 μM], J-protein [1 μM], 0.5 J-protein [0.5 μM] | 120 min/ 30°C |
| EDFig.3f | 15 min/ 45°C | 100 nM/ 500 nM (100 nM/ 500 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 120 min/ 30°C |
| EDFig.3i | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM] | 120 min/ 30°C |
|
| ||||
| EDFig.4b | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], GroELD87K [10 μM], J-protein [380 nM], 0.5 J-protein [190 nM] | 40 min/ 30°C |
| EDFig.4e | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], 0.5 J-protein [190 nM] | 240 min/ 4–30°C |
| EDFig.4f | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], J-protein [380 nM], 0.5 J-protein [190 nM], 0.2 J-protein [76 nM], 0.8 J-protein [304 nM] | 40–120 min/ 30°C |
|
| ||||
| EDFig.5c | NA | NA | J-protein [190 nM] | 10 min/ 30°C |
|
| ||||
| EDFig.6b | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], 0.5 J-protein [190 nM] | 120 min/ 30°C |
| EDFig.6d | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], 0.5 J-protein [190 nM] | 90 min/ 30°C |
| EDFig.6e | 15 min/ 45°C | 20 nM/ 100 nM (25 nM/ 125 nM) | HSP70 [750 nM], NEF [40 nM], 0.5 J-protein [190 nM], 1x JD [380 nM], 5x JD [1.9 μM], 10x JD [3.8 μM] | 120 min/ 30°C |
| Panel | Heat treatment time/ Temp | α-glucosidase/ Hsp26 rxn concentrations (aggregating conditions) | Chaperone mixture | Reaction time/ Temp |
|---|---|---|---|---|
| EDFig.3c | 15 min/ 50°C | 40 nM/ 400 nM (50 nM/ 500 nM) | HSP70 [3 μM], NEF [0.1 μM], J-protein [1 μM], 0.5 J-protein [0.5 μM] | 300 min/ 30°C |
| EDFig.4c,d | 15 min/ 50°C | 40 nM/ 400 nM (50 nM/ 500 nM) | HSP70 [3 μM], NEF [0.1 μM], J-protein [1 μM], 0.5 J-protein [0.5 μM] | 60 min/ 30°C |
| Panel | Heat treatment time/ Temp | malate dehydrogenase/ Hsp26 rxn concentrations (aggregating conditions) | Chaperone mixture | Reaction time/ Temp |
|---|---|---|---|---|
| EDFig.3d | 30 min/ 47°C | 150 nM/ 750 nM (150 nM/ 750 nM) | HSP70 [2 μM], NEF [0.1 μM], 1 J-protein [0.063 μM], 2 J-protein [0.126 μM], 4 J-protein [0.252 μM], GroEL [1 μM], GroES [1 μM] | 220 min/ 30°C |
| Panel | Light scattering | Luciferase | Chaperone mixture | Reaction time/ Temp |
|---|---|---|---|---|
| EDFig.3g,h | 600 nm | 200 nM | 4x J-protein [800 nM], 4x BSA (control) [800 nM] | 30 min/ 42°C |
| EDFig.7c,d | 600 nm | 200 nM | 4x J-protein [800 nM], 20x JD [4 μM], 40x JD [8 μM] | 30 min/ 42°C |
Precise concentrations and conditions tabulated for reactions in Figures as indicated in first column (Panel). EDFig., Extended Data Figure.
Acknowledgements
We thank A. Mogk for critical reading of the manuscript and S. Ungelenk for Hsp26. This work was funded by the Deutsche Forschungsgemeinschaft (SFB1036, BU617/19-1 to B.B.; EXC257, SFB740 to J.K.), Alexander von Humboldt Foundation Postdoctoral Fellowships (to N.B.N. and A.Sz.), National Institutes of Health (the NIGMS, NIA, NIMS), Ellison Medical Foundation and Daniel F. and Ada L. Rice Foundation (to R.I.M.), German Federal Ministry of Education and Research (BMBF) Virtual Liver Network and EU FEP Flagship Programme Human Brain Project (0315749, 604102 to R.C.W.), Klaus Tschira Foundation (to M.B., A.St. and R.C.W.), Sir Henry Wellcome Postdoctoral Fellowship (to F.S.), ETH Zurich and ERC advanced grant Proteomics v3.0 (233226 to R.A.).
Footnotes
Author Contributions N.B.N. and B.B. conceived the study. N.B.N., J.K., A.Sz., M.B., A.St., F.S., D.L.G., R.C.W., M.P.M. and B.B. designed the experiments. N.B.N., J.K., A.Sz., M.B., A.St., F.S., K.A., X.G. and A.Sc. performed the experiments. N.B.N., J.K., A.Sz., M.B., A.St., F.S., R.A., R.C.W., R.I.M., D.L.G., M.P.M. and B.B. analysed the data. N.B.N., D.L.G., M.P.M. and B.B. wrote the manuscript.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirstein-Miles J, Scior A, Deuerling E, Morimoto RI. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 2013;32:1451–1468. doi: 10.1038/emboj.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rampelt H, et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl Acad. Sci. USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein HSP104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 7.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nature Rev. Mol. Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cyr DM, Ramos CH. Specification of Hsp70 function by type I and type II HSP40. Subcell. Biochem. 2015;78:91–102. doi: 10.1007/978-3-319-11731-7_4. [DOI] [PubMed] [Google Scholar]
- 11.Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl Acad. Sci. USA. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzankov S, Wong MJ, Shi K, Nassif C, Young JC. Functional divergence between co-chaperones of Hsc70. J. Biol. Chem. 2008;283:27100–27109. doi: 10.1074/jbc.M803923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauch JN, Gestwicki JE. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J. Biol. Chem. 2014;289:1402–1414. doi: 10.1074/jbc.M113.521997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Z, Cyr DM. Protein folding activity of HSP70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 15.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 16.Ramos CH, Oliveira CL, Fan CY, Torriani IL, Cyr DM. Conserved central domains control the quaternary structure of type I and type II HSP40 molecular chaperones. J. Mol. Biol. 2008;383:155–166. doi: 10.1016/j.jmb.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borges JC, Fischer H, Craievich AF, Ramos CH. Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J. Biol. Chem. 2005;280:13671–13681. doi: 10.1074/jbc.M408349200. [DOI] [PubMed] [Google Scholar]
- 18.Tsai J, Douglas MG. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 19.Suh WC, Lu CZ, Gross CA. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J. Biol. Chem. 1999;274:30534–30539. doi: 10.1074/jbc.274.43.30534. [DOI] [PubMed] [Google Scholar]
- 20.Genevaux P, Schwager F, Georgopoulos C, Kelley WL. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics. 2002;162:1045–1053. doi: 10.1093/genetics/162.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc. Natl Acad. Sci. USA. 2006;103:6166–6171. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 23.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 26.Andréasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 2008;283:8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- 27.Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J. Biol. Chem. 2005;280:23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J. Biol. Chem. 2005;280:23861–23868. doi: 10.1074/jbc.M502697200. [DOI] [PubMed] [Google Scholar]
- 29.Carra S, et al. Different anti-aggregation and pro-degradative functions of the members of the mammalian sHSP family in neurological disorders. Phil. Trans. R. Soc. Lond. B. 2013;368:20110409. doi: 10.1098/rstb.2011.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vos MJ, et al. HSPB7 is the most potent polyQ aggregation suppressor within the HSPB family of molecular chaperones. Hum. Mol. Genet. 2010;19:4677–4693. doi: 10.1093/hmg/ddq398. [DOI] [PubMed] [Google Scholar]
- 31.Skouri-Panet F, Michiel M, Ferard C, Duprat E, Finet S. Structural and functional specificity of small heat shock protein HspB1 and HspB4, two cellular partners of HspB5: role of the in vitro hetero-complex formation in chaperone activity. Biochimie. 2012;94:975–984. doi: 10.1016/j.biochi.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 32.Peschek J, et al. Regulated structural transitions unleash the chaperone activity of αB-crystallin. Proc. Natl Acad. Sci. USA. 2013;110:E3780–E3789. doi: 10.1073/pnas.1308898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leitner A, et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014126. M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinner O, et al. Identification of cross-linked peptides from large sequence databases. Nature Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walzthoeni T, et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nature Methods. 2012;9:901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- 36.Adams SR, et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 37.Spagnuolo CC, Vermeij RJ, Jares-Erijman EA. Improved photostable FRET-competent biarsenical-tetracysteine probes based on fluorinated fluoresceins. J. Am. Chem. Soc. 2006;128:12040–12041. doi: 10.1021/ja063212q. [DOI] [PubMed] [Google Scholar]
- 38.Dixit A, Ray K, Lakowicz JR, Black LW. Dynamics of the T4 bacteriophage DNA packasome motor: endonuclease VII resolvase release of arrested Y-DNA substrates. J. Biol. Chem. 2011;286:18878–18889. doi: 10.1074/jbc.M111.222828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(suppl. 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 40.Kopp J, Schwede T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki H, et al. Peptide-binding sites as revealed by the crystal structures of the human Hsp40 Hdj1 C-terminal domain in complex with the octapeptide from human Hsp70. Biochemistry. 2010;49:8577–8584. doi: 10.1021/bi100876n. [DOI] [PubMed] [Google Scholar]
- 44.Qian YQ, Patel D, Hartl FU, McColl DJ. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J. Mol. Biol. 1996;260:224–235. doi: 10.1006/jmbi.1996.0394. [DOI] [PubMed] [Google Scholar]
- 45.Martinez M, et al. SDA7: a modular and parallel implementation of the simulation of diffusional association software. J. Comput. Chem. 2015;36:1631–1645. doi: 10.1002/jcc.23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gabdoulline RR, Wade RC. Simulation of the diffusional association of barnase and barstar. Biophys. J. 1997;72:1917–1929. doi: 10.1016/S0006-3495(97)78838-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 48.Madura JD, et al. Electrostatics and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics Program Comp. Phys. Comm. 1995;91:57–95. [Google Scholar]
- 49.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 50.Gabdoulline RR, Wade RC. Effective charges for macromolecules in solvent. J. Phys. Chem. 1996;100:3868–3878. [Google Scholar]
- 51.Elcock AH, Gabdoulline RR, Wade RC, McCammon JA. Computer simulation of protein-protein association kinetics: acetylcholinesterase-fasciculin. J. Mol. Biol. 1999;291:149–162. doi: 10.1006/jmbi.1999.2919. [DOI] [PubMed] [Google Scholar]
- 52.Gabdoulline RR, Wade RC. On the contributions of diffusion and thermal activation to electron transfer between Phormidium laminosum plastocyanin and cytochrome f: Brownian dynamics simulations with explicit modeling of nonpolar desolvation interactions and electron transfer events. J. Am. Chem. Soc. 2009;131:9230–9238. doi: 10.1021/ja809567k. [DOI] [PubMed] [Google Scholar]
- 53.Barends TR, et al. Combining crystallography and EPR: crystal and solution structures of the multidomain cochaperone DnaJ. Acta Crystallogr. D. 2013;69:1540–1552. doi: 10.1107/S0907444913010640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J, Walter S, Horwich AL, Smith DL. Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nature Struct. Biol. 2001;8:721–728. doi: 10.1038/90443. [DOI] [PubMed] [Google Scholar]
- 57.Linke K, Wolfram T, Bussemer J, Jakob U. The roles of the two zinc binding sites in DnaJ. J. Biol. Chem. 2003;278:44457–44466. doi: 10.1074/jbc.M307491200. [DOI] [PubMed] [Google Scholar]
- 58.Garimella R, et al. Hsc70 contacts helix III of the J domain from polyomavirus T antigens: addressing a dilemma in the chaperone hypothesis of how they release E2F from pRb. Biochemistry. 2006;45:6917–6929. doi: 10.1021/bi060411d. [DOI] [PubMed] [Google Scholar]
- 59.Greene MK, et al. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl Acad. Sci. USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]