Abstract
Cells move through perpetual protrusion and retraction cycles at the leading edge. These cycles are coordinated with substrate adhesion and retraction of the cell rear. Here, we tracked spatial and temporal fluctuations in the molecular activities of individual moving cells to elucidate how extracellular regulated kinase (ERK) signaling controlled the dynamics of protrusion and retraction cycles. ERK is activated by many cell-surface receptors and we found that ERK signaling specifically reinforced cellular protrusions so that they translated into rapid, sustained forward motion of the leading edge. Using quantitative fluorescent speckle microscopy (qFSM) and cross-correlation analysis, we showed that ERK controlled the rate and timing of actin polymerization by promoting the recruitment of the actin nucleator Arp2/3 to the leading edge. Arp2/3 activity generates branched actin networks that can produce pushing force. These findings support a model in which surges in ERK activity induced by extracellular cues enhance Arp2/3-mediated actin polymerization to generate protrusion power phases with enough force to counteract increasing membrane tension and to promote sustained motility.
Introduction
Cell movement is essential to many biological phenomena, including embryogenesis, wound healing, and cancer metastasis. The motility process involves cycles of membrane protrusion and retraction at a leading edge, which are coordinated in space and time with adhesion dynamics and cell rear retraction (1).
In migrating epithelial sheets, the rate of edge protrusion is driven by the rate of F-actin assembly (2). A dendritically-branched polymer network grows against the leading edge plasma membrane and turns over within 1 to 4 micrometers from the cell edge, which defines the lamellipodium (3, 4). The seven subunit Arp2/3 protein complex mediates nucleation of this branched actin filament assembly. The WAVE regulatory complex activates Arp2/3 (5, 6) and is recruited along with Arp2/3 to the edge of expanding protrusions (7–9). Rac and phospholipid binding recruit the WAVE regulatory complex to the plasma membrane (10–13).
We have previously proposed a model in which protrusion initiation is followed by a power phase of increased actin filament assembly (we calculated power output from the product of the cell boundary force and the cell edge motion) (14). We have proposed that as membrane tension increases during edge advancement, the power phase is terminated by a maximal tension level that exceeds the amount of propulsion and adhesion stress produced by the combined assembly of actin filaments and nascent adhesions. In this scenario, protrusion cycle duration is directly related to the efficiency with which actin filament assembly is increased after protrusion initiation. Biochemical mechanisms involving signaling proteins likely contribute to the force and tension-based control. For example, the Rac exchange factor β-PIX and the Rac-recruited Arp2/3 inhibitory molecule Arpin create positive and negative feedback loops for lamellipodial actin polymerization that control protrusion and retraction cycles (15, 16). How extracellular signals feed into and perturb the force and control of protrusion cycle timing is largely unexplored.
Myriad signaling inputs from growth factors, hormones, neurotransmitters, and chemokines feed into the cell migration machinery. One of the chief transducers of signals is Extracellular Regulated Kinase (ERK), a Mitogen Activated Protein Kinase (MAPK) (17, 18). ERK is activated by the small GTPase Ras, which recruits the Ser/Thr kinase Raf to the plasma membrane for activation. Raf phosphorylates and activates the kinases MEK1/2, which activate ERK1/2 (17, 18). Hereafter, we use MEK to refer to MEK1/2 and ERK to refer to the ERK1/2 isoforms. ERK activity is necessary for epithelial sheet and tubule movement, forms of cell migration common during embryogenesis, wound healing and cancer metastasis (19–21). Reports on ERK’s role in migration include transcription-dependent induction of EMT (22, 23) to direct regulation of actin polymerization and focal adhesions (24–26).
We have previously found that ERK phosphorylation of the WAVE regulatory complex promotes the interaction of WAVE with Arp2/3 (25). ERK inhibition for several hours reduces spontaneous protrusion velocity in model migrating epithelial sheets (25). Here, we asked if the role of ERK in protrusion could be separated from its transcriptional activity by assaying the immediate effects of acute ERK inhibition. We analyzed fluctuations in edge motion during steady-state motility and discovered that ERK promoted a gain in protrusion velocity and duration. We spatiotemporally resolved ERK’s point of action and found that following protrusion initiation, ERK promoted Arp2/3-accumulation at the cell edge, which drove the increase in actin polymerization for protrusion reinforcement. Thus, ERK signaling creates the assembly power needed to overcome increasing membrane tension as protrusions progress and cells move forward.
Results
ERK promotes cell motility
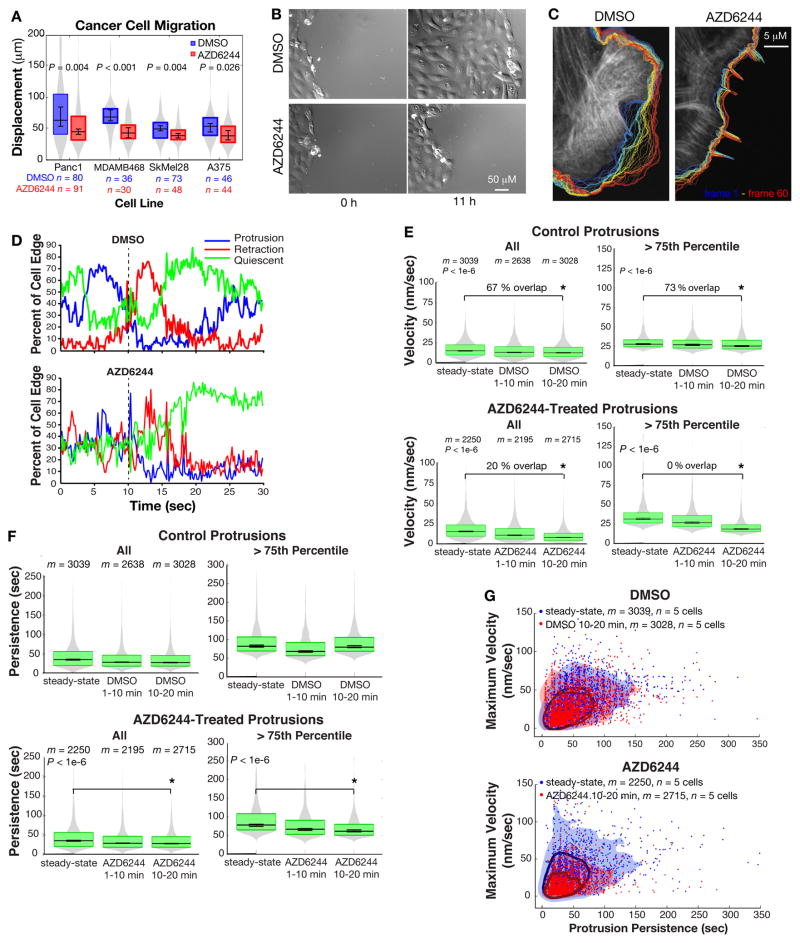
We assessed the effects of MEK1/2 inhibition on cell movements using AZD6244, a non-competitive MEK1/2 inhibitor with improved potency and specificity over U0126, an older inhibitor that also inhibits MEK5 signaling to ERK5 (27–29). Phosphatases constantly remove MEK’s activating phosphorylations on ERK (30), so that treatment of PtK1 epithelial cells with the MEK inhibitor AZD6244 reduced ERK activation within 1 min (Fig. S1A). Phosphorylation of the ERK substrates p90 ribosomal S6K (RSK) (31) and WAVE2 (32) was lost in 2–3 min (Fig. S1A, B). Inhibiting MEK with the compound AZD6244 reduced single-cell migration distance in many cell types, including Panc1 pancreatic cancer cells harboring an activating Kras mutation, MDA-MB-468 breast cancer cells harboring amplification of the EGFR gene (which encodes the epidermal growth factor receptor), and Sk-Mel-28 and A375 melanoma lines harboring the activating BrafV600E mutation (Fig. 1A). MEK inhibition also reduced the epithelial sheet migration of untransformed PtK1 cells closing a wound (Fig. 1B), a process driven by edge protrusion and adhesion rather than actomyosin contraction (2). DMSO-treated sheets moved at a median velocity of 0.328 μm/min while AZD-treated sheets moved at 0.198 μm/min (p = 0.0081).
Fig. 1. ERK signaling extends membrane protrusion velocity and persistence time.
(A) Displacement distributions of cancer cells migrating after treatment with DMSO or AZD6244 (MEK inhibitor). n = number of cells tracked in 3 independent experiments. (B) Representative images of the migration of a DMSO- or AZD6244-treated PtK1 monolayer after scratch wounding. (C) Representative overlay of single cell edge dynamics tracked for 10–20 min post-treatment. Blue: early time points; red: late time points. (D) Representative profile (1 of 5 cells in each treatment group) of the fraction of the cell edge in protrusion, retraction, quiescent states. (E) Velocity and (F) persistence time distributions of m significant protrusion events in n = 5 DMSO- and AZD6244-treated cells tracked in 3–4 independent experiments. The “> 75th Percentile” graphs plot the distribution of the top 25% of events in the corresponding “All” graphs. For A, E and F, gray area indicates total smoothed distribution and the boxes’ upper and lower edges represent the 75th and 25th distribution percentiles, respectively. Central horizontal line indicates the median. Error bars about the median indicate 95% confidence interval. (G) Maximum velocity compared to persistence time of all protrusion events tracked in 3–4 independent experiments. Encircled areas indicate events within the 1–75th percentile of the joint velocity and persistence distribution. Shaded areas indicate protrusions within the 1–99th percentile.
ERK signaling increases membrane protrusion velocity and persistence time
We imaged the spontaneous edge protrusion and retraction cycles of steady-state (log-phase) PtK1 epithelial cells expressing Emerald-LifeAct as a generic actin cytoskeleton marker (Movies S1, 2). We identified the cell edge and tracked it over time (Fig. 1C) and from these traces we calculated the velocity along the entire cell edge (Movies S3, 4) (33). We developed a signal detection algorithm to extract the significant protrusion and retraction events from the noisy velocity profile of each edge segment (Materials and Methods). From each detected event, we extracted the event duration and calculated the mean and maximum velocities of that duration. Quantification of the percentage of cell edge segments in states of protrusion, retraction, and quiescence (when velocity was at 0) revealed a retraction response induced by the physical perturbation of adding reagent to both control and treated cells. DMSO-treated cells recovered their native motion state within 5–10 min after treatment, while AZD6244-treated cells did not (Fig. 1D). We removed the retraction artifact from the edge velocity analysis by dividing the data into three time intervals: steady-state, 1–10 min and 10–20 min post-treatment. We found that short-term MEK inhibition reduced the mean protrusion velocity 10–20 min post-treatment (Fig. 1E, All, Table S1). Although the protrusion velocity of the control cells was statistically different between steady-state and 10–20 min after treatment, the distributions overlapped by 70%. In contrast, the distribution overlap between steady-state and MEK inhibitor-treated cells after 10–20 min was only 20%. The reduction in protrusion velocity after DMSO treatment was likely a residual effect of the retraction artifact induced by physically perturbing the cells. When compared to the protrusion dynamics of control cells treated with DMSO for 10–20 min (median of the mean velocities: 12.4 nm/sec), MEK inhibition significantly reduced protrusion velocity (median of the mean velocities: 8.1 nm/sec, Fig. S1C, Table S1). MEK inhibition also reduced protrusion persistence time (Fig. 1F, All, Table S1). Thus, MEK inhibition shifted the distribution of edge protrusions to events with slower velocities and shorter durations.
We hypothesized that ERK activity was particularly necessary for the production of faster, more persistent protrusions that have entered the power phase. Indeed, the reduction in both protrusion velocity and persistence upon MEK inhibition became more evident when only the 25% fastest and longest protrusion events were analyzed (Fig. 1E, 1F, and S1C, 75th Percentile, Table S1). Cells treated for 10–20 min with DMSO exhibited a 75th percentile mean protrusion velocity of 19.3 nm/sec, whereas cells treated with the MEK inhibitor exhibited protrusions with a 75th percentile mean velocity of 13.6 nm/sec. Similar results were found when the data were analyzed using the maximum protrusion velocity reached per protrusion cycle (Table S1) and when cells were treated with the structurally-distinct MEK inhibitor U0126 and imaged on an independent microscope configuration (Fig. S1E–G). In the latter case, the overall protrusion velocity was slower, which we attributed to lot-specific variability in the imaging media and changes in the microscope configuration. Nonetheless, U0126 significantly reduced protrusion velocity and persistence. To test whether ERK was necessary for the maximum range of protrusion velocity and persistence, we plotted the maximum velocity against the duration of each protrusion event in steady-state cells and 10–20 min after treatment (Fig. 1G). In control cells, 25% of protrusion events reached velocities greater than 50 nm/sec and lasted for more than 90 sec. Treatment with the vehicle DMSO did not change this distribution, while MEK inhibition caused a shift to slower, shorter protrusions, such that only 1% of the treated events reached peak velocities greater than 50 nm/sec. These findings were further confirmed in PtK1 cells stably expressing hyperactive BrafV600E, which causes a moderate increase in ERK activity (Fig. S2A). The BrafV600E cells exhibited more frequent protrusion events that reached greater mean and maximum velocities than cells expressing empty vector (Fig. S2B and C). The persistence time was reduced in the 75th percentile BrafV600E events (Fig. S2D), suggesting that the more frequent and faster protrusions might be running out of other necessary actin assembly factors to maintain their persistence.
We also found that MEK inhibition reduced and MEK activation promoted retraction velocity (Fig. S3A, B, E). This is in contrast to our previous data (25), in which we treated cells with the MEK inhibitor for 4 hours and did not remove noise in the velocity analysis. ERK may also promote retraction persistence. Retractions of control cells exhibited an insignificant reduction in persistence time from 32.5 sec to 30.9 sec upon treatment with DMSO for 10–20 min, while retractions in MEK inhibitor-treated cells exhibited a significant reduction in persistence from 34.6 to 27.4 sec (Fig. S3C, Table S1). However, due to cell-to-cell heterogeneity, the steady-states in control and treated groups were not equivalent and the retraction persistence following DMSO and AZD6244-treatments were not significantly different (Fig. S3D). Increased ERK activation by BrafV600E expression reduced retraction persistence, (Fig. S3F), similar to the BrafV600E-induced reduction in protrusion persistence. The regulation of retraction by ERK could be secondary to reduced protrusion, because protrusion and retraction forces are in equilibrium during motility (2) or through direct modulation of the retraction machinery, such as myosin light chain kinase or the adhesion component Paxillin (34–37).
ERK promotes actin assembly dynamics in protrusions
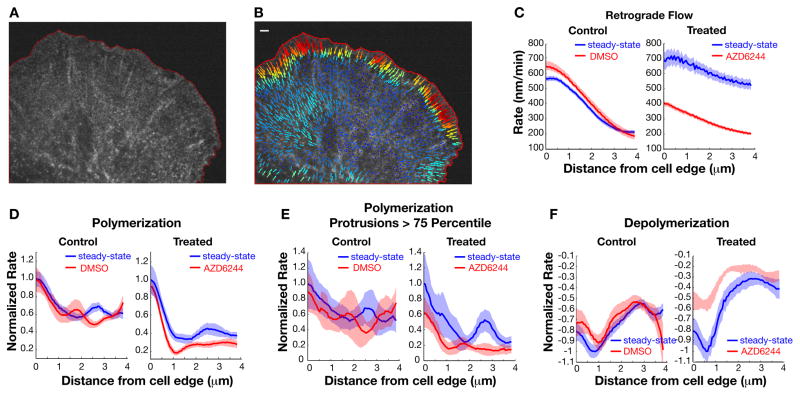
We hypothesized that ERK regulates protrusion by directly regulating actin assembly in the power phase. Because ERK activation at adhesions contributes to cell migration velocity (34), we aimed to directly assess actin dynamics in situ. We microinjected fluorescent actin and used quantitative fluorescent speckle microscopy (qFSM) to identify speckles of assembled actin monomers and track their motion (38). We imaged cells for 9 min to obtain a measure of steady-state dynamics, added the MEK inhibitor, and imaged the cells again after a 10 min recovery period (Movies S5 and S6). We confirmed that these cells had reduced protrusion velocity and persistence time upon MEK inhibition, similar to the cells labelled with LifeAct (39) (Fig. S4A–C).
Using actin qFSM software, we identified speckles in the raw images (Fig. 2A) and their displacements tracked (Fig. 2B, Movies S7 and S8) and used to identify local events of actin filament polymerization and depolymerization (40). In this method, speckles are assigned assembly and disassembly scores, which are a function of the rate of change in speckle intensity (net number of fluorescent actin monomers that are incorporated or unincorporated into the actin filament) and background fluorescence. In our PtK1 cells, heterogenous and rapidly changing cell protrusion and retraction states along the cell edge meant that actin assembly and assembly-driven actin retrograde flows were spatially segregated and transient. As previously reported, averaging the actin dynamics over time concealed spatio-temporal definition (41), but revealed typical lamellipodial actin characteristics: rapid retrograde flow (600–800 nm/min) towards the cell center and a 1–2 micron band of assembly followed by a 1–2 micron band of disassembly (4, 42, 43) (Fig. S4D, E). FRAP of EGFP-actin in B16-F1 melanoma cells has also shown that actin turnover is restricted to within ~2 μm of the cell edge (7). We then removed the non-specific spatial and temporal averaging and averaged the actin assembly and disassembly scores as a function of distance from the cell edge. We found that during protrusion events, flow rates peaked at the cell edge and continuously decreased over the 2–3 μm width of the lamellipodium (Fig. 2C). Actin polymerization peaked at, but was not restricted to the cell edge, and depolymerization peaked 0.5 μm inside the edge. Treatment with the MEK inhibitor reduced actin retrograde flow and actin assembly and disassembly rates at the cell edge in protruding edges (Fig. 2C–F). The effect of MEK inhibition on actin flow was likely secondary to its effect on actin polymerization, because less polymerization against the plasma membrane would generate less force converted into retrograde flow (14). The MEK inhibitor-induced reduction in actin disassembly is likely a combination of a shift in the steady-state balance of lamellipodial actin assembly and disassembly and reduced incorporation of Arp2/3, because ATP hydrolysis of incorporated Arp2 and Arp3 contribute to actin network disassembly (44).
Fig. 2. ERK promotes actin flow and assembly in protrusions.
(A) Raw image of fluorescent actin speckles. The red outline defines the computationally segmented cell mask. (B) Instantaneous flow vectors of speckles identified in A, calculated with qFSM software (38). Longer and red vectors indicate faster flow rate. Scale bar, 5 μm. (C to F) Normalized instantaneous rates of actin retrograde flow (C), polymerization (D, E), and depolymerization (F) calculated as average rate over all identified speckles as a function of distance from the cell edge. Data from a single representative control cell and a MEK-inhibitor-treated cell before and after treatment. Shaded areas are 95% confidence interval. On average, within a micron from the cell edge, DMSO-treatment reduced flow 18% +/− 1.1%, actin assembly 2% +/− 3.3%, and actin disassembly 9% +/− 7.3% (n = 6 cells tracked in 4 independent experiments). AZD6244 treatment reduced flow rates 28% +/− 0.8%, actin assembly 31% +/− 5.7%, and actin disassembly 38% +/− 3.1% (n = 4 cells tracked in 4independent experiments).
ERK regulates actin dynamics during the protrusion power phase
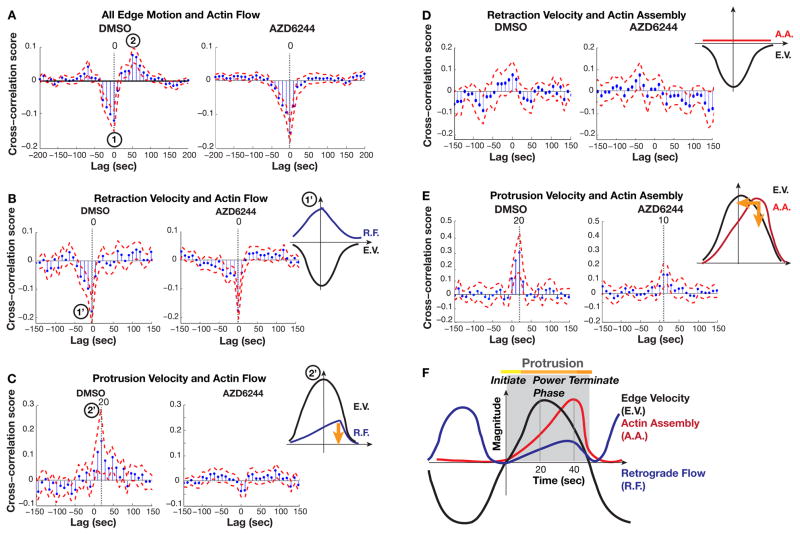
Because cells treated with ERK inhibitor exhibit reduced protrusion velocity and persistence and reduced actin filament assembly rates, we postulated that these cells generated a weaker actin assembly-driven “power phase” and their filament propulsion was outcompeted by plasma membrane tension earlier in the protrusion cycle than in cells with normal ERK signaling. To test if ERK controlled the timing of actin dynamics with respect to edge motion, we computed the cross-correlation of these two waveforms. Cross-correlation scores indicated the strength of coupling between two waveforms and the lag of the peak in the score indicated the timing of the strongest correlation. Consistent with our previous results (14), we found that in DMSO-treated cells, actin flow negatively correlated with edge dynamics at time lag = 0 (Fig. 3A, mark 1). Furthermore, actin flow positively correlated with edge motion in a MEK inhibitor-sensitive manner when flow was shifted 50–60 sec forward in time (Fig. 3A, mark 2 in left panel; this correlation disappears in the right panel).
Fig. 3. ERK regulates actin dynamics during protrusion power phase.
(A) Temporal cross-correlation of actin flow with edge velocity along entire edge, including retractions and protrusions. Marks 1 and 2 indicate negative and positive extrema, respectively. (B and C) Temporal cross-correlation of actin flow specifically in retractions (B) and protrusions (C). Marks 1′ and 2′ indicate that the negative and positive extrema of the combined cross-correlation in (A) are associated with retraction and protrusion events, respectively. (D and E) Temporal cross-correlation of actin assembly with edge velocity in retractions (D) and protrusions (E). Retraction velocity was computed with negative values. Positive time-lags in x-axis indicate that actin dynamics were delayed relative to edge motion. The corresponding model diagrams interpret the cross-correlation scores as readouts of temporal coordination between actin dynamics and edge velocity. E.V., edge velocity. R.F., retrograde flow. A.A., actin assembly. Orange arrows indicate how the indicated parameter of actin dynamics is altered relative to the edge velocity upon MEK inhibition. In control DMSO-treated cells, actin assembly correlates with protrusive edge motion with a cross-correlation score of 0.4 and a delay of 20 sec. In cells treated with AZD6244 and lacking ERK activity, actin assembly dynamics are less correlated with edge motion (cross-correlation score of 0.2) and this correlation occurs with a delay of only 10 sec. (F) Revised model of the sequence of mechanical processes during protrusion and retraction events. Actin retrograde flow correlates with the cell edge twice during a protrusion-retraction cycle: once when peak retraction velocities are attained and once 20 seconds after protrusion initiation, which coincides with peak actin assembly.
We then deconvolved the correlation plots by calculating the correlation scores in retraction against protrusion events separately. We found that the negative correlation between edge motion and flow was present in retractions, but not in protrusions (Figs. 3B, C), indicating that retrograde flow was fastest when retraction velocity was peaking (Fig. 3B, mark 1′). In contrast, edge motion and flow positively correlated in the protrusions of DMSO-treated cells, but with a shorter time delay (time lag = 20 sec) than when total edge motion was analyzed (Fig. 3C, mark 2′ compared to Fig. 3A, mark 2). We analyzed a synthetic dataset (Fig. S5A) to test if edge motion in retractions artificially delayed the positive correlation between actin flow and edge motion in protrusions when both forms of edge motion were included in the analysis. The synthetic total edge motion and flow and retraction motion and flow were negatively correlated at lag = 0 (Fig. S5B, C). However, the positive correlation between edge motion and flow was shifted towards longer time lags when total edge motion was correlated with protrusion motion (Fig. S5B, D). Thus, because the negative correlation was stronger than the positive, its inclusion in the analysis artificially delayed the positive correlation between protrusion motion and actin retrograde flow. We concluded that the rate of retrograde flow peaked 20 sec after the maximal protrusion velocity, at the end of the power phase (Fig. 3C).
Because our previous model of the protrusion and retraction cycle was based on correlations with total edge motion (14), we computed the cross-correlation between edge motion and actin assembly rates separately during protrusion and retraction events. No correlation was found during retraction events (Fig. 3D). In protrusion events, the correlation maximum occurred at lag = 20 sec (Fig. 3E, left panel), indicating that the highest rate of actin assembly occurred 20 sec after maximal protrusion velocity is reached, consistent with our previous report (14). Our new analysis indicated that maximum retrograde flow occurred at the same time as the peak actin assembly during protrusion events. Both were delayed relative to the peak protrusion velocity. Retrograde flow then peaked again when retraction velocity was maximal, likely as a result of a release of adhesions and an increase in actomyosin contraction (14, 45) (Fig. 3F).
We next tested if ERK activity controls the timing of actin dynamics in the protrusion cycle. MEK inhibition had no effect on the negative correlation between edge motion and actin flow during retraction (Fig. 3B, right panel). In contrast, MEK inhibition abrogated the correlation between edge dynamics and actin retrograde flow during protrusions (Fig. 3C). AZD6244 treatment also reduced the correlation score and shortened the time delay between edge dynamics and actin assembly during protrusion (Fig. 3E). Thus, ERK specifically coordinated the timing of actin polymerization and retrograde flow during edge protrusion, but not retraction. Actin assembly and retrograde flow were reduced in cells with ERK inhibition (Fig. 2), and actin assembly peaked sooner in the protrusion cycle and was less correlated with edge motion than in cells with intact ERK signaling (Fig. 3). We conclude that ERK was necessary for reinforcing actin assembly against increasing membrane tension during the protrusion power phase.
ERK controls the protrusion power phase by modulating Arp2/3 recruitment
We wanted to determine if the effect of ERK on protrusion speed and persistence was through regulation of Arp2/3-mediated actin polymerization. We hypothesized that ERK controls the intensity of activation of pre-localized WAVE regulatory complex to increase the recruitment and activation of Arp2/3. This is because ERK does not regulate WAVE recruitment to the membrane (25) and although ERK is sequestered in different subcellular locales through scaffolding proteins, passive diffusion allows activated Ras to rapidly spread over 10 micrometers within seconds (18, 46). Thus, ERK is unlikely to be a localization signal for Arp2/3 recruitment or protrusion initiation. In our previous experiments in which serum starved cells had been stimulated with EGF, we observed localization of phosphorylated ERK with WAVE2 along the cell edge (25). Here, we examined log-phase migrating cells harboring a low ratio of phosphorylated/unphosphorylated ERK. At any given time, these steady-state cells have edges in states of protrusion, retraction, and quiescence, but immunofluorescence did not show a strong cell-edge localization of active ERK. Rather, phosphorylated ERK was found at the edge and throughout the lamella and cell body (Fig. S6A). Phosphorylated ERK was not spatially restricted to areas of active protrusion or retraction, as gauged by edge morphology and Arp2/3 levels (Fig. S6A). The specificity of the phospho-ERK-488 signal was demonstrated by the lack of staining in cells treated with MEK inhibitor (Fig. S6B, D) or in cells that were not incubated with the anti-phospho-ERK antibody (Fig. S6C).
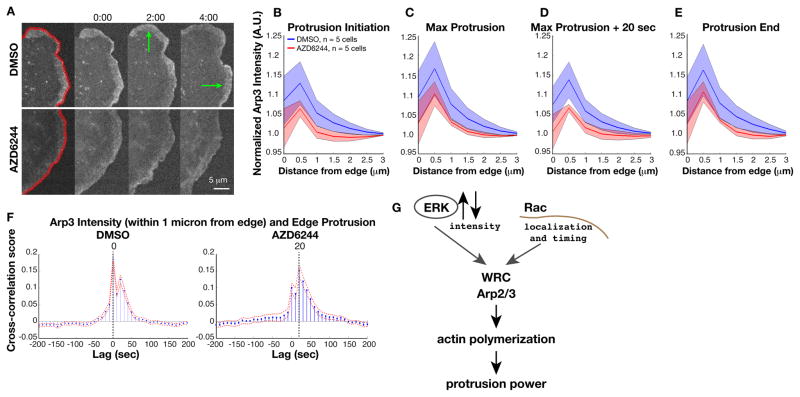
We used GFP-tagged Arp3 to determine the effects of ERK inhibition on Arp2/3 localization to the cell edge during steady-state motility (Fig. 4A). We quantified the GFP intensity in temporally aligned profiles (Fig. 4B–E). MEK inhibition reduced the amount of Arp3 recruited along the profiles, but the reduction was only significant in the profile temporally aligned 20 sec after maximum protrusion, during the power phase (Fig. 4D). Thus, the presence of active ERK in initiated protrusions increased Arp2/3 recruitment during the remainder of the protrusion cycle. We used immunofluorescence of ARPC2, a distinct Arp2/3 subunit, as an independent method of validating that ERK regulates Arp2/3 recruitment. We did not expect to find a complete abrogation of Arp2/3 localization to the cell edge upon ERK inhibition, because the edges of fixed cells are in various states of protrusion initiation, power phase, retraction, and quiescence. However, we did find a trend in which MEK inhibition reduced overall ARPC2 localization to the cell edge (Fig. S6A, B, E). We propose that in cells with ERK inhibition, reduced Arp2/3 recruitment led to a loss of correlation between actin and edge dynamics during protrusion.
Fig. 4. ERK controls actin assembly and protrusion power phase by enhancing Arp2/3 recruitment.
(A to E) GFP-Arp3 localization at cell edge. Green arrows indicate protrusion events (A). Normalized mean Arp3 intensity (m = 2779 protrusion events from n = 5 cells tracked in 3 independent experiments) in cells treated with DMSO (blue) or MEK inhibitor (red) at protrusion initiation (B), point of maximum velocity (C), power phase (~20 s after maximal protrusion velocity) (D), and protrusion end (E). GFP intensity at the cell edge (within 0–0.5 microns) was normalized to intensity in the lamella (3–3.5 microns). Shaded areas indicate 95% confidence intervals. (F) Cross-correlation of Arp2/3 intensity and cell edge velocity shows reduced correlation and delayed recruitment with MEK inhibitor. (G) Model of the role of ERK in controlling edge protrusion. Following protrusion initiation, ERK controls an intensity rheostat of WAVE regulatory complex (WRC) activation and Arp2/3 recruitment to induce actin polymerization power for sustained events.
Cross-correlation of GFP-Arp3 intensity with edge velocity during protrusion events showed 2 peaks in Arp2/3 recruitment (Fig. 4F). The first peak indicated that a pool of Arp2/3 was recruited to the cell edge at the same time as the peak in edge protrusion velocity. The second peak indicated that a second, weakly correlated pool of Arp2/3 was recruited 20 sec later, which coincided with the peak in actin polymerization rates. The timing of the second Arp2/3 peak might depend upon the first. In addition, we expect that both of these pools of Arp2/3 contribute to the peak in actin assembly in the power phase.
In cells treated with the MEK inhibitor, the peak in Arp2/3 correlation was delayed by 20 sec (Fig. 4F). A smaller peak coincided with the peak in edge protrusion, a finding that indicated that recruitment of Arp2/3 to the cell edge took longer when ERK was inhibited. Furthermore, the first pool of Arp2/3 that was recruited at the same time as the increase in protrusion velocity was less correlated with edge motion than in uninhibited cells (cross-correlation score is 0.2 in control cells and 0.1 with AZD6244 treatment). The residual Arp2/3 that was recruited during the shortened, weakened power phase was slightly more correlated with edge motion than the power phase peak in uninhibited cells. This could be explained by the homogenization of protrusion behavior under MEK inhibition. Uninhibited cells had a wide range of protrusion behavior, ranging from slower protrusions that lack a “power” phase to faster, longer protrusions with substantial power phases. In cells treated with the MEK inhibitor, the distribution collapsed to a uniform protrusion cycle of weakened, short protrusions. In conclusion, ERK regulated both the extent and the correlation of Arp2/3 recruitment to edge motion.
Discussion
In this work, we have deduced the mechanisms by which the ERK signaling pathway altered the protrusion and retraction cycles to promote productive migration. We found that the ERK pathway was not essential for protrusion initiation, but rather promoted protrusion reinforcement by Arp2/3-mediated actin polymerization. We uncovered this mechanistic insight by cross-correlating signaling read-outs with the mechanical processes of steady-state cell migration in defined protrusion and retraction events.
Phospholipids and locally-activated Rac recruit and basally activate the WAVE regulatory complex, which activates Arp2/3 (10–12). Arp2/3 nucleates actin dendritic branches to accelerate membrane protrusion. Meanwhile, a positive feedback loop between Rac and the actin cytoskeleton recruits more Rac, WAVE regulatory complex, and basally active Arp2/3 (15, 47). We have previously found that peak Rac recruitment occurs 20 sec after protrusion initiation (48), which corresponds to our finding here that actin polymerization peaked 20 sec after protrusion initiation. Yet, we also found here that a relatively small percentage of protrusion events were “stabilized” with increased protrusion velocity and duration to compel cell displacement. We propose that in these events, ERK localized to the cell edge phosphorylates and activates the WAVE regulatory complex for further Arp2/3 recruitment and activation (Fig. 4G) (25). This effect generates the actin polymerization power needed to overcome increasing membrane tension and to augment protrusion velocity and persistence (Fig. 3F) (14). We speculate that the role of ERK-mediated enhancement of actin polymerization may be more important in 3D migration, a process in which protrusion events tend to experience resistance from the surrounding matrix in addition to membrane tension.
We have previously found that upon growth factor stimulation, active ERK co-localizes with WAVE2 at the cell edge at the time of active protrusion, but does not control WAVE2 localization (25). Here, we found that in steady-state migrating cells, active ERK was distributed throughout the cell edge and cytoplasm. These results are consistent with the diffuse cytoplasmic localization of active ERK in migrating epithelial sheets (20). Thus, growth factor stimulation induces active ERK to transiently accumulate at the cell edge before moving to the nucleus, while steady-state conditions maintain low levels of active ERK localized at the cell edge and throughout the cell (18, 25, 49–51). Our finding that ERK regulated Arp2/3 recruitment is consistent with our previous study showing that ERK-mediated phosphorylation of WAVE2 and of Abi1 controls the ability of Arp2/3 to interact with the WAVE regulatory complex (25). We propose that ERK signaling to the WAVE regulatory complex and Arp2/3 modulates an intensity rheostat of actin polymerization to increase the volume of treadmilling branched actin at the cell edge. In the setting of oncogenic BrafV600E or an influx of growth factor signals, a global increase in ERK activity will lead to an increase in cell edge-localized active ERK. In this model, the presence of active ERK at the cell edge when Rac and the WAVE regulatory complex are recruited would increase the WAVE-mediated recruitment of Arp2/3 and the likelihood that protrusions are stabilized into productive, persistent events that generate motility. In this manner, the WAVE regulatory complex functions as a coincidence detector for ERK and Rac signals to recruit sufficient Arp2/3 for protrusion power.
Our finding that inhibition of a central signaling pathway during steady-state motility causes specific effects on protrusion mechanics suggests that protrusion is controlled by multiple, overlapping points of regulation by additional pathways. Indeed, when these other regulatory pathways are suppressed by starvation, ERK inhibition completely blocks EGF-induced protrusion, including protrusion initiation (25). In the human mammary epithelial cells (HMECs) used in those experiments, EGF is not expected to activate PI3K/Akt, Abl, or Src signaling (32), which also participate in WAVE activation, or Rho GTPases, which activate the formin family of actin nucleators and elongators (52). In that less physiological situation involving serum starvation and EGF stimulation, ERK signaling drives protrusion. We found here that in log-phase migrating cells, steady-state ERK signaling specifically regulated protrusion reinforcement. We observed migration defects upon MEK inhibition in various cancer cell lines and embryonic ERK inhibition causes neural crest cell migration defects that result in craniofacial and heart abnormalities (53, 54). However, the weak, short protrusions that remain after ERK inhibition may be sufficient for motility in some cell types and environments. For example, Arp2/3 deficient fibroblasts lack lamellipodia and undergo filopodia-based protrusion with reduced migration speed (55).
In conclusion, this work elucidates the role of ERK’s direct activity on the actin polymerization machinery: to reinforce the progression of initiated protrusions into rapid, sustained forward motion of the leading edge. ERK-mediated control occurs specifically in protrusion events, not retractions, and involves the recruitment of Arp2/3 for actin polymerization and treadmilling at the cell edge. This point of regulation explains the previously predicted and observed timing sequence in which actin assembly peaks after protrusion onset and after maximal protrusion velocity is reached (14).
Materials and Methods
Cell Culture, Imaging Media, Inhibitor Treatment
Panc1, MDA-MB-468, Sk-Mel28, and A375 cells were cultured and imaged in DMEM with 10% FBS, 20 mM Hepes. PtK1 cells were cultured in F-12 with 10% FBS and imaged in L-15 with 10% FBS, 20 mM Hepes. 5 μM AZD6244 was used for all treatments.
Migration and Wound Healing Assay
Cells were plated in their normal growth medium on glass-bottomed dishes 2 days prior to assay. Cells were treated with DMSO or AZD6244 and then immediately imaged with phase contrast at 37 °C, 5% CO2 using a Nikon Ti inverted microscope with perfect focus, Prior Proscan motorized stage, and incubation chamber. Images were collected every 10 min with Metamorph software for 12 hours. For migration assay, displacement or the distance migrated over a 6 hour period was calculated using the Track Points function in Metamorph. To ensure bias was not introduced into the analysis, every cell that did not divide or move out of the field of view over the 6 hour period was tracked. The results were obtained from 3 independent experiments. Normality was assessed using the Anderson-Darling test in Matlab. The data were not normally distributed. Therefore, the permutation test was used to calculate statistical significance of changes in the distributions. Error bars about the median were estimated from the 95% confidence interval of the bootstrapped median distribution.
For wound healing, cells were treated with DMSO or AZD644 immediately following wound scratching. Start and end-point of assay were 0 h and 11 h after scratching, respectively. Results were obtained from 2 independent experiments. Wounds in which the edge did not uniformly migrate were excluded (for example, when only part of the edge migrated or the wound closed from opposite or different part of edge). To avoid issues with the unequal sample size causing type I error inflation (56), we used the Wilcoxon rank sum test to assess the significance of the difference between the control and treated sample medians.
LifeAct Imaging and Image Processing
PtK1 cells were electroporated with EmeraldGFP-tagged LifeAct using the Neon transfection system and plated on acid-treated glass coverslips. Cells were cultured for 48-hours at 37 °C, 5% CO2 and imaged using a Nikon Ti motorized inverted microscope with Perfect Focus System, Yokagawa CSU-X1 spinning disk confocal and Spectral Applied Research Borealis modification, 491 solid-state laser, 100× 1.45 N.A. objective, Hamamatsu ORCA-AG cooled CCD camera, and Metamorph software. Cells were in L-15 medium containing 10% FBS, 20 mM HEPES, and 0.03 U/ml Oxyrase. Experiments using U0126 were carried out using a Nikon TE2000 inverted microscope with Perfect Focus System, Yokogawa CSU-10 spinning disk confocal, 488 solid-state laser, 60× 1.4 N.A. objective, Andor Clara cooled CCD camera, and NIS Elements software.
Cells were imaged for 10 minutes before perturbation and 20 minutes following perturbation (30 min total). PtK1 cell boundaries were detected from the fluorescence microscopy data using a threshold method. Once identified, the edge was then subdivided into 1 micron long segments and their respective velocities were calculated as described previously (25). For inhibitor-treated experiments, results were obtained from 3–4 independent experiments. For BrafV600E overexpression experiments, results were obtained from 2 independent experiments.
Edge Velocity Analysis
The velocity traces for each segment were processed using a signal detection algorithm based on empirical mode decomposition (57). Velocities within the noise level were classified as insignificant and the respective edge segment as inactive. Once a significant event was detected, it was either classified as protrusion or retraction and the mean and maximum values extracted. The event duration was also measured. The Anderson Darling test indicated the distributions were not Gaussian. Rather, protrusion and retraction velocities followed a log-normal distribution, which suggested the use of the median value instead of mean for comparisons among different experimental conditions. The median error bars were estimated from the 95% confidence interval of the bootstrapped median distribution.
Protrusion and retraction velocity data exhibited log-normal distributions. We applied the Anderson-Darling test to confirm that the velocity and persistence time for all cells in each protrusion and retraction treatment group came from the same distribution, indicating they were representative for the same general cell population. The fast and slow, long and short events were confirmed to be equally distributed among the different cells in all conditions. The permutation test was used to calculate statistical significance of changes in the distributions of the mean velocity and persistence time for the different conditions.
Actin qFSM
Actin microinjection, imaging and qFSM analysis was carried out as described previously (38). Cells were microinjected with Alexa-568 actin monomers (Molecular Probes) using a Sutter microinjection system. Briefly, cells were allowed to recover for 30 min and were then imaged in L-15 media with 10% FBS in a 37°C, 5% CO2 incubation chamber on a Nikon Ti motorized inverted microscope with Perfect Focus and Yokagawa CSU-X1 spinning disk confocal with a Spectral Applied Research Borealis modification. Images were acquired every 10 sec using a 561nm (200mW) solid state laser excitation and a Hamamatsu ORCA-AG cooled CCD camera controlled by MetaMorph image acquisition software. The actin speckles provided enough out-of-focus light for computational detection of the cell edge and the partitioning of into cell edge segments and associated probing windows. Steady-state actin flow maps depict the raw speckle displacements spatially averaged in 0.7 micron regions and temporally averaged over 4 minutes.
We identified and tracked > 103 speckles per FSM movie. The autocorrelation function of the PtK1 cell edge dynamics was 6 frames (60 seconds). Correlation-based flow tracking revealed the raw speckle displacements spatially averaged in 0.7 micron regions and temporally integrated over 40 seconds. We used then used these flow templates to track the individual speckles and performed statistical processing of the speckle intensity fluctuations to map F-actin flow and assembly and disassembly (kinetics). We averaged 9 frames for steady-state flow analysis and 55 for kinetic maps. Average flow at the cell edge varied between 400–1000 nm/min between cells, likely because of variability in the fraction of protruding and retracting cell edge and in the protrusion velocity during the captured time series. Reduced flow upon MEK inhibition was reproduced in n = 5 cells, obtained from 4 independent experiments.
GFP-Arp3 analysis
PtK1 cells were infected with Retrovirus carrying pBabeNeo-GFP-Arp3 and selected with G418. Cells were imaged by Yokagawa CSU-X1 spinning disk confocal with Spectral Applied Research Borealis modification, in L-15 medium containing 20 mM HEPES and 0.03 U/ml Oxyrase, with a 100× 1.45 N.A. objective and a Hammamatsu R2 camera with 2×2 binning.
Cells were segmented using a thresholding method and the Arp2/3 fluorescence intensity was then averaged in 0.5 micron-thick bands parallel to the cell edge at increasing distances from the cell edge. To handle the variability of fluorescence intensity among cells, we normalized the average of each band by the average of the band 3 microns away from the cell edge. This normalization allowed us to average the signal from multiple cells without introducing artifacts from cells with too high or too low intensities. The final confidence interval was calculated by propagating the error from the normalized mean calculated for each cell. Images were obtained from 3 independent experiments.
Immunofluorescence
PtK1 cells were grown on glass coverslips for 48 hours, fixed with 3.7% formaldehyde with 10 nM Caliculyn A, in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1 mM KH2PO4, pH 7). Cells were permeabilized and blocked with 0.2% Triton and 3% BSA. Cells were stained with purified mouse monoclonocal anti-phospho-ERK (Sigma), rabbit anti-ArpC2 (Millipore), and phalloidin-647 (Molecular Probes) in 1% BSA, 0.2% Tween-20 in TBS. Images were acquired on a Nikon Ti motorized inverted microscope with Yokogawa CSU-X1 spinning disk confocal, using 405, 488, 561nm and 647 solid state laser excitation and an Andor Clara cooled CCD camera controlled by Nikon Elements image acquisition software.
Cells were segmented by thresholding in the phalloidin-647 channel. Because the peak Arp2/3 fluorescence was in front of the phalloidin signal, the segmented masks were then dilated out 0.5 microns from the edge. These dilated masks were then applied to the 488 and 568 channels, and the signal intensities were integrated over the cell area (mask). The ARP2C-568 fluorescence intensity was further averaged in a 1 micron-thick band parallel to the cell edge (0.5 microns to 0.5 microns) and the percentage of total signal present in this band was averaged for each group. The standard error of the mean and a 95% confidence interval was graphed.
Cross-correlation
Cross-correlation analysis can indicate relationships and timing between two events (58). As previously described, correlation scores between the edge motion and sampled signals (for example, Arp3 intensity) were calculated for each window of the cell edge for all cells and then averaged using bootstrap and a variance stabilization method (58). Cross-correlation scores were graphed in the y-axis, with the score value indicating how well the two activities were correlated with a specific time lag (x-axis) between them. The uncertainty on each correlation value was defined as the 95% confidence interval of the bootstrapped distribution at each time lag.
In silico experiments
We performed in silico experiments to confirm the artificial shift on the positive correlation peak between edge motion and retrograde flow. Time series for both variables were generated using a periodic sawtooth wave form with added Gaussian noise (signal to noise = 2). These waves had 180° phase difference and a Gaussian random sequence of one quarter of the period was introduced between the peaks for the synthetic retrograde flow. A thousand replicas of both time series were generated and the correlation scores were calculated as described above. Then, the correlation scores were separately recalculated for the negative and positive component of the edge motion.
Supplementary Material
Spontaneous protrusion and retraction cycles.
MEK inhibition dampens spontaneous edge motion.
Edge motion analysis.
Analysis of spontaneous edge motion upon MEK inhibition.
Actin speckles.
Actin speckles with MEK inhibition.
Flow tracking of actin qFSM.
Actin flow under MEK inhibition.
Fig. S1. ERK inhibition in PtK1 cells.
Fig. S2. ERK hyperactivation induces protrusion velocity.
Fig. S3. ERK increases retraction velocity.
Fig. S4. ERK promotes peak membrane protrusion velocity and persistence time in PtK1 cells microinjected with Alexa-568 actin monomers.
Fig. S5. Simulation of cross-correlation of actin retrograde flow with cell edge velocity.
Fig. S6. ERK regulates Arp2/3 localization to the cell edge.
Table S1. Event velocity averages.
Acknowledgments
The authors thank the Nikon Imaging Center at Harvard Medical School and Torsten Wittman and Anita Sil at the University of California, San Francisco for help with light microscopy.
Funding: This work was funded by the NIH, K01 CA168850 to MCM, F32 GM103278 to MV, R01 GM071868 and U01 GM068230 to GD, and R01 CA046595 to JB.
Footnotes
Author contributions: The project was conceived by MCM and JB. The research design was regularly adjusted in discussions between MCM, MV, and GD. MCM performed all live-cell imaging experiments, Western blotting for the AZD6244 treatments, and routine data analysis using speckle analysis software. JJ performed the Western blotting for the U0126 treatments and the ARPC2 immunofluorescence. MV extended the speckle analysis software, wrote new programs for correlation analysis, and together with MCM interpreted the results in the context of the proposed model. MCM and MV jointly prepared the figures; MCM drafted the manuscript. All authors contributed to the final editing.
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 2.Lim JI, Sabouri-Ghomi M, Machacek M, Waterman CM, Danuser G. Protrusion and actin assembly are coupled to the organization of lamellar contractile structures. Experimental cell research. 2010;316:2027–2041. doi: 10.1016/j.yexcr.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 4.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 5.Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Developmental cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nature reviews Molecular cell biology. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 7.Lai FP, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TE, Dunn GA, Small JV, Rottner K. Arp2/3 complex interactions and actin network turnover in lamellipodia. The EMBO journal. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millius A, Watanabe N, Weiner OD. Diffusion, capture and recycling of SCAR/WAVE and Arp2/3 complexes observed in cells by single-molecule imaging. Journal of cell science. 2012;125:1165–1176. doi: 10.1242/jcs.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stradal T, Courtney KD, Rottner K, Hahne P, Small JV, Pendergast AM. The Abl interactor proteins localize to sites of actin polymerization at the tips of lamellipodia and filopodia. Current biology : CB. 2001;11:891–895. doi: 10.1016/s0960-9822(01)00239-1. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, Stradal TE, Di Fiore PP, Carlier MF, Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nature cell biology. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 12.Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Molecular cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, Suetsugu S, Takenawa T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nature cell biology. 2004;6:420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- 14.Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nature cell biology. 2008;10:1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castro-Castro A, Ojeda V, Barreira M, Sauzeau V, Navarro-Lerida I, Muriel O, Couceiro JR, Pimentel-Muinos FX, Del Pozo MA, Bustelo XR. Coronin 1A promotes a cytoskeletal-based feedback loop that facilitates Rac1 translocation and activation. The EMBO journal. 2011;30:3913–3927. doi: 10.1038/emboj.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang I, Gorelik R, Sousa-Blin C, Derivery E, Guerin C, Linkner J, Nemethova M, Dumortier JG, Giger FA, Chipysheva TA, Ermilova VD, Vacher S, Campanacci V, Herrada I, Planson AG, Fetics S, Henriot V, David V, Oguievetskaia K, Lakisic G, Pierre F, Steffen A, Boyreau A, Peyrieras N, Rottner K, Zinn-Justin S, Cherfils J, Bieche I, Alexandrova AY, David NB, Small JV, Faix J, Blanchoin L, Gautreau A. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature. 2013;503:281–284. doi: 10.1038/nature12611. [DOI] [PubMed] [Google Scholar]
- 17.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends in biochemical sciences. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roskoski R., Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacological research : the official journal of the Italian Pharmacological Society. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Locascio A, Nieto MA. Cell movements during vertebrate development: integrated tissue behaviour versus individual cell migration. Current opinion in genetics & development. 2001;11:464–469. doi: 10.1016/s0959-437x(00)00218-5. [DOI] [PubMed] [Google Scholar]
- 20.Matsubayashi Y, Ebisuya M, Honjoh S, Nishida E. ERK activation propagates in epithelial cell sheets and regulates their migration during wound healing. Current biology : CB. 2004;14:731–735. doi: 10.1016/j.cub.2004.03.060. [DOI] [PubMed] [Google Scholar]
- 21.O’Brien LE, Tang K, Kats ES, Schutz-Geschwender A, Lipschutz JH, Mostov KE. ERK and MMPs sequentially regulate distinct stages of epithelial tubule development. Developmental cell. 2004;7:21–32. doi: 10.1016/j.devcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Doehn U, Hauge C, Frank SR, Jensen CJ, Duda K, Nielsen JV, Cohen MS, Johansen JV, Winther BR, Lund LR, Winther O, Taunton J, Hansen SH, Frodin M. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Molecular cell. 2009;35:511–522. doi: 10.1016/j.molcel.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Molecular cell. 2010;38:114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishibe S, Joly D, Liu ZX, Cantley LG. Paxillin serves as an ERK-regulated scaffold for coordinating FAK and Rac activation in epithelial morphogenesis. Molecular cell. 2004;16:257–267. doi: 10.1016/j.molcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Mendoza MC, Er EE, Zhang W, Ballif BA, Elliott HL, Danuser G, Blenis J. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Molecular cell. 2011;41:661–671. doi: 10.1016/j.molcel.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y, Xia Y, Hawke D, Halle M, Tremblay ML, Gao X, Zhou XZ, Aldape K, Cobb MH, Xie K, He J, Lu Z. FAK phosphorylation by ERK primes ras-induced tyrosine dephosphorylation of FAK mediated by PIN1 and PTP-PEST. Molecular cell. 2009;35:11–25. doi: 10.1016/j.molcel.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamakura S, Moriguchi T, Nishida E. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. The Journal of biological chemistry. 1999;274:26563–26571. doi: 10.1074/jbc.274.37.26563. [DOI] [PubMed] [Google Scholar]
- 28.Mody N, Leitch J, Armstrong C, Dixon J, Cohen P. Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS letters. 2001;502:21–24. doi: 10.1016/s0014-5793(01)02651-5. [DOI] [PubMed] [Google Scholar]
- 29.Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, Parry J, Smith D, Brandhuber BJ, Gross S, Marlow A, Hurley B, Lyssikatos J, Lee PA, Winkler JD, Koch K, Wallace E. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:1576–1583. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
- 30.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 31.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nature reviews Molecular cell biology. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 32.Mendoza MC. Phosphoregulation of the WAVE regulatory complex and signal integration. Seminars in cell & developmental biology. 2013;24:272–279. doi: 10.1016/j.semcdb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machacek M, Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophysical journal. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boeckeler K, Rosse C, Howell M, Parker PJ. Manipulating signal delivery - plasma-membrane ERK activation in aPKC-dependent migration. Journal of cell science. 2010;123:2725–2732. doi: 10.1242/jcs.062299. [DOI] [PubMed] [Google Scholar]
- 35.Klemke RL, Cai S, Giannini AL, Gallagher PJ, de Lanerolle P, Cheresh DA. Regulation of cell motility by mitogen-activated protein kinase. The Journal of cell biology. 1997;137:481–492. doi: 10.1083/jcb.137.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu ZX, Yu CF, Nickel C, Thomas S, Cantley LG. Hepatocyte growth factor induces ERK-dependent paxillin phosphorylation and regulates paxillin-focal adhesion kinase association. The Journal of biological chemistry. 2002;277:10452–10458. doi: 10.1074/jbc.M107551200. [DOI] [PubMed] [Google Scholar]
- 37.Radtke S, Milanovic M, Rosse C, De Rycker M, Lachmann S, Hibbert A, Kermorgant S, Parker PJ. ERK2 but not ERK1 mediates HGF-induced motility in non-small cell lung carcinoma cell lines. Journal of cell science. 2013;126:2381–2391. doi: 10.1242/jcs.115832. [DOI] [PubMed] [Google Scholar]
- 38.Mendoza MC, Besson S, Danuser G. Quantitative fluorescent speckle microscopy (QFSM) to measure actin dynamics. In: Paul Robinson J, et al., editors. Current protocols in cytometry. Unit2. Chapter 2. 2012. p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nature methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danuser G, Waterman-Storer CM. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annual review of biophysics and biomolecular structure. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- 41.Ponti A, Matov A, Adams M, Gupton S, Waterman-Storer CM, Danuser G. Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative Fluorescent Speckle Microscopy. Biophysical journal. 2005;89:3456–3469. doi: 10.1529/biophysj.104.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Current biology : CB. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Symons MH, Mitchison TJ. Control of actin polymerization in live and permeabilized fibroblasts. The Journal of cell biology. 1991;114:503–513. doi: 10.1083/jcb.114.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ingerman E, Hsiao JY, Mullins RD. Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. The Journal of cell biology. 2013;200:619–633. doi: 10.1083/jcb.201211069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Dobereiner HG, Freund Y, Borisy G, Sheetz MP. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harvey CD, Yasuda R, Zhong H, Svoboda K. The spread of Ras activity triggered by activation of a single dendritic spine. Science. 2008;321:136–140. doi: 10.1126/science.1159675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunida K, Matsuda M, Aoki K. FRET imaging and statistical signal processing reveal positive and negative feedback loops regulating the morphology of randomly migrating HT-1080 cells. Journal of cell science. 2012;125:2381–2392. doi: 10.1242/jcs.096859. [DOI] [PubMed] [Google Scholar]
- 48.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen RH, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Molecular and cellular biology. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez FA, Seth A, Raden DL, Bowman DS, Fay FS, Davis RJ. Serum-induced translocation of mitogen-activated protein kinase to the cell surface ruffling membrane and the nucleus. The Journal of cell biology. 1993;122:1089–1101. doi: 10.1083/jcb.122.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khokhlatchev AV, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb MH. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 52.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nature reviews Molecular cell biology. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 53.Nakamura T, Gulick J, Colbert MC, Robbins J. Protein tyrosine phosphatase activity in the neural crest is essential for normal heart and skull development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11270–11275. doi: 10.1073/pnas.0902230106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart RA, Sanda T, Widlund HR, Zhu S, Swanson KD, Hurley AD, Bentires-Alj M, Fisher DE, Kontaridis MI, Look AT, Neel BG. Phosphatase-dependent and -independent functions of Shp2 in neural crest cells underlie LEOPARD syndrome pathogenesis. Developmental cell. 2010;18:750–762. doi: 10.1016/j.devcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Y, Xu H, Calian V, Hsu JC. To permute or not to permute. Bioinformatics. 2006;22:2244–2248. doi: 10.1093/bioinformatics/btl383. [DOI] [PubMed] [Google Scholar]
- 57.Huang NE, Shen SSP. Hilbert-Huang Transform and its Applications. World Scientific; London: 2005. [Google Scholar]
- 58.Vilela M, Halidi N, Besson S, Elliott H, Hahn K, Tytell J, Danuser G. Fluctuation analysis of activity biosensor images for the study of information flow in signaling pathways. Methods in enzymology. 2013;519:253–276. doi: 10.1016/B978-0-12-405539-1.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spontaneous protrusion and retraction cycles.
MEK inhibition dampens spontaneous edge motion.
Edge motion analysis.
Analysis of spontaneous edge motion upon MEK inhibition.
Actin speckles.
Actin speckles with MEK inhibition.
Flow tracking of actin qFSM.
Actin flow under MEK inhibition.
Fig. S1. ERK inhibition in PtK1 cells.
Fig. S2. ERK hyperactivation induces protrusion velocity.
Fig. S3. ERK increases retraction velocity.
Fig. S4. ERK promotes peak membrane protrusion velocity and persistence time in PtK1 cells microinjected with Alexa-568 actin monomers.
Fig. S5. Simulation of cross-correlation of actin retrograde flow with cell edge velocity.
Fig. S6. ERK regulates Arp2/3 localization to the cell edge.
Table S1. Event velocity averages.