Abstract
Increasing evidence suggests that defective RNA processing contributes to the development of amyotrophic lateral sclerosis (ALS). This may be especially true for ALS caused by a repeat expansion in C9orf72 (c9ALS), in which the accumulation of RNA foci and dipeptide-repeat proteins are expected to modify RNA metabolism. We report extensive alternative splicing (AS) and alternative polyadenylation (APA) defects in the cerebellum of c9ALS cases (8,224 AS, 1,437 APA), including changes in ALS-associated genes (e.g. ATXN2 and FUS), and cases of sporadic ALS (sALS; 2,229 AS, 716 APA). Furthermore, hnRNPH and other RNA-binding proteins are predicted as potential regulators of cassette exon AS events for both c9ALS and sALS. Co-expression and gene-association network analyses of gene expression and AS data revealed divergent pathways associated with c9ALS and sALS.
Amyotrophic lateral sclerosis (ALS) is a devastating and fatal neurodegenerative disease that predominantly affects upper and lower motor neurons, although evidence suggests it is a multisystem disease involving other brain regions1. Approximately 90% of ALS cases are sporadic (sALS), with the remaining 10% being inherited familial ALS (fALS). Mutations in several genes are known to cause fALS and a small proportion of seemingly sporadic cases. This underscores the etiological diversity of ALS despite clinical and neuropathological similarities among familial and sporadic forms, and calls attention to the need for treatments based on specific forms of ALS. With this in mind, the discovery of a G4C2 repeat expansion in C9orf72 as the most common known cause of ALS2, 3 offers an exciting opportunity to determine aberrant events initiated by this genetic abnormality.
The C9orf72 repeat expansion accounts for approximately 34% of fALS cases and 6% of sALS cases4. Several pathological mechanisms have been postulated for “c9ALS”, including the involvement of RNA toxicity resulting from the accumulation of repeat-containing transcripts (r(G4C2)exp and r(G2C4)exp) bidirectionally transcribed from the C9orf72 expansion. Both r(G4C2)exp and r(G2C4)exp are subject to repeat-associated non-ATG (RAN) translation, which leads to the production of dipeptide repeat (DPR) proteins5–8. Inclusions immunopositive for these DPR proteins are numerous in cerebellum, neocortical regions and hippocampus of c9ALS cases, and several studies provide evidence of their toxicity9–15, including a potential effect on RNA biogenesis12.
r(G4C2)exp and r(G2C4)exp may also contribute to neurodegeneration through the formation of nuclear RNA foci that sequester, and cause loss of function, of key RNA-binding proteins (RBPs)16–20. Several RBPs, including adenosine deaminase RNA-specific B2 (ADARB2) and the heterogeneous nuclear ribonucleoprotein hnRNPH1, co-localize with RNA foci in c9ALS brain tissues and/or neurons differentiated from induced pluripotent stem cells16, 19, 20. The impaired ability of RBPs to regulate their targets and the resulting defects in RNA processing are putative contributors to c9ALS pathogenesis. This notion is supported by the fact that misregulated RNA processing is widely implicated in ALS. For instance, cytoplasmic inclusions of transactive response DNA binding protein 43 kDa (TDP-43) are present in the majority of ALS cases21, and mislocalization of TDP-43 from the nucleus to the cytoplasm is believed to result in misregulated splicing of TDP-43 RNA targets22. Additionally, ALS-associated mutations have been identified in several RBP-encoding genes, including TARDBP23 (encoding TDP-43), fused in sarcoma (FUS)24, 25, ataxin 2 (ATXN2)26, EWS RNA-binding protein 1 (EWSR1)27, TAF15 RNA polymerase II (TAF15)28, HNRNPA1 and HNRNPA2B129. Given the growing body of evidence that the C9orf72 repeat expansion, by virtue of foci and/or DPR proteins, results in aberrant RNA processing, we sought to investigate alterations in the c9ALS transcriptome.
RESULTS
Next generation RNA sequencing of brain tissues
To evaluate the c9ALS transcriptome, we performed RNA sequencing (RNAseq) using RNA (N=8) from cerebellum and frontal cortex, two neuroanatomical regions showing neuropathological hallmarks of c9ALS. To identify c9ALS-specific transcriptome changes, we also analyzed RNA from sALS cases (N=10) having no mutations in the most common ALS-associated genes, and non-neurological disease cases (controls, N=8–9). Age at symptom onset of motor neuron disease did not differ between c9ALS (median=49.6 years [25th, 75th%=46.0, 55.1]) and sALS (58.9 years [47.9, 64.7]) cases (P=0.118). Disease duration did not differ between c9ALS (30.4 months [16.9, 49.0]) and sALS (38.5 months [22.2, 55.2]) (P=0.769). Motor neuron disease was noted to affect upper and lower motor neurons with observed depopulation of Betz cells in the motor cortex of c9ALS and sALS cases. TDP-43 pathology was found in the motor cortex, hypoglossal nucleus, and anterior horn cells with glial cytoplasmic inclusions and skein-like neuronal cytoplasmic inclusions characteristic of ALS. Alzheimer’s type pathology did not differ among c9ALS, sALS, and controls (all Braak tanglestage=I [0, II]) (P=0.964). Clinical and neuropathological information can be found in Supplementary Table 1. Paired-end RNAseq was performed using the Illumina Hiseq 2000 platform, and the sequencing reads were aligned to the hg19 human genome (see Online Methods). The average number of reads was ~83 million (91.5 million for cerebellum, 73.6 for frontal cortex), with approximately 72% of total reads mapping to the human transcriptome (72.4% for cerebellum, 71.4% for frontal cortex). As described below, computational analyses were performed to evaluate differentially expressed (DE) genes, alternative splicing (AS) and alternative polyadenylation (APA) in c9ALS and sALS transcriptomes.
Gene expression is differentially misregulated in ALS
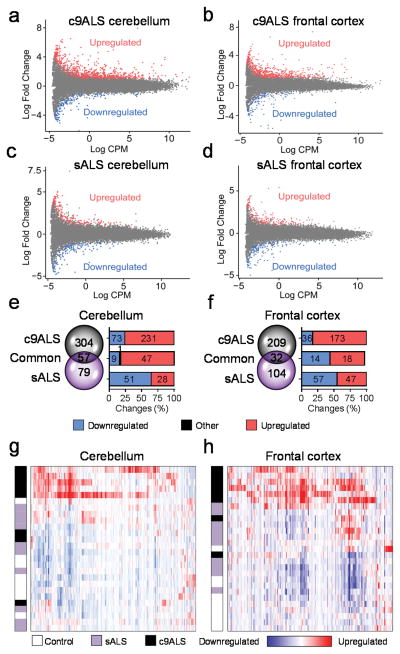
To identify DE genes in c9ALS or sALS in comparison to controls (P < 0.05), EdgeR analyses were performed on the aligned RNAseq data. MA plots shown in Figures 1a–d highlight genes that were up- or down-regulated in ALS samples at least 2-fold (|Log2FC| ≥ 1). c9ALS cases showed a greater number of up-regulated than down-regulated genes, with far more DE genes in the cerebellum than in the frontal cortex (Fig. 1a,b). In contrast, the number of up- and down-regulated DE genes in sALS was similar and no marked differences were observed between the cerebellum and frontal cortex (Fig. 1c,d). When exclusively examining genes DE at least 4-fold (|Log2FC| ≥ 2), 361 genes were identified in the cerebellum of c9ALS cases compared to only 136 in sALS (Fig. 1e). In the frontal cortex, 241 DE genes were found in c9ALS compared to 136 in sALS (Fig. 1f).
Figure 1.
Differential regulation of gene expression in c9ALS and sALS. (a–d) MA plots showing up- and down-regulated gene expression (P < 0.05, |Log2FC| ≥ 1) in c9ALS (a,b) and sALS (c,d) vs. controls in cerebellum (a,c) and frontal cortex (b, d). (e,f) Venn diagrams and graphs depicting the number up- and down-regulated transcripts (P < 0.05, |Log2FC| ≥ 2,) that are unique or common to c9ALS and sALS cerebellum (e) or frontal cortex (f). The black section in graph (e) shows a DE gene common to both c9ALS and sALS, but with the expression change going in opposite directions between both forms of ALS. (g,h) Hierarchical clustering representation of c9ALS DE genes (P < 0.05, |Log2FC| ≥ 2) in cerebellum (g) and frontal cortex (h). Each row of the heat maps corresponds to either a control, sALS or c9ALS case, as designated by the color-coded bar on the left. A legend is provided below the heat maps.
Fifty-seven genes in cerebellum and 32 genes in frontal cortex were abnormally expressed in both c9ALS and sALS. These similarities are perhaps indicative of common molecular pathways. Indeed, gene ontology (GO) analyses show a predominance of genes involved in inflammatory and defense responses in both cerebellum and frontal cortex in c9ALSand sALS (Supplementary Tables 2,3). Nevertheless, that the number of misregulated genes in c9ALS is approximately double that in sALS suggests differences between these two forms of ALS. Of note, the unfolded protein response (UPR) was a GO pathway identified from differentially upregulated genes in c9ALS but not sALS (Supplementary Tables 4,5). The divergence between c9ALS and sALS or controls is corroborated by findings from hierarchical clustering (Fig. 1g,h) and principal component analyses (Supplementary Fig. 1) of DE genes in c9ALS cerebellum or frontal cortex. Indeed, c9ALS cases clustered together and were separated, not only from controls, but also from sALS cases, an effect independent of clinical features (Supplementary Figs. 2,3).
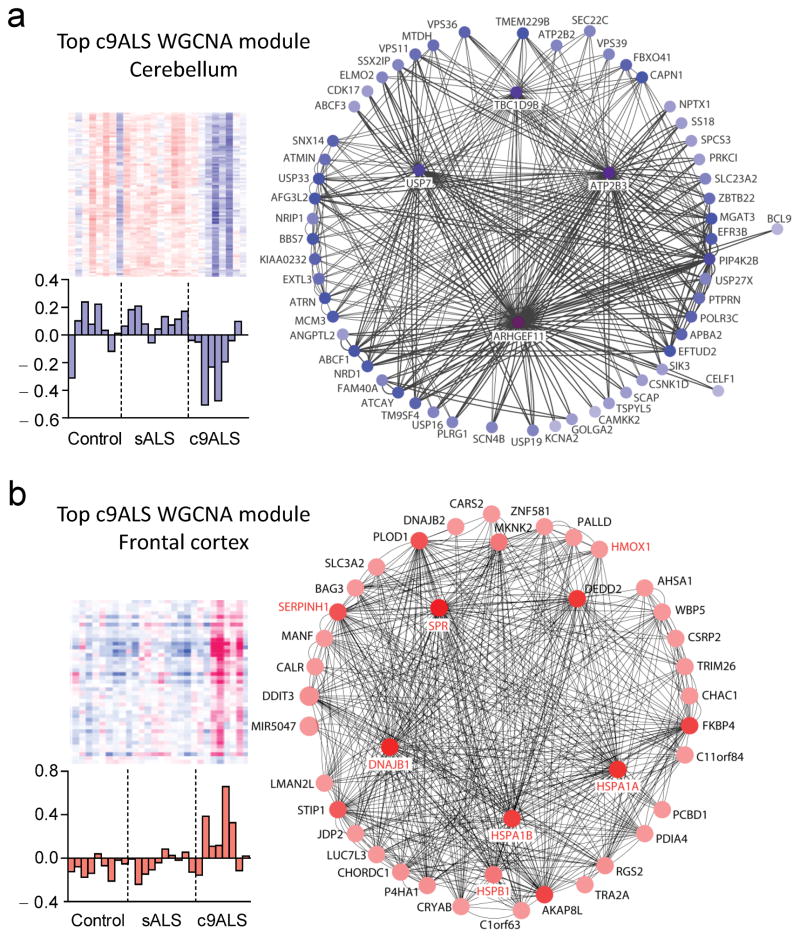
To evaluate expression differences between c9ALS or sALS and controls at a systems level, weighted gene co-expression correlation network analyses (WGCNA) were completed. Several significant modules were identified and GO annotation of each module determined the functional pathways of genes within a given module (see Online Methods). In c9ALS, the top module identified in cerebellum is enriched in genes involved in neuron development, protein localization and transcription (Table 1, Fig. 2a), while the top module identified in the frontal cortex is enriched in UPR-related genes (Table 1, Fig. 2b). Of note, genes involved in the UPR pathway were among the top DE genes both in the c9ALS cerebellum and frontal cortex based on EdgeR analyses, as mentioned above, and validated by qRT-PCR (Supplementary Fig. 4). In sALS, the top modules identified involved genes in calcium transport and synaptic transmission in cerebellum, and in oxidative phosphorylation in frontal cortex (Supplementary Table 6). These studies reveal marked differences in gene expression patterns between c9ALS and sALS, with substantially more changes observed in c9ALS, especially in the cerebellum.
Table 1.
Weighted gene co-expression correlation network analyses (WGCNA) reveal significantly altered gene modules in cerebellum and frontal cortex of c9ALS.
| c9ALS | Cerebellum | ||||
| Module name | P value | Gene count | Notable GO terms | Enrichment P value | |
| MEpink | 0.0137 | 148 | Neuron development, protein localization, transcription | 3.40E-03 | |
| MEgrey60 | 0.0350 | 93 | Vesicle transport, protein transport | 1.90E-02 | |
| MEdarkred | 0.0359 | 68 | rRNA processing, lysosome organization, RNA processing | 3.30E-02 | |
| MEgreen | 0.0493 | 210 | Intracellular transport | 1.60E-04 | |
| MEblue | 0.0497 | 299 | Chromatin modification | 4.60E-09 | |
| Frontal cortex | |||||
| Module name | P value | Gene count | Notable GO terms | Enrichment P value | |
| MEsalmon | 0.0217 | 43 | Response to unfolded protein | 1.40E-08 | |
| MEturquoise | 0.0413 | 1,678 | Protein localization | 8.10E-25 | |
| MEblack | 0.0428 | 66 | Oxidative phosphorylation | 7.50E-08 | |
| MEgreen | 0.0457 | 98 | rRNA processing | 8.20E-03 | |
WGCNA co-expression analyses were performed with data from c9ALS cerebellum and frontal cortex (versus sALS and controls, P < 0.05). All significant modules are shown. The most notable gene ontology terms and their enrichment P value are included for each module. Note MEpink is shown in blue in Figure 2a, while MEsalmon is shown in red in Figure 2b.
Figure 2.
Weighted gene co-expression correlation network analyses (WGCNA) of top identified modules in cerebellum and frontal cortex of c9ALS. (a,b) Shown on the left side of panels are heat maps of genes and bar plots for eigengene values of top c9ALS cerebellum (MEpink, shown in blue) and frontal cortex (MEsalmon, shown in red) WGCNA co-expression modules. Network diagrams for these two modules are shown (right), with node color darkness proportional to the number of connections (degree). Note that eight genes within the top co-expression module in c9ALS frontal cortex were differentially expressed (those in red font as well as BAG3). The increase in expression for seven of the genes (those in red font) was validated by qRT-PCR in cerebellum and frontal cortex tissues (see Supplementary Fig. 4).
Extensive misregulation of alternative splicing in ALS brain
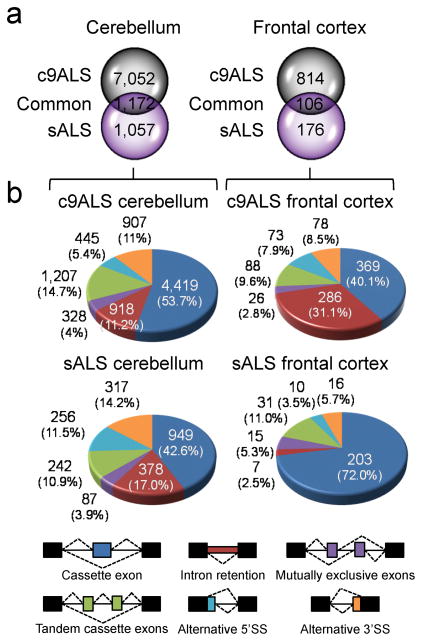
To evaluate AS changes in ALS, OLego software30 was used to align RNAseq reads to the hg19 genome assembly and splice junctions. The total number of AS events [false discovery rate (FDR) < 0.05] was over 3 times higher in c9ALS than sALS cases in the cerebellum (8,224 in c9ALS vs. 2,229 in sALS) and frontal cortex (920 in c9ALS vs. 282 in sALS) (Fig. 3a, Supplementary Tables 7,8). Also, the number of AS events was approximately 8–9 times higher in the cerebellum than the frontal cortex in c9ALS and sALS (Fig. 3a). While 1,172 and 106 of the AS events that occur in the cerebellum and frontal cortex, respectively, were shared between c9ALS and sALS cases, they represent a relatively small percentage (~11–15%) of the total AS changes seen in c9ALS.
Figure 3.
Widespread alternative splicing defects are found in c9ALS and sALS. (a) Venn diagrams depicting the number of unique and common total AS events in c9ALS and sALS cerebellum (left) and frontal cortex (right) (FDR < 0.05). (b) Pie charts showing the percentage (%) of different types of AS events in cerebellum and frontal cortex of c9ALS (top) and sALS (bottom) (FDR < 0.05). A color-coded legend of the different AS events is provided below the pie charts.
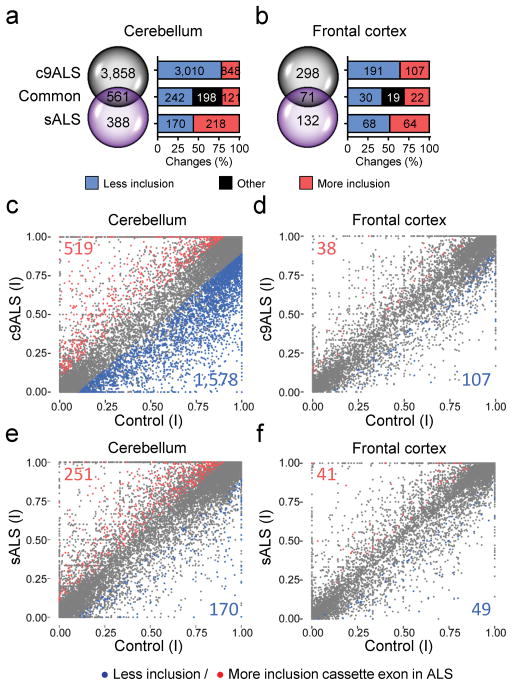
Among the different types of AS changes in ALS, cassette exon (CE) events were the most common and intron retention events represented a significant proportion of total changes (FDR< 0.05, Fig. 3b). A total of 918 intron retention events occurred in the c9ALS cerebellum whereas 286 were found in the c9ALS frontal cortex. In sALS, there were 378 alternative intron retention events in the cerebellum but only 7 in the frontal cortex (Fig. 3b, Supplementary Fig. 5). With regards to AS CEs (FDR < 0.05) in c9ALS, approximately 12 times more events were found in the cerebellum (4,419) than the frontal cortex (369) (Fig. 3b, Fig. 4a,b). In sALS, there were 949 AS CE events in the cerebellum, which exceeded the 203 events in the frontal cortex by more than 4 times (Fig. 3b, Fig. 4a,b).
Figure 4.
Extensive misregulation of cassette exon splicing occurs in the c9ALS cerebellum. (a, b) Venn diagrams showing the number of unique and common AS cassette exon events in c9ALS and sALS cerebellum (a) and frontal cortex (b) (FDR < 0.05). (c–f) Scatter plots showing exclusion (blue) and inclusion (red) of AS cassette exons in c9ALS (c,d) and sALS (e,f) in comparison to controls (FDR < 0.05, |dI| ≥ 0.1) in cerebellum (c,e) or frontal cortex (d,f). dl: differential index value. Exon inclusion indices (I) are plotted for control group (x-axis) vs. c9ALS (c,d, y-axis) or vs. sALS (e,f, y-axis).
The majority of CE events in c9ALS are the result of exon skipping, whereas a similar proportion of CE exclusions and inclusions were observed in sALS (Fig. 4c–f). Likewise, hierarchical clustering of c9ALS differentially spliced CEs in cerebellum and frontal cortex not only demonstrate clear differences between c9ALS and sALS cases as well as controls, but also illustrate the abundance of AS CEs in c9ALS independent of clinical features (Supplementary Figs. 6,7).
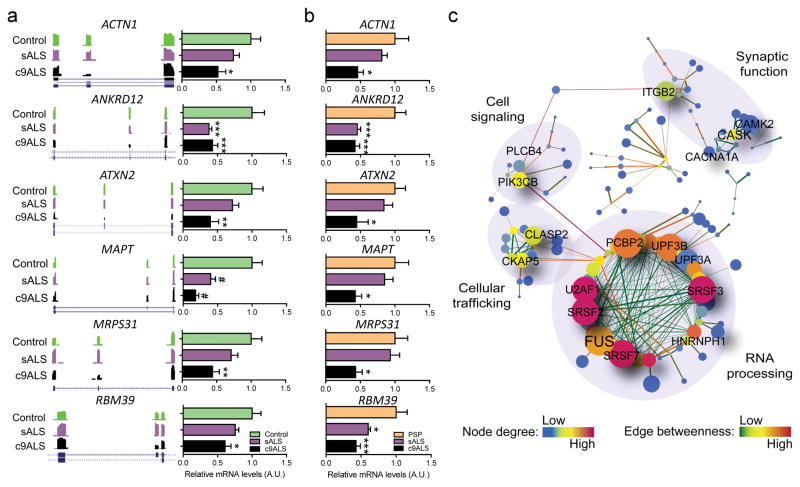
Systematic qRT-PCR validation of some of the top AS CEs in c9ALS cerebellum (Fig. 5a, Supplementary Fig. 8) and frontal cortex (Supplementary Fig. 9) confirmed the occurrence of CE skipping events, some of which were also observed in sALS, including those in known neurodegenerative disease-related genes. For instance, exon 21 (chr12:111,902,466–111,902,519) was more prone to be spliced out of ATXN2 transcripts, as was exon 10 (chr17:44,087,676–44,087,768) of microtubule associated protein tau (MAPT) transcripts (Fig. 5a, Supplementary Fig. 9). ATXN2 lacking exon 21 corresponds to ATXN2 isoform type IV, which is found in neuronal and non-neuronal tissues31, whereas AS of exon 10 in MAPT disrupts the ratio of tau proteins containing 3 or 4 microtubule-binding domains32. Other validated AS CEs included alpha actinin 1 (ACTN1) smooth muscle exon (chr14:69,345,175–69,345,240) that dictates calcium sensitivity33; poly(ADP-ribosyl) polymerase family member 6 (PARP6) exon 17 (chr15:72,543,186–72,543,295), which is part of the catalytic domain and may affect PARP6’s role in cell cycle regulation34; hypoxia inducible factor 1 alpha subunit (HIF1A) exon 14 (chr14:62,212,409–62,212,535), an AS isoform of a transcriptional regulator for hypoxia response35; and RNA binding motif protein 39 (RBM39) (chr20:34,317,384–34,317,449) encoding a protein involved in AS and transcription (Fig. 5a, Supplementary Figs. 8,9). We further demonstrate that these exon skipping events are ALS-specific and not a consequence of neurodegeneration, as they are not observed in postmortem brain of progressive supranuclear palsy cases (Fig. 5b).
Figure 5.
Misregulation of cassette exon splicing in c9ALS affects transcripts with roles in diverse molecular pathways. (a) Wiggle plots from RNAseq data and qRT-PCR bar graphs (mean ± s.e.m.) of top differentially misregulated cassette exons (FDR < 0.05, |dI| ≥ 0.1) in c9ALS cerebellum (see additional examples in Supplementary Fig. 8). Shown is an example of the 3 technical qRT-PCR replicates performed for each AS event in c9ALS (N=9) and sALS (N=10) and compared to controls (N=9). (b) qRT-PCR bar graphs of the same differentially misregulated cassette exons shown in a in the cerebellum of c9ALS (N=9) and sALS (N=10) compared to a disease control group (PSP: progressive supranuclear palsy, N=13). Relative mRNA levels shown in all bar graphs were normalized to the endogenous control, RPLP0, and respective controls (mean value set to 1). The full list of primers used in this study can be found in Supplementary Table 11.Statistical differences were calculated by one-way ANOVA with Bonferroni post-hoc test (*P < 0.05, **P < 0.01, ***P < 0.005, #P < 0.001). Exact P values can be found in Supplementary Fig. 8. (c) Gene-association network of the top most significant misregulated cassette exon events in c9ALS cerebellum (FDR < 0.05, |dI| ≥ 0.1). dl: differential index value. Genes are represented by nodes of different colors, which vary according to degree. The size of the node denotes neighborhood connectivity: nodes are bigger if they are connected to other nodes with higher connectivity. Edges are colored according to edge betweenness to indicate the proximity to other nodes, with low betweeness (closer proximity) meaning larger influence to other nodes. GO annotations for different interconnected cellular pathways are indicated. A more complete figure containing all gene names for each of the nodes can be found in Supplementary Figure 11.
As ALS cases present motor neuron involvement, we examined whether the splicing events we identified also occur in c9ALS motor cortex. Of a total of 14 c9ALS exon skipping events validated in cerebellum and frontal cortex, 6 events were also observed in motor cortex of the same cases (Supplementary Fig. 10), further supporting the importance of these transcript changes to disease.
To assess whether specific RBPs may regulate AS of CEs discussed above, motifs enriched within AS CEs and flanking intronic regions were identified and cross-referenced with a RBP motif database (http://rbpdb.ccbr.utoronto.ca,36). Multiple references to hnRNPH were found for c9ALS and sALS (Supplementary Fig. 11). This finding is congruent with a previous report of hnRNPH sequestration by foci in c9ALS19, which is expected to influence hnRNPH function, and also uncovers a potential role for hnRNPH in sALS. Other binding motifs enriched in the cerebellum included those recognized by serine/arginine-rich splicing factor 1 (SRSF1). The binding motif recognized by matrin 3 (MATR3) was enriched in sALS AS cassette exons, notable because mutations in MATR3 are causative of ALS37.
To identify cellular pathways affected by CE missplicing in c9ALS and sALS, gene-association network analyses were conducted using the STRING web tool along with Cytoscape network analyzer (see Online Methods). No significant networks were generated for frontal cortex tissues, likely due to the relatively low number of AS events in this brain region; however, several misregulated networks were identified in the cerebellum. GO network annotation predicted RNA-processing as the most altered pathway in c9ALS cerebellum (Fig. 5c, Supplementary Fig. 12). Cytoskeleton and cellular trafficking pathways were largely altered in sALS cerebellum (Supplementary Fig. 13).
Widespread alternative polyadenylation events in ALS
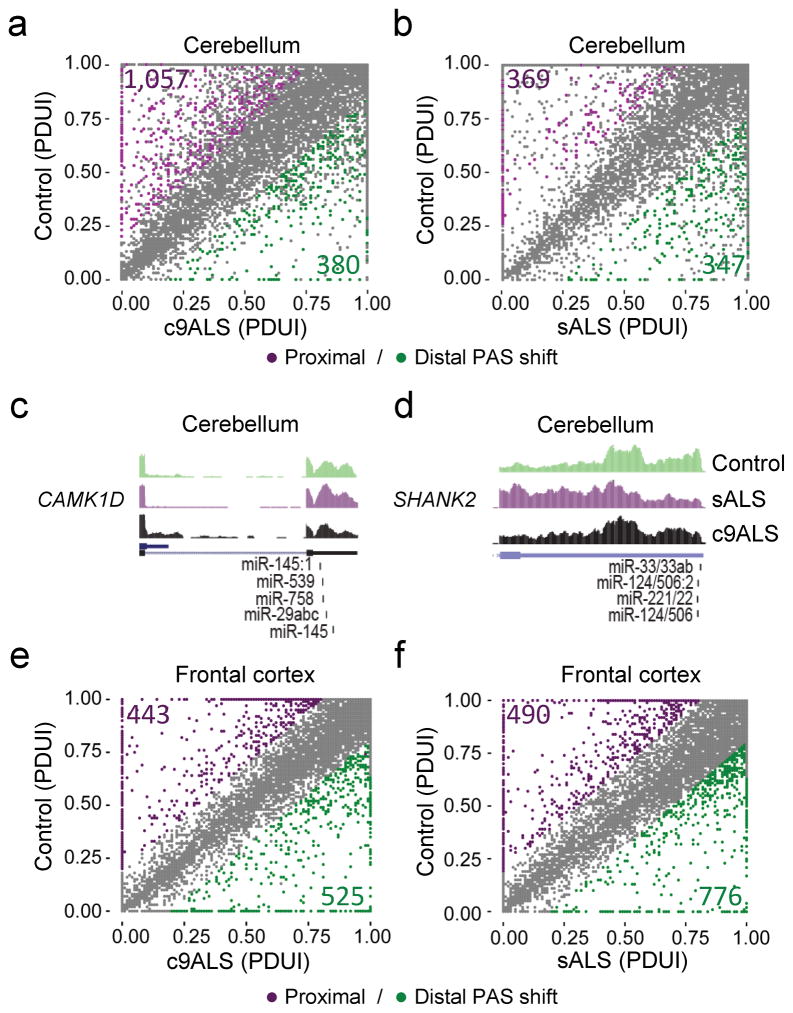
APA events can arise when more than one functional polyadenylation site (PAS) is present in the 3′UTR of an mRNA. APA events were examined in ALS using DaPars software38. Relative to control cases, the total number of PAS shifts (FDR < 0.05, |ΔPDUI| ≥ 0.2, |dPDUI| ≥ 0.2) in the cerebellum was higher in c9ALS (1,437) than sALS (716), with c9ALS displaying an increased usage of proximal (1,057) compared to distal (380) PAS, while sALS showed a similar distribution between proximal (369) and distal (347) PAS usage (Fig. 6a,b). GO analyses of transcripts with PAS shifts in cerebellum revealed top pathways of RNA processing in c9ALS (Supplementary Table 9) and response to oxidative stress in sALS (Supplementary Table 10). Figures 6c,d show examples of proximal PAS shifts in c9ALS and sALS cerebellum, respectively, with miRNA binding sites on the 3′UTRs depicted. Note that, in the event that a more proximal PAS is used, miRNA binding to the 3′UTR would be circumvented. In contrast to the cerebellum, the number of APA events in the frontal cortex was lower in c9ALS (968) compared to sALS (1,266), with c9ALS cases displaying a similar proportion of proximal (443) and distal (525) PAS shifts, while sALS cases show an increased usage of distal (776) compared to proximal (490) PAS shifts (Fig. 6e,f). Interestingly, the main GO pathways of transcripts characterized by alternative PAS usage in both c9ALS and sALS frontal cortex present roles in transcription (Supplementary Tables 9,10).
Figure 6.
Alternative polyadenylation changes are prominent in cerebellum of c9ALS and sALS cases. (a,b) Scatter plots representing differential PAS usage index (PDUI) in cerebellum of c9ALS (a) or sALS (b) compared to controls. PAS shifts are color coded to indicate significant distal (green) and proximal (purple) PAS usage (FDR < 0.05, |ΔPDUI| ≥ 0.2, |dPDUI| ≥ 0.2). (c,d) Wiggle plots of RNAseq cerebellum data for calcium/calmodulin-dependent protein kinase 1D (CAMK1D), which shows increased proximal and decreased distal PAS usage in c9ALS (c), and for SH3 and multiple ankyrin repeat domains 2 (SHANK2), in which a decreased usage of distal PAS is observed in sALS (d). (e,f) Scatter plots representing differential PAS usage index (PDUI) in frontal cortex of c9ALS (e) or sALS (f) compared to controls. PAS shifts are color coded to indicate significant distal (green) and proximal (purple) PAS usage (FDR < 0.05, |ΔPDUI| ≥ 0.2, |dPDUI| ≥ 0.2).
DISCUSSION
Misregulated RNA processing and metabolism are recurrent themes in many neurological disorders but their involvement in c9ALS is not clearly established. The present study reports widespread transcriptome changes in ALS cases carrying a C9orf72 repeat expansion, as well as in those with sALS. Our analyses found major misregulated RNA processing events in ALS, several affecting genes previously associated with ALS such as ATXN2 and FUS24–26, 39. Overall, the c9ALS transcriptome was affected to a greater degree than that of sALS, with little overlap. This limited overlap may result from the presence of pathological features unique to c9ALS in the frontal cortex and cerebellum, as well as heterogeneity among sALS cases (e.g. mutation in an unknown gene), which may mask transcriptional changes that occur in subsets of cases. Nonetheless, several aberrant transcriptome changes were shared between c9ALS and sALS, perhaps indicative of common pathological mechanisms or of pathways altered as an indirect consequence of the disease process. While TDP-43 inclusions are present in both c9ALS and sALS, the frontal cortex and cerebellum are relatively spared of this pathology rendering the contribution of misregulated TDP-43 to common changes unlikely.
The pronounced number of transcriptional changes in c9ALS is presumably due to the presence of RNA foci and DPR protein pathology as a consequence of r(G4C2)exp and r(G2C4)exp accumulation. For example, several RBPs (e.g. hnRNPH) interact with DPR proteins12, 15, 40 or bind RNA foci19, associations that may influence RNA processing and account for many of the RNA defects in c9ALS. With this in mind, it is of interest that reverse engineering analysis of over-represented RBP motifs in sequences surrounding misregulated AS cassette exons in c9ALS cerebellum and frontal cortex showed an enrichment of binding motifs for hnRNPH.
Differences in RNA processing in c9ALS could be caused by multiple other factors. For example, it was recently shown that DPR proteins cause endoplasmic reticulum (ER) stress in primary neurons11. UPR genes, which are activated by ER stress, were among the top DE genes in c9ALS cerebellum and frontal cortex. Notably, the UPR pathway was not observed as a top GO category in sALS. While pathological TDP-43, like DPR proteins, may cause dysfunction of the UPR, little TDP-43 pathology is observed in the fontal cortex and cerebellum of ALS cases. Several studies have also shown that some DPR proteins cause nucleolar stress, as well as impair RNA biogenesis and RNA granule formation, suggesting these DPR proteins lead to a global perturbation in RNA processing12, 14, 15, 41. Despite these impressive advances, it remains unclear which neuropathological features of c9ALS contribute to disease pathogenesis and/or correlate with the transcriptome defects observed.
In addition to differences in gene expression profiles between c9ALS and sALS, evaluation of AS and APA events further establishes the distinction between c9ALS and sALS. While defects in RNA processing and metabolism are observed in both c9ALS and sALS brains, our studies highlight major differences in functional pathways that may be affected in c9ALS and sALS. Indeed, in addition to the UPR, the intracellular protein transport and localization pathway are affected in c9ALS. Some of the genes within this pathway encode for proteins involved in transport/docking of synaptic vesicles (syntaxins), and in nuclear transport (nucleoporins, exportins). The latter is particularly intriguing given the increasing interest in impaired nuclear transport in c9ALS. In contrast, the majority of pathways that may be affected in sALS include cytoskeleton organization, defense response and synaptic transmission.
Approximately ~70% of human genes undergo APA in a tissue-specific manner42. Given that the 3′UTR of genes contain binding sites for several RBPs and miRNAs, APA plays important roles in regulating gene expression, RNA localization and RNA translation. Thus, the presence of distinct PAS shifts in the brain of both c9ALS and sALS cases are of particular interest. Indeed, widespread PAS shifts are associated with several human diseases43, 44, with this study adding ALS to the list.
Unexpectedly, more robust transcriptome changes were found in the cerebellum than the frontal cortex. Although the role of the cerebellum in coordination and motor control is well-documented, its involvement in ALS has been largely overlooked. Cerebellar atrophy is observed in ALS and behavioral variant frontotemporal dementia (bvFTD), which is believed to be part of the same spectrum of neurodegenerative diseases as ALS45–48. In fact, differences between ALS and bvFTD syndromes may depend partly on the cerebellar subregions affected, with ALS cases showing atrophy in the inferior cerebellum and vermis, which associates with motor dysfunction, and bvFTD cases showing widespread atrophy predominantly in the superior cerebellum47. Furthermore, studies prompted by the discovery of the C9orf72 repeat expansion in c9ALS coupled with the fact that these cases show marked cerebellar pathology, have revealed that cerebellar atrophy is also common to these patients49–50.
Overall, data presented herein provide evidence that disruption of normal RNA processing is part of the pathological signature of c9ALS and sALS. Additionally, these findings show distinct transcriptome profiles between c9ALS and sALS, and call attention to the potential contribution of the cerebellum to disease.
ONLINE METHODS
Standard protocol approvals and patient consents
Protocols were approved by the Mayo Clinic Institutional Review Board and Ethics Committee on human experimentation (09-008148, 12-007795). All participants or authorized family members gave written informed consent after which participant and family information was gathered, autopsies were performed and postmortem analyses were conducted.
Clinical, genetic and pathological assessments
All ALS patients in this study were recruited at Mayo Clinic Jacksonville and were independently ascertained by trained neurologists as having ALS per El Escorial criteria upon neurological examinations, electromyography, laboratory testing and, if deemed necessary, neurological imaging51, 52. ALS was confirmed neuropathologically by the Mayo Clinic Jacksonville brain bank for neurodegenerative disorders, and the presence or absence of a C9orf72 repeat expansion was determined using a previously described repeat-primed polymerase chain reaction method2. Mutations were absent from the coding regions of SOD1, TARDBP and FUS genes for ALS cases carrying a C9orf72 repeat expansion. Mutations were also absent in SOD1, TARDBP, FUS, and PFN1 for sporadic ALS cases, and the size of the C9orf72 hexanucleotide repeat was in the normal range. The majority of sALS cases lack any family history of neurodegenerative diseases with the exception of the maternal grandmother of sample #71, who was suspected to suffer from Parkinson’s disease, and the paternal first cousin and maternal grandfather of sample #72, who died from ALS and suffered from dementia (type unknown), respectively. Since no first or second degree relatives of cases #71 and #72 were affected with ALS, and since no genetic cause was identified, they were classified as sporadic ALS. Hippocampal and cortical regions were evaluated for co-existing Alzheimer’s type pathology and Braak tangle stage53 using thioflavin-S microscopy, as previously described54, 55. TDP-43 immunohistochemistry was performed on formalin-fixed tissue of cases used in the study, which are listed in Supplementary Table 1, using the phosphorylated Ser409/410 mouse monoclonal TDP-43 antibody, as previously described54. TDP-43 immunoreactivity was qualified given the presence or absence of neuronal inclusions and/or dystrophic neurites.
Human tissue and RNA processing
Frozen human postmortem brain tissue was sampled from the middle lobe (non-vermis) of the lateral hemisphere of the cerebellum, prefrontal cortex at the level of the nucleus accumbens (Brodmann area 9/44), and motor cortex at the level of the pulvinar (Brodmann area 4). RNA was then extracted using 20–40 mg of sampled frozen tissue. A two-step tissue homogenization method was performed using pestle tips and 21G syringes. Total RNA was extracted using the RNAeasy Plus Mini Kit (QIAGEN, Germantown, MD, USA), as per manufacturer’s instructions. RNA integrity (RIN) was verified on an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and only samples with RIN ≥ 7.0 were used for subsequent analyses. Note that no sample size calculations were performed, but our sample size numbers are consistent with many recent human brain transcriptome studies.
RNAseq library preparation and sequencing
A total of 100–200 ng of RNA in a 5 μl volume was used for library preparation and RNA sequencing (RNAseq). Paired-end sequencing libraries were prepared using the TruSeq Stranded Total Sample Preparation kit (Illumina) by the Mayo Clinic sequencing core facilities (Rochester, MN) followed by quality control, cluster generation and sequencing on the Illumina HiSeq 2000 platform. The reads were de-multiplexed and converted to FASTQ format using CASAVA software from Illumina (by the Mayo Clinic core). All RNAseq data for each individual RNA sample of cerebellum and frontal cortex tissues are available at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database under accession number GSE67196.
FASTQ alignment
All FASTQ files were aligned to the hg19 reference human genome. For expression analyses, FASTQ files were aligned using the Mayo MAP-RSeq pipeline v1.356. The alignments for alternative splicing analyses were done separately by using OLego, a seed and extend aligner that has high-sensitivity for splice-junction mapping of very small seeds (14 nt seed size), as previously described30, 57.
Bioinformatics analyses
A series of bioinformatics analyses were performed on aligned data, as described below. No randomization was used to assign samples to the experimental groups and data analyses were not performed blind. Note that, to decrease false negatives (type-II errors) and keep the criteria for detection of disease-specific events relatively stringent, the Bonferroni correction was avoided, using instead P value and FDR parameters for transcriptome-wide data analyses.
Expression analyses
FASTQ-derived read counts were analyzed for differential expression in R using Bioconductor edgeR version 3.4.2 using TMM normalization58. Venn diagrams were obtained by counting genes with a P value < 0.05 followed by selection of genes with|log2FC| ≥ 2 (FC: fold change). Pathway enrichment analyses of all genes showing differential expression (P < 0.05, |log2FC| ≥ 2) were performed using DAVID v6.759 on: 1) up- versus down-regulated genes; and 2) biological function GO terms for all DE genes (both up- and down-regulated). Expression MA scatter plots representing Log2FC (M) versus Log2 of counts per million (CPM; A) were generated using the R-studio program with ggplot2 library.
Hierarchical clustering and principal component analyses (PCA) of differentially expressed genes
Log2(n+1) transformed reads per kilobase per million (RPKM) for DE genes in c9ALS (P < 0.05, |log2FC| ≥ 2, as previously analyzed by EdgeR) were clustered using Cluster 3.0 software, heat maps were generated using Treeview60, and PCA plots were generated for top 5,000 expressed genes using the rgl library in R environment (http://rgl.neoscientists.org/about.shtml).
Weighted Gene Co-expression Network Analyses (WGCNA)
RPKM values of genes (log2RPKM > 1) from EdgeR data (described previously in expression analyses) were used to construct signed co-expression networks using the WGCNA package in R61. Low expression genes were excluded from the analyses to remove noise. Network construction was performed using the manual function in the WGCNA package, which calculates a dissimilarity matrix based on the topological overlap, which is then used for hierarchical clustering. For each set of genes, a pair-wise correlation matrix is computed and an adjacency matrix is calculated by raising the correlation matrix to a power of 20, which was chosen to attain scale-free topology. Module definitions were calculated by using the hybrid treecutting option with deepsplit parameter = 1–3 depending upon the dataset. All the modules were summarized by module eigengenes (ME) which explain the maximum amount of variation of the module expression levels. Heat maps for modules were generated using RPKM values for genes represented in the module, using Cluster 3.0. Gene module membership (kME) was calculated as the correlation between gene expression values and the module eigengene. Modules were visualized using Cytoscape 3.1.062, and annotated for gene ontology using DAVID v6.759.
Bioinformatic analyses of RNAseq data for identification of alternatively splicing (AS) events
Following FASTQ alignment using OLego, differential exon inclusion and exclusion events were determined and quantified by using the Quantas module, as described in OLego30. A Fisher’s exact test was used to evaluate the statistical significance of splicing changes using both exon and exon-junction fragments, followed by Benjamini multiple testing correction to estimate the false discovery rate (FDR). In addition, inclusion or exclusion junction reads were used to calculate the proportional change of exon inclusion (I). Differential splicing events were identified by requiring FDR < 0.05, or more stringent criteria (FDR < 0.05, |dI| ≥ 0.1,). Wiggle plots were generated for visualization of splicing events on UCSC genome browser (http://genome.ucsc.edu). Splicing scatter plots were generated using the R-studio program with ggplot2 library. Pathway enrichment analyses of genes showing AS were performed on biological function GO terms using DAVID v6.759.
Hierarchical clustering for alternatively spliced cassette exons
Splicing index data obtained from the AltAnalyze package63 for cassette exons, which showed alternative splicing changes in c9ALS (FDR < 0.05, |dI| ≥ 0.1,), were clustered using Cluster 3.0 program and heat maps were generated using Treeview60.
Gene-association networks
Gene-association networks or protein-protein interaction networks were constructed with String v9.164 using splicing results from Quantas. Gene names were input and the resulting networks were based on existing co-expression, gene interaction, and experimental evidence data. Networks were visualized and analyzed for degree and edge betweenness using Cytoscape 3.1.0, and annotated for gene ontology using DAVID v6.759.
RNA-binding motif determination
FASTA sequences containing the identified AS cassette exons (from Quantas, FDR < 0.05, |dI| ≥ 0.1) and flanking introns were extracted from the UCSC genome browser. The consensus motifs were identified using the MEME-ChIP program65 of the MEME suite package. The identified motifs were then cross-referenced with the known binding motifs of RNA-binding proteins (RBP) and literature36, 66(RBP database: http://rbpdb.ccbr.utoronto.ca/), to identify RBPs that may be responsible for AS cassette exon events significantly represented in our dataset.
Alternative polyadenylation analyses
Following FASTQ alignment using OLego, the aligned SAM files were converted into BED files and then to wiggle coverage files. The bioinformatics algorithm DaPars67, which uses regression model to locate end points of alternative polyadenylation signals (PAS), was used to identify alternative polyadenylation (APA) events between ALS samples and controls. To identify significant APA events, we used the following cutoffs: FDR < 0.05, |ΔPDUI| (absolute PDUI or ratio of distal and proximal PAS usage, for each gene in one condition) ≥ 0.2, and |dPDUI| (differential PDUI or PAS usage between control and disease) ≥ 0.2. APA scatter plots were generated using the R-studio program with ggplot2 library. Pathway enrichment analyses of genes showing PAS shifts were performed on biological function GO terms using DAVID v6.759.
Validation of transcriptome changes by RT-PCR and qRT-PCR
RNA was extracted from cerebellar, frontal cortex, and motor cortex tissues as explained above (Human tissue and RNA processing) and 500 ng of RNA was used to generate cDNA by reverse transcription polymerase chain reaction (RT-PCR) using the High Capacity cDNA Transcription Kit (Applied Biosystems, Foster City, CA, USA), as per manufacturer’s instructions. Following standard protocols, quantitative real-time PCRs (qRT-PCRs) were conducted using SYBR GreenER qPCR SuperMix (Invitrogen, Carlsbad, CA, USA) for all samples, in triplicates. Note all primer pairs used are listed in Supplementary Table 11. qRT-PCRs were run on an ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Relative quantification was determined using the ΔΔCt method and normalized to the endogenous control RPLP0. Note, we evaluated several endogenous controls by qRT-PCR (GAPDH, GUSB, HMBS and RPLP0), and RPLP0 consistently presented the lowest Ct values and Ct variation (not shown) across cerebellar and frontal cortex tissues, and we have previously used it as reference transcript11. One-way ANOVA followed by Bonferroni post-hoc test was used for multiple comparisons with a 95% confidence level. We considered the difference between comparisons to be significant when P < 0.05 (*P < 0.05; **P < 0.01; ***P < 0.005, #P < 0.001). All graphs represent mean +/− s.e.m.. A Supplementary Methods Checklist is available
Supplementary Material
Acknowledgments
We are extremely grateful to all patients who agreed to donate their brain to research. This work was supported by the National Institutes of Health/National Institute on Aging [R01AG026251 (LP), P50AG016574 (LP)], National Institutes of Health/National Institute of Neurological Disorders and Stroke [R21NS089979 (TFG, KBB), R21NS084528 (LP), R01NS088689 (LP), R01NS063964 (LP, CDL); R01NS077402 (LP); P01NS084974 (LP, DD, RR, KBB)], National Institute of Environmental Health Sciences [R01ES20395 (LP)], Department of Defense [ALSRP AL130125 (LP)], Mayo Clinic Foundation (LP), Mayo Clinic Center for Individualized Medicine (LP, KBB), ALS Association (KBB, LP, TFG), Robert Packard Center for ALS Research at Johns Hopkins (LP), Target ALS (LP), Milton Safenowitz post-doctoral fellowships from the ALS Association (VVB; MP), post-doctoral fellowship from the Canadian Institutes of Health Research (VVB), The Siragusa Foundation career development award for young investigators (VVB), and Robert and Clarice Smith & Abigail Van Buren Alzheimer’s Disease Research Foundation postdoctoral fellowship (VVB). HL and MEM are supported by the Mayo Clinic Center for Individualized Medicine and the Donors Cure Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
M.P., V.V.B., R.B., and C.A.R. contributed equally to this work. M.P., V.V.B. and L.P. contributed to the conception and design, and to tissue selection and collection. M.P. performed RNA extractions. M.P. and V.V.B. made cDNA. M.P. ran qRT-PCRs for expression and AS validations. R.B., C.A.R. and H.L. performed expression and WGCNA bioinformatics analyses. R.B. conducted AS, APA and system network analyses. M.P. and R.B. carried-out gene ontology analyses. H.L. supervised the bioinformatics analyses. All co-authors contributed to the critical revision of the manuscript for important intellectual content. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content. All authors gave final approval of the version to be published.
References
- 1.Geser F, et al. Evidence of multisystem disorder in whole-brain map of pathological TDP-43 in amyotrophic lateral sclerosis. Archives of Neurology. 2008;65:636–641. doi: 10.1001/archneur.65.5.636. [DOI] [PubMed] [Google Scholar]
- 2.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renton AE, et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011 doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Blitterswijk M, DeJesus-Hernandez M, Rademakers R. How do C9ORF72 repeat expansions cause amyotrophic lateral sclerosis and frontotemporal dementia: can we learn from other noncoding repeat expansion disorders? Curr Opin Neurol. 2012;25:689–700. doi: 10.1097/WCO.0b013e32835a3efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ash PE, et al. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mori K, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta neuropathologica. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 7.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 8.Gendron TF, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta neuropathologica. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizielinska S, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014 doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.May S, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta neuropathologica. 2014 doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YJ, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta neuropathologica. 2014 doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon I, et al. Poly-dipeptides encoded by the C9ORF72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014 doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamakawa M, et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Human Molecular Genetics. 2015;24:1630–1645. doi: 10.1093/hmg/ddu576. [DOI] [PubMed] [Google Scholar]
- 14.Wen X, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao Z, et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv005. [DOI] [PubMed] [Google Scholar]
- 16.Donnelly CJ, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagier-Tourenne C, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proceedings of the National Academy of Sciences of the United States of America; 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sareen D, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YB, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper-Knock J, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain: a journal of neurology. 2014;137:2040–2051. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 22.Gendron TF, Josephs KA, Petrucelli L. Review: transactive response DNA-binding protein 43 (TDP-43): mechanisms of neurodegeneration. Neuropathol Appl Neurobiol. 2010;36:97–112. doi: 10.1111/j.1365-2990.2010.01060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwiatkowski TJ, Jr, et al. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 25.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Couthouis J, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Human molecular genetics. 2012;21:2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ticozzi N, et al. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2011;156B:285–290. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- 29.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J, Anczukow O, Krainer AR, Zhang MQ, Zhang C. OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic acids research. 2013;41:5149–5163. doi: 10.1093/nar/gkt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Affaitati A, de Cristofaro T, Feliciello A, Varrone S. Identification of alternative splicing of spinocerebellar ataxia type 2 gene. Gene. 2001;267:89–93. doi: 10.1016/s0378-1119(01)00402-4. [DOI] [PubMed] [Google Scholar]
- 32.Yen SH, Hutton M, DeTure M, Ko LW, Nacharaju P. Fibrillogenesis of tau: insights from tau missense mutations in FTDP-17. Brain pathology. 1999;9:695–705. doi: 10.1111/j.1750-3639.1999.tb00551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kremerskothen J, et al. Brain-specific splicing of alpha-actinin 1 (ACTN1) mRNA. Biochemical and biophysical research communications. 2002;295:678–681. doi: 10.1016/s0006-291x(02)00734-9. [DOI] [PubMed] [Google Scholar]
- 34.Tuncel H, et al. PARP6, a mono(ADP-ribosyl) transferase and a negative regulator of cell proliferation, is involved in colorectal cancer development. Int J Oncol. 2012;41:2079–2086. doi: 10.3892/ijo.2012.1652. [DOI] [PubMed] [Google Scholar]
- 35.Gothie E, Richard DE, Berra E, Pages G, Pouyssegur J. Identification of alternative spliced variants of human hypoxia-inducible factor-1alpha. The Journal of biological chemistry. 2000;275:6922–6927. doi: 10.1074/jbc.275.10.6922. [DOI] [PubMed] [Google Scholar]
- 36.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JO, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Blitterswijk M, et al. Ataxin-2 as potential disease modifier in C9ORF72 expansion carriers. Neurobiol Aging. 2014;35:2421 e2413–2427. doi: 10.1016/j.neurobiolaging.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori K, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta neuropathologica. 2013;125:413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 41.Mizielinska S, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batra R, et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell. 2014;56:311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenal M, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 45.Thivard L, et al. Diffusion tensor imaging and voxel based morphometry study in amyotrophic lateral sclerosis: relationships with motor disability. J Neurol Neurosurg Psychiatry. 2007;78:889–892. doi: 10.1136/jnnp.2006.101758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeley WW, et al. Frontal paralimbic network atrophy in very mild behavioral variant frontotemporal dementia. Arch Neurol. 2008;65:249–255. doi: 10.1001/archneurol.2007.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan RH, et al. Cerebellar integrity in the amyotrophic lateral sclerosis-frontotemporal dementia continuum. PLoS One. 2014;9:e105632. doi: 10.1371/journal.pone.0105632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bede P, et al. Patterns of cerebral and cerebellar white matter degeneration in ALS. Journal of neurology, neurosurgery, and psychiatry. 2015;86:468–470. doi: 10.1136/jnnp-2014-308172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahoney CJ, et al. Longitudinal neuroimaging and neuropsychological profiles of frontotemporal dementia with C9ORF72 expansions. Alzheimers Res Ther. 2012;4:41. doi: 10.1186/alzrt144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boeve BF, et al. Characterization of frontotemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72. Brain. 2012;135:765–783. doi: 10.1093/brain/aws004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. Journal of the neurological sciences. 1994;124(Suppl):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 52.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 53.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta neuropathologica. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 54.Murray ME, et al. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122:673–690. doi: 10.1007/s00401-011-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murray ME, et al. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: a retrospective study. The Lancet. Neurology. 2011;10:785–796. doi: 10.1016/S1474-4422(11)70156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalari KR, et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics. 2014;15:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charizanis K, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 61.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nature protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emig D, et al. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic acids research. 2010;38:W755–762. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huelga SC, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Masamha CP, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.