Abstract
Intra-tumor heterogeneity (ITH) drives neoplastic progression and therapeutic resistance. We used EXPANDS and PyClone to detect clones >10% frequency within 1,165 exome sequences from TCGA tumors. 86% of tumors across 12 cancer types had at least two clones. ITH in nuclei morphology was associated with genetic ITH (Spearman ρ: 0.24–0.41, P<0.001). Mutation of a driver gene that typically appears in smaller clones was a survival risk factor (HR=2.15, 95% CI: 1.71–2.69). The risk of mortality also increased when >2 clones coexisted (HR=1.49, 95% CI: 1.20–1.87). In two independent datasets, copy number alterations affecting either <25% or >75% of a tumor’s genome predicted reduced risk (HR=0.15, 95% CI: 0.08–0.29). Mortality risk also declined when more than four clones coexisted in the sample, suggesting a tradeoff between costs and benefits of genomic instability. ITH and genomic instability have the potential to be useful measures universally applicable across cancers.
Cancers are a mosaic of clones of varying population sizes, different genetic makeup and distinct phenotypic characteristics1–4. This intra-tumor heterogeneity provides the fuel for the engine of natural selection that drives the process of carcinogenesis and acquired therapeutic resistance in neoplasms1,5. When analyzing genome sequencing data derived from single tumor samples, it is important to recognize that technically, sequences obtained from each tumor sample encode a tumor-metagenome, since they represent the aggregate genomes of all clones that coexist within the sample6,7,10–12. Recently, McGranahan et al. used exome-sequencing data derived from single tumor samples to determine the clonal status of known, actionable drivers across 9 cancer types and to identify events that trigger clonal expansions, causing ITH6. However, the availability of just one sample per tumor and moderate sequencing depth has limited the opportunity for systematic analysis of extent and the clinical consequences of ITH, in previous pan-cancer studies12–14,3,7–9. To overcome these limitations, a variety of different algorithms have been developed to deconvolute tumor-metagenomes. These algorithms estimate the cellular prevalence of mutations and quantify ITH15–19. We leveraged two of these tumor mixture separation algorithms, EXPANDS18 and PyClone17, to quantify ITH from TCGA exome sequencing data, and to validate the robustness of our results.
RESULTS
Intra-tumor genetic heterogeneity exists in all tumor types
We measured the number and size of genetically diverse clones of 1,165 primary tumor samples across 12 cancer types from The Cancer Genome Atlas (TCGA), using paired tumor-normal exome sequencing data. These samples originated from a single sequencing center (the Broad Institute) and were chosen because they fulfilled established strict criteria to obtain uniform sequence data quality and depth (Supplementary Fig. 1.1). As clone detection sensitivity is highly dependent on genomic depth and breadth of coverage, these criteria are necessary to ensure that measures of ITH derived from these sequences are comparable. Detailed inclusion criteria are available in Supplementary Note 1.2.
Somatic single nucleotide variants (SNVs) and copy number variants (CNVs) were called using MuTect20 and ExomeCNV21 respectively (Supplementary Fig. 1.3). We distinguished non-synonymous SNVs and splice site or regulatory region SNVs (generally referred to as non-silent) from synonymous SNVs and SNVs within intergenic and intronic regions (referred to as silent). The incidence of CNVs and somatic non-silent SNVs varied considerably within and between tumor types (Fig. 1a–c), similar to results obtained from other genome wide sequencing studies13,22,23.
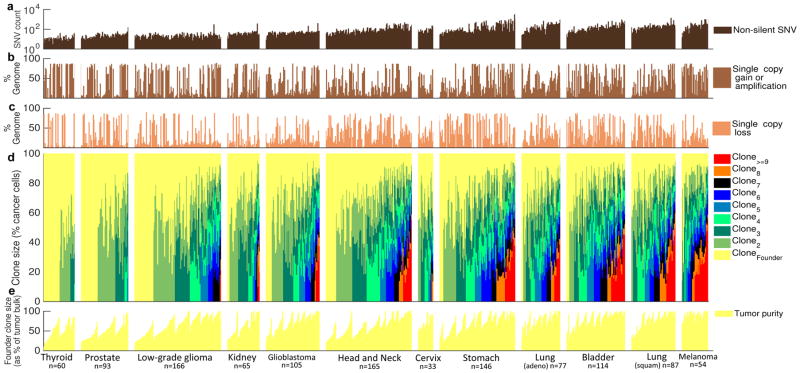
Figure 1. Tumor-metagenomes and subclonal genomes in 12 tumor types from TCGA.
(a) Prevalence of non-silent somatic SNVs per tumor. Percentage of tumor-metagenome affected by (b) single copy gains/amplifications and (c) single copy losses. (d) Clonal composition inferred from SNVs and copy numbers. Every sample contains a founder tumor population (yellow), identified as the largest clone within the sample. Each change in color marks the presence of an additional clone at the indicated size, calculated as % of the founder population size (y-axis). Color-variety within each tumor-type panel reflects the extent of intra-tumor heterogeneity in the corresponding tumor type. The average number of detectable (>10% frequency) clones increases from thyroid carcinoma (left) to melanoma (right). (e) The size of the founder clone is a measure of tumor purity. The exact number of tumors of each type (n) is indicated at the bottom of each panel.
EXPANDS was applied to all detected somatic SNVs (including silent SNVs), loss of heterozygosity (LOH) and copy number estimates to infer the number, size, and genetic content of subpopulations of cells that coexisted in the tumor (Fig. 1d). Briefly, EXPANDS models the cellular prevalence of each SNV as a copy-number dependent probability distribution. Subsequently, these cellular prevalence distributions are clustered to obtain the genetic content of each subpopulation, i.e. the set of SNVs and CNVs that accumulated in ancestral cells prior to each clonal expansion. Previous results18 indicate that the sequencing data available per tumor (on average 5,221 Mb reads) translates to an accuracy of 50–80% at which EXPANDS detects genetic heterogeneity at a macroscopic resolution24 (i.e. clones present in ≥ 10% of the sample). An independent algorithm, PyClone17, was used to validate the conclusions derived from EXPANDS. PyClone infers the cellular prevalence of SNVs differently from EXPANDS. In particular, PyClone does not model subclonal CNVs and leverages high depth rather than high breadth of sequencing17 (Supplementary Note 2.1).
In general, the cellular prevalence of SNVs assigned by PyClone and EXPANDS was concordant for SNVs located within segments of clonal copy number (Spearman ρ=0.77). However, for regions in which CNVs affect only a subset of tumor cells, EXPANDS and PyClone tended to make different inferences for cellular prevalence of SNVs within those regions (ρ=0.25; Supplementary Fig. 2.1b,c).
Subpopulations detected within the same tumor sample may have sizes that cumulatively exceed 100%, as a subpopulation may be nested in a parental population that carries earlier mutations. Both algorithms detect such nested subpopulation compositions. We will refer to these inferred subpopulations as clones, and to the cellular prevalence of a subpopulation within the tumor sample as its clone size. As noted previously, we define the term ‘tumor-metagenome’ as the aggregate genomes of all co-existent clones within a tumor.
Assuming a monoclonal tumor origin, the largest inferred clone in each sample corresponds to the first (founder) clonal expansion. This holds true, regardless of the fitness difference between the founder and descending clones. The cellular prevalence of founder-mutations will always be greater than or equal to the cellular prevalence of mutations acquired by descendant subclones, even if these later subclones proliferate faster than the founder. This implies that the size of the largest clone is also a measure of tumor purity (Fig. 1e); this was confirmed by an independent study that compared the performance of EXPANDS to four other methods that predict tumor purity25. The size of the largest clone was correlated to tumor purity as predicted from expression profiling with ESTIMATE26 (EXPANDS: Pearson r=0.43; P≪1E–6; PyClone: r=0.63; P≪1E–6; Supplementary Fig. 2.2a).
We observed that the number of somatic SNVs in large clones correlated with age at diagnosis (ρ=0.3; P≪1E–6), a result previously reported for chronic lymphocytic leukemia (CLL)10. In addition, the number of SNVs in small clones also correlated with age (ρ=0.18; P=5E–6; Supplementary Fig. 2.2b,c).
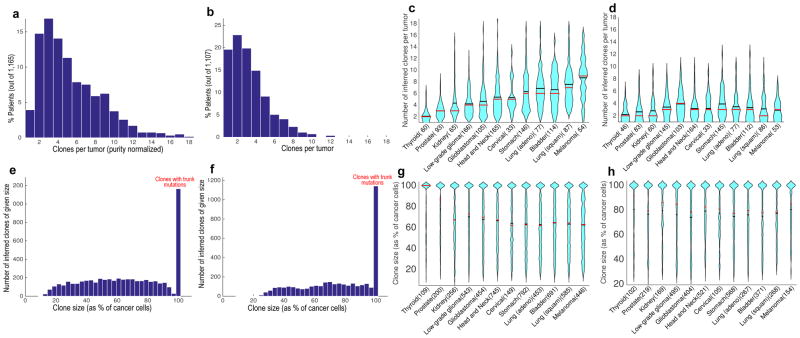
We compared the extent of genetic ITH across and within tumor types (Fig. 2a–d). The difference between tumor types in the number of clones they harbor was similar before (Supplementary Fig. 2.3a) and after correcting for tumor purity (Fig. 2c, Online Methods). On average four clones were estimated to coexist in a tumor at the time of biopsy or surgical resection (median clone number EXPANDS: 5; PyClone: 3; Fig. 2a,b). There was a median of 10 (EXPANDS estimate) to 16 (PyClone estimate) non-silent somatic SNVs per clone and the distribution of clone sizes across tumor types was relatively uniform (Supplementary Fig. 2.4a and Fig. 2e–h). Notably, reduced detection sensitivity (due to low tumor purity) was not sufficient to explain the smaller number of clones observed in low-purity tumors (Supplementary Fig. 2.4b). In 14% (EXPANDS estimate) to 20% (PyClone estimate) of the analyzed tumor samples, only a single, genetically homogeneous cell population was detected. Even for thyroid carcinoma – the least heterogeneous tumor type – two or more clones were predicted to coexist in >50% of the samples (EXPANDS estimate: 52%; PyClone estimate: 65%). Therefore, we concluded that genetic ITH occurs in the vast majority of cancers represented among the 12 types that we included in this study.
Figure 2. Intra-tumor genetic heterogeneity in 12 tumor types.
Clone number distribution predicted by EXPANDS (a) and PyClone (b) across tumor types. Violin plots of clone number distribution predicted by EXPANDS (c) and PyClone (d) within tumor types. Clone size distribution predicted by EXPANDS (e) and PyClone (f) across tumor types. Violin plots of clone size distribution predicted by EXPANDS (g) and PyClone (h) within tumor types. EXPANDS derived clone numbers (a, c) and all clone sizes (e-h) have been normalized by tumor purity. For PyClone derived clone numbers, normalization by tumor purity was not necessary. Violin plots contain marks for the mean (black lines) and median (red lines). [Thyroid = Thyroid Carcinoma; Prostate = Prostate Adenocarcinoma; Kidney = Kidney Renal Clear Cell Carcinoma; Head and Neck = Head and Neck Squamous Cell Carcinoma; Cervical = Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma; Stomach = Stomach Adenocarcinoma; Lung (adeno) = Lung Adenocarcinoma; Bladder = Bladder Urothelial Carcinoma; Lung (squam) = Lung Squamous Cell Carcinoma; Melanoma = Skin Cutaneous Melanoma].
Driver genes are mutated in clones of characteristic sizes
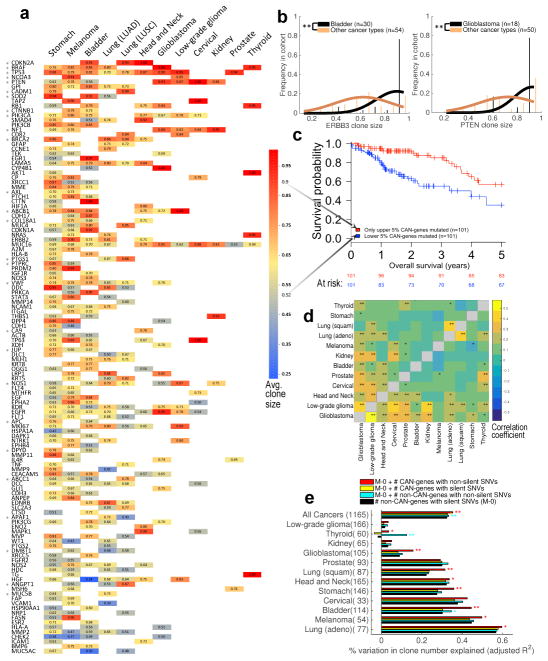
To investigate the influence of driver gene mutation incidence on genetic ITH, we analyzed 259 cancer driver genes (CAN-genes; Supplementary Table 3.1). A gene was included as significantly associated with a given cancer type based on: i) prior experimental evidence; ii) frequency of gene-mutations in our sample cohort and iii) mutation deleteriousness (Online Methods). Shown in Fig. 3a are the 124 non-private CAN-genes (48%) that are mutated in a minimum of two cancer types.
Figure 3. Association of driver gene mutations to clone size and clone number.
(a) Clone sizes were predicted by EXPANDS for mutations in 124 CAN-genes and normalized by purity. For each cancer type CAN (x-axis) and gene G (y-axis), average clone size was calculated across all CAN clones that harbor non-silent SNVs in G. Blank entries denote that G was not significantly associated to CAN. SNVs in CAN-genes often have the tendency to occur in clones of characteristic sizes, independent of cancer type (one-sided t-test: *P<0.05). (b) Mutations in some CAN-genes tend to drive large clonal expansions in certain cancer types, for example ERBB3 mutations in bladder carinoma and PTEN mutations in Glioblastoma (one-sided t-test: **P<1E–4). (c) SNVs in CAN-genes that characteristically grew to smaller clones predicts poor prognosis across tumor types (Log-rank test: P=2.9E–4; HR=2.72). (d) Clones with CAN-gene mutations have similar sizes across certain tumor types, suggesting the order/selective advantage of CAN-gene mutations is often not tissue-specific. Pairwise similarity between tumor types is calculated as Spearman correlation (**P<0.01; *P<0.05) based on EXPANDS (above diagonal) and PyClone results (below diagonal). (e) The number of clones identified in a sample depends on SNV incidence, but not all SNV categories are equally associated with the resulting number of clones. Non-silent SNV incidence in CAN-genes (red; mean = 2 genes) explain variability in clone number better than silent SNV incidence in CAN-genes (yellow; mean = 1 gene) or non-silent SNV incidence in non-CAN-genes (cyan; mean =128 genes). Log-likelihood test: **P<0.01; *P<0.05.
Next, we tested whether clones differ in their size depending on which CAN-genes are mutated in the corresponding clones. The size of a clone depends on its selective fitness (how fast it expands relative to the other clones within a tumor) and on its formation time (when the underlying clonal expansion started). CAN-gene SNVs specific to a given clone may therefore have a direct impact on its size. To test this possibility, we first normalized clone sizes by tumor purity. We then calculated the mean and variance in clone-size among all clones with non-silent mutations in a given CAN-gene and compared them to the variance calculated from random samples of clone sizes from our data (Supplementary Fig. 3.2).
The size of clones harboring CAN-gene mutations varied across CAN-genes (Fig. 3a) and was correlated to both, the relative order of driver gene mutations reported in earlier studies13,28–31 and to the clone sizes predicted by PyClone (0.43<r<0.94; 3.4E–18<P<0.12; Supplementary Fig. 3.3a,b). Across tumor samples, and even across tumor types, CAN-genes were often mutated in clones of a characteristic size, i.e. the variance in clone size was significantly lower than expected by chance (one-sided t-test: P<0.05; Fig. 3a). Citing an example, TP53 SNVs were found in larger clones (EXPANDS: 0.811; PyClone: 0.746 average cancer-cell fraction) in all nine cancers significantly associated with TP53 genetic aberrations. In contrast, somatic SNVs in DMBT1 were found in smaller clones (EXPANDS: 0.641; PyClone: 0.652 average cancer cell fraction) in the three cancers in which DMBT1 was among the drivers (Fig. 3a).
For a subset of CAN-genes however, we found significant differences in the dominance of mutated clones depending on cancer type. For example, clones with ERBB3 mutations were larger in bladder cancer than they were in any other cancer type, while clones with PTEN mutations grew particularly large in Glioblastoma (Fig. 3b). Clones with SNVs in CAN-genes that are druggable (n=98) did not have significantly different sizes as compared to clones with SNVs in the remaining CAN-genes (n=161; T-test: P=0.77).
Furthermore, clone size inferiority of a mutated CAN-gene (as shown in Fig. 3a) was correlated to the propensity of the CAN-gene as risk factor (univariate Cox EXPANDS: P=3.2E–07, HR=2.86; PyClone: P=0.005, HR=1.67). For instance, mutations in CAN-genes within the lower 5% average clone size were associated with poor outcome (Fig. 3c). This relation was also significant in low-grade gliomas, kidney carcinoma and glioblastoma (Supplementary Table. 4.3a). Several cellular functions/pathways were associated exclusively with small-size and medium-size clones, but not with large-size clones including tyrosine-protein kinase activity (P=1.39E–12) and positive regulation of locomotion (P=3.39E–9) (Supplementary Tables 3.1 and 3.4).
Next, we used the size-rankings of clones with CAN-gene mutations to compare cancer types (Fig. 3d). For CAN-genes that are critical among different tumor types, we measured whether clones containing mutations in these genes are of similar size, regardless of tumor type. Head and neck cancer, low-grade glioma and glioblastoma showed significant clone-size similarities to most other cancer types (0.12≤ρ≤0.43; P<0.05), though all cancers were similar to at least one other cancer type (Fig. 3d; PyClone and EXPANDS: 0.12≤ρ≤0.58; P≤0.05).
Finally, we tested whether distinct SNV categories differ in how well they model the number of detected clones per tumor. Per cancer type, silent SNVs in non-CAN-genes accounted for an average of 25% of the variability in the number of clones. Including silent SNVs in CAN-genes as predictors of clone number did not improve the model. In contrast, including non-silent SNVs in CAN-genes improved the predictions, accounting for 30% of the variability in the number of clones (log-likelihood test: P<0.05; Fig. 3e). These results suggest that mutations driving clonal expansions are more common among non-silent SNVs in CAN-genes, than among other SNV categories (Online Methods and Supplementary Note 2.1).
Histologic ITH and proliferation rate reflect genetic ITH
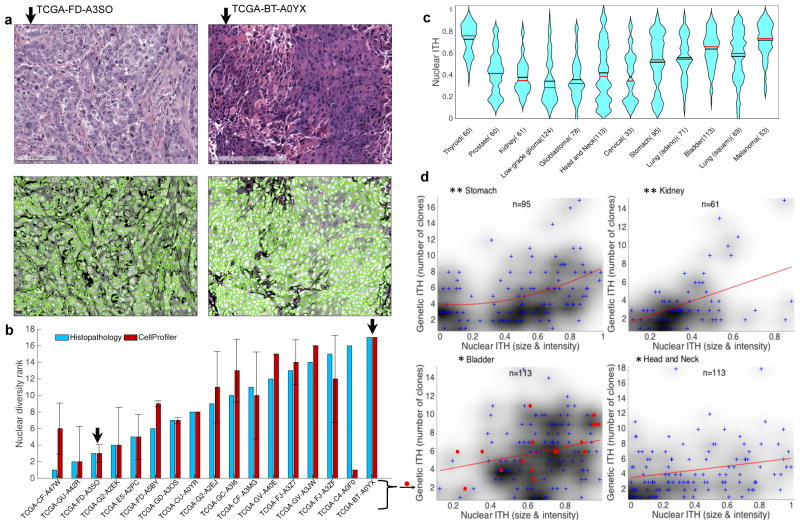
Nuclear size and staining variability is a standard histomorphologic metric of tumor differentiation, facilitating comparisons across cancers independent of tissue origin. A total of 2,231 H&E images were available at TCGA for 930 (80%) of the analyzed tumor samples (Supplementary Fig. 1.3). To quantify histologic ITH from these images, we measured the variability in nuclei size and staining intensity (Supplementary Note 2.5). For each tumor, the established image-analysis software CellProfiler34 was used to measure the size and staining intensity of every nucleus detected on the tumor’s H&E images35,36. A histopathologist conducted an independent and blinded scoring of a subset of 17 H&E images (Supplementary Fig. 2.6), which confirmed the accuracy of nuclear diversity scoring by CellProfiler (ρ=0.64, P=0.007; Fig. 4a,b).
Figure 4. Intra-tumor nuclear diversity accompanies intra-tumor genetic diversity.
(a) Quantitation of intra-tumoral nuclear diversity from H&E images. Conventional H&E stainings (upper panels) of two bladder cancer specimens are shown. The lesion on the left (TCGA-GD- A3SO) demonstrates monomorphic high-grade nuclei with open chromatin and prominent nucleoli, while the lesion on the right (TCGA-BT-A0YX) demonstrates nuclei that vary from small with condensed chromatin to very large with open chromatin (anisochromasia). CellProfiler outlines nuclei (lower panels) and quantifies nuclear variability from the H&E images. (b) Quantitation of nuclear diversity is shown for the two bladder cancer specimens in panel a (black arrows) along with 15 other bladder cancer specimens. Independent ranking of intra-tumor nuclear diversity across these 17 bladder cancer specimens by an expert histopathologist (blue) validates the automated nuclear diversity measures (red) (ρ=0.64; P=0.007). (c) Violin plots of nuclear diversity within tumor types. Nuclear diversity was normalized to account for differences in tumor purity. Tumor types are ordered according to their extent of genetic ITH (Fig. 2b). (d) Nuclear diversity per tumor (x-axis; quantified based on nuclear intensity and size diversity) increases with increasing clone number per tumor (y-axis). This is true for all cancers combined (ρ=0.243; P=6.30E–14) as well as for the specific types shown (* ρ>0.25; P<0.01; ** ρ>0.4; P<0.001). The p-values shown here have not been corrected for multiple hypothesis testing.
The extent of nuclear ITH varied between tumor types (Fig. 4c). Greater nuclear diversity was observed with increasing clone number (Fig. 4d) in kidney cancer (ρ=0.413; FDR adjusted P=0.004), stomach cancer (ρ=0.406; FDR adjusted P=2.97E–4), head and neck cancer (ρ=0.278; FDR adjusted P=0.009), bladder cancer (ρ=0.246; FDR adjusted P=0.022) as well as across all 12 cancer types (ρ=0.243; FDR adjusted P=8.15E–13). Increased nuclear diversity with increasing clone number was observed for both PyClone and EXPANDS based clone number predictions, as well as after normalizing nuclear and genetic ITH measures to account for tumor purity (Supplementary Table 3.5).
We used mRNA expression levels of the proliferation marker KI67, available for 854 (73%) of the samples, to measure proliferation rate37. Clone number was significantly correlated to proliferation rate within low-grade glioma (ρ=0.18; P=0.021) and prostate cancer (ρ=0.21; P=0.046) as well as across cancers (ρ=0.31; P=2.69E–20). However, tumor-type specific p-values did not remain significant after FDR correction for multiple testing (P>0.05). For a subset of cancer types (the three squamous cell carcinomas of the head and neck, lung and cervix), very heterogeneous tumors (>8 clones) had low KI67 expression (Supplementary Figure 2.7).
Overall, these results show that nuclear and cellular features typically associated with aggressive disease correlate with greater genetic ITH across cancer types.
Prognostic value of genomic instability and genetic ITH
We tested whether measures of genomic instability and genetic ITH (Supplementary Table 4.1) could predict overall and progression free survival. We constructed univariate Cox models for each cancer type separately as well as pan-cancer Cox models (Supplementary Tables 4.2 and 4.3). Prostate adenocarcinoma and thyroid carcinoma were excluded from the cancer type specific survival analysis due to insufficient availability of uncensored survival information (Supplementary Table 1.4).
When considering each cancer type separately, no significant monotonic association between clone number and survival was evident (P>0.05; Supplementary Table 4.3), apart from gliomas (EXPANDS: P=0.03, HR=3.25; PyClone: P=0.04, HR=2.34). Across cancer types, the presence of more than two clones was associated with worse overall survival as compared to tumors in which either one or two clones were detected (Log-rank test EXPANDS: P=8.6E–4, HR=1.49; PyClone: P=0.09, HR=1.21; Fig. 5a and Supplementary Figs. 4.4a,d).
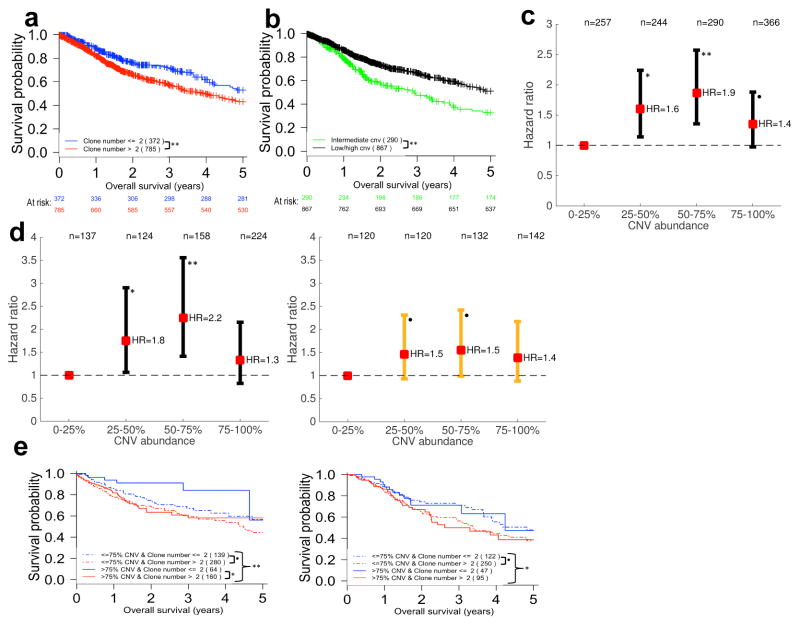
Figure 5. Clone number and CNV burden appear to be universal prognostic biomarkers.
(a) The presence of more than two clones detected by EXPANDS in a tumor sample predicts poor overall survival across all 12 tumor types (HR=1.497). (b) Survival curves are stratified by the fraction of the tumor-metagenome affected by CNVs (CNV abundance) across 12 tumor types. Intermediate levels of CNV abundance predict poor outcome (HR=0.597). (c) Hazard ratios as a function of CNV abundance. The hazard ratio for each of the upper three CNV abundance quartiles is calculated relative to the hazard of individuals in the lowest quartile (0–25% CNV abundance) and displayed along with 95% confidence interval. (d) Individuals treated with chemo- or radiotherapy (right panel) and untreated individuals (left panel) are stratified by CNV abundance. Individuals with low (<25%) or high CNV abundance (>75%) progress more slowly than individuals with intermediate CNV abundance levels (25–75%), especially within the group that did not receive adjuvant chemo- or radiotherapy. (e) Untreated individuals (left panel) with few clones in their tumors (blue lines) survive longer than untreated individuals with a large number of clones detected in their tumors (red lines), especially when these few clones share a large CNV burden (blue continuous line). This is not the case for treated individuals (right panel). All hazard ratios were calculated with log-rank tests (** P<0.005; * P<0.05; • P<0.1). For each stratum in panels (a,b,e) at least 50% of the 12 analyzed tumor types were represented at >5% frequency.
The association between clone number and survival was non-linear. An increased risk with increasing clone number was only observed for up to 4 clones. Additional diversification, beyond 4 clones, did not impart further risk. In fact, a tendency for reduced risk was observed among highly diverse tumors. This risk reduction did not reach significance in the univariate setting (Supplementary Fig. 4.5a), although it was significant in the multivariate analysis described below. The non-linear relationship between ITH and survival was also apparent when using alternative measures of ITH (e.g. nuclear diversity or accounting for differential tumor purity; Supplementary Fig. 4.5b,d,e).
A measurement of genomic instability is the fraction of the tumor-metagenome affected by CNVs (CNV abundance)38. Because genomic instability correlates with ITH (Supplementary Fig. 4.5f), we hypothesized that increased genomic instability necessary to produce a high level of ITH (i.e. >4 detected clones) may adversely affect tumor cell fitness following the generation of deleterious CNVs. We therefore analyzed the impact of somatic CNV abundance in the tumor-metagenomes and its relation to ITH. We find that low or high CNV abundance, i.e. CNVs affecting either a very low or a very high fraction of the tumor-metagenome, was predictive of improved survival (Log-rank test: P=5E–6; HR=0.15; Fig. 5b). This was not the case for low/high somatic SNV abundance (adjusted P>0.05; Supplementary Table 4.2a,b). We validated this result using CNVs measured by genome-wide SNP-arrays from: i) the same tumor samples and ii) an independent dataset consisting of 2,010 tumor samples, across seven distinct cancer types. Both validation analyses confirmed that intermediate CNV abundance is associated with poor survival (Supplementary Table 4.6 and Fig. 4.7a,b).
The highest risk was observed among individuals with 50–75% of their tumor-metagenome affected by CNVs in both the original and the independent datasets (Fig. 5c and Supplementary Figs. 4.7a,b). In fact, tumors with 50–75% CNV abundance did represent the highest risk group among individuals with bladder cancer, head and neck cancer, lung adenocarcinoma, stomach adenocarcinoma, cervical cancer and low-grade gliomas (Supplementary Fig. 4.8). These observations suggest the existence of an optimal degree of genomic instability that is independent of tumor-type.
Of the 12 tumor types, Glioblastoma was the only cancer for which >75% CNV abundance was associated with the worst prognosis (Supplementary Fig. 4.8g). Notably, with 85% of individuals diagnosed with Glioblastoma undergoing chemo- and/or radiotherapy, DNA damaging therapy is administered more frequently for Glioblastoma than for any of the other analyzed tumor types (Supplementary Table 1.4). Therefore we verified whether or not adjuvant chemo- or radiotherapy affected the non-linear association between CNV abundance and survival. In contrast to the 643 individuals who did not undergo chemo- or radiotherapy, the association between intermediate CNV abundance and poor survival was not significant amongst the 514 individuals treated with DNA-damaging agents (Fig. 5d). This finding was confirmed in the independent SNP-array dataset, where tumors with intermediate CNV abundance did represent the highest risk group among untreated, but not among individuals treated with chemo- or radiotherapy (Supplementary Fig. 4.7c,d).
A tumor with a critical level of >75% CNV abundance per tumor-metagenome may either be composed of many clones with low CNV abundance per clone, or few clones, each carrying high CNV abundance (Supplementary Fig. 4.9b,c). We used a clone number of 2 and 75% CNV abundance as thresholds to stratify untreated individuals into four groups with: i) CNV abundance below 75% and maximum 2 clones; ii) CNV abundance below 75% and minimum 3 clones; iii) CNV abundance above 75% and maximum 2 clones; and iv) CNV abundance above 75% and minimum 3 clones. Overall survival between these four groups was significantly different (Log- rank test EXPANDS: P=0.0015; HR=1.4). In general, as before, low clone number was associated with good outcome. In particular, the best outcome among the four groups was observed when a high CNV burden was shared among ≤2 clones (group iii; Fig. 5e and Supplementary Fig. 4.4e). When stratifying individuals who had undergone chemo- and/or radiotherapy in the same way, differences in clone number, rather than CNV burden, were associated with differences in overall survival between the four groups (Log-rank test EXPANDS: P=0.038; HR=1.4; Fig. 5e). Stratification based on PyClone derived clone numbers also supported these conclusions, albeit with borderline significance (P≤0.07, Supplementary Fig. 4.4b,c).
To account for factors that may confound the associations observed between clinical outcome and genetic ITH, the prognostic significance of clone number and low/high CNV abundance were evaluated with multivariate Cox models (Online Methods). All tumor types were included in a pan-cancer Cox model, except for gliomas, as the staging system is not applicable to gliomas. Across cancers, both, genomic instability and genetic ITH remained significantly associated to survival in the multivariate setting (Table 1). As concluded from univariate analysis (Supplementary Fig. 4.5a,b), the relation between clone number and survival was non-linear: an increased risk with increasing clone number was only observed for up to 4 clones. Additional diversity beyond 4 clones, was associated with an increase in overall CNV burden and a significant decrease in risk of mortality (Supplementary Fig. 4.5c,f). A similar scenario was observed within 8 out of the 10 analyzed cancer types, where the highest hazard was associated with an intermediate number of clones (between 3 and 5). ITH levels above/below an intermediate number of clones were associated with significantly reduced risk (multivariate Cox: HR=0.01–0.21; P≤0.05) relative to the intermediate group in head and neck cancer, melanoma and kidney cancer (Supplementary Fig. 4.5g and Table 4.10).
Table 1.
Pan-cancer multivariate Cox model of overall survival.
| P-value | Hazard ratio | Standard error (Coefficient) | Z-score | |
|---|---|---|---|---|
| 4 clones (Ref.) | NA | 1.000 | NA | NA |
| 1 or 2 clones vs. Ref. | 0.006 | 0.442 | 0.300 | −2.723 |
| 3 clones vs. Ref. | 0.076 | 0.618 | 0.271 | −1.776 |
| 5 clones vs. Ref. | 0.007 | 0.450 | 0.296 | −2.703 |
| 6 or 7 clones vs. Ref. | 0.014 | 0.503 | 0.279 | −2.463 |
| 8 or 9 clones vs. Ref. | 0.014 | 0.489 | 0.290 | −2.469 |
| 10 or more clones vs. Ref. | 0.003 | 0.389 | 0.314 | −3.011 |
| Age at diagnosis | 0.002 | 5.938* | 0.579 | 3.078 |
| Low/high CNV abundance | 1.81E–04 | 0.129 | 0.548 | −3.744 |
| Pathologic stage | 2.90E–08 | 3.339 | 0.217 | 5.548 |
| MKI67 mRNA expression | 2.21E–04 | 5.236 | 0.448 | 3.694 |
| % Lymphocytes | 0.141 | 0.310 | 0.796 | −1.473 |
| Model summary | Likelihood ratio test=92 on 11 degrees of freedom, P=6.88E–15, n= 610, number of events= 157 | |||
The hazard ratio for ‘age at diagnosis’ may not be reliable (Test of Proportional Hazards: P=0.007).
DISCUSSION
Quantification of ITH is a key measure of tumor evolution. We performed a cross-sectional analysis of ITH in 1,165 cancers from 12 cancer types, revealing the extent of ITH, and supporting its potential as a universal, though perhaps non-linear, prognostic biomarker. Evidence from two tumor mixture separation algorithms and from H&E imaging analysis, collectively indicate that ITH is a feature of the vast majority of cancers diagnosed.
To our knowledge, this is the first report of a cross-cancer correlation between genetic ITH and histopathologic ITH, suggesting that measures of tumor H&E sections can provide a proxy for genetic ITH. Currently, single tumor samples provide the only opportunity to study genetic ITH in a large pan-cancer cohort6. Measuring ITH from single tumor samples benefits from high depth and high genomic breadth of coverage. Exome-sequencing data represents the best tradeoff between these two sequencing- design parameters that is currently available at TCGA, across a broad range of tumors and cancer types. Using exome-sequencing to quantify ITH implies that clone distinction is confined to coding regions. Two clones that only differ in non-coding regions would be indistinguishable. Whole genomes sequenced at higher depth and multiple geographical tumor-samples will further improve our sensitivity to detect small clones and increase our resolution on clonal composition and its variability across cancer types.
Our results show that mutations in particular driver genes are associated with clones of a characteristic size, often independent of tumor type. This observation suggests that there appear to be constraints on the order in which neoplastic cells acquire driver events43 or that these events differ in the magnitude of the fitness-advantages they provide to neoplastic cells (Fig. 3a). Small clones may be fit, but evolve late in tumor progression. Alternatively, they may be less fit, but function as a “cornucopia of evolution” from which new clones frequently emerge. Both alternatives explain both, why these clones are so small and why their presence is associated with poor outcome (Fig. 3c). Importantly, the number of mutated CAN-genes did not predict outcome, suggesting that the relation between survival and presence of small clones was not confounded by CAN-gene mutation incidence.
The significant association between high clone number and poor survival detected in the combined analysis of low-grade glioma and glioblastoma may be interesting in the context of the highly variable clinical behavior of low-grade gliomas. A recent study found that histopathologic classification may overlook a subset of glioblastoma tumors, labeling them as low-grade gliomas7. Knowing the extent of ITH may help improve differential diagnosis between glioblastoma and low-grade glioma44.
As previously observed in ovarian, gastric, non-small cell lung cancers and ER− breast cancers45,46, individuals with intermediate CNV burdens detected in their primary untreated tumor had the worst overall survival. We find this association is present across several tumor types, but its strength varies with the type of therapy the individuals received subsequently. Our results suggest a potential advantage when tumors with intermediate levels of CNVs are treated with adjuvant chemo- and radiotherapy (Fig. 5d). Chemo- and radiotherapy may be particularly effective against tumors with intermediate CNV burdens, by pushing them past the limit of ‘tolerable’ genomic instability. Our results from two distinct high-throughput technologies measuring CNV abundance in two independent pan-cancer cohorts suggest that this limit is exceeded when >75% of a tumor’s metagenome is affected by CNVs, independent of cancer type. Given that >37% of cancers have been shown to undergo whole genome doubling events12, in will be of interest to see whether these tumors have the same phenotype as tumors with >75% CNV burden.
In light of recent evidence supporting a stronger role of CNVs than SNVs in developing and maintaining ITH11, this upper limit of tolerable genomic instability may be responsible for the non-linear association we observed between genetic ITH and survival. Previously, low ITH has been found to predict favorable prognosis in Barrett’s esophagus49,50, head and neck cancer11, as well as leukemia10,51. Consistent with these studies we found that the presence of only one or two clones is in general prognostic of favorable outcome, especially when these few clones share a high CNV burden. However, diversification beyond four clones was associated with decreased risk.
The decrease in risk may be because large numbers of clones can attract more immune-cells. Alternatively, the decrease in risk may be in part due to the technical difficulty of distinguishing between ITH and genomic instability, in particular as both measures increase. Finally, it may be a consequence of a tradeoff that exists between the chance of acquiring an advantageous alteration initiating a new clonal expansion and the risk of generating inviable daughter cells. The observed synchronous increase of ITH and CNV burden suggests that efforts aimed at modulating this tradeoff may represent a new therapeutic avenue to slow tumor evolution and improve clinical outcomes.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants P01 CA91955, R01 CA149566, R01 CA170595, R01 CA185138 and R01 CA140657 as well as CDMRP Breast Cancer Research Program Breakthrough Award BC132057 to CCM; NIH grants P01 HG000205, U01CA151920, U01CA17629901 and R01 HG006137 to HPJ. Additional support to HPJ came from the Doris Duke Clinical Foundation Clinical Scientist Development Award, Research Scholar Grant, RSG-13-297-01-TBG from the American Cancer Society and a Howard Hughes Medical Institute Early Career Grant. NA was supported by the awards from the Don and Ruth Seiler Fund and the NCI Cancer Target Discovery and Development (CTDD) Consortium (U01CA17629901). LCX was supported by R01 HG006137. TAG was supported by the higher education founding council for England (HEFCE).
We are grateful to Dr. Hans W. Mewes for advise on presentation of our results and for insightful discussions about their implications. We thank Dr. Shane T. Jensen for advise on statistical data analysis. We also thank Drs. Chris W. Turck and Martin Oft for reviewing the manuscript. The results presented here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. We thank Hoffmann H. from University of Bonn, Germany for the availability of MATLAB-function “violin”, used to generate the violin plots of clone number and -size distribution.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Code availability. CellProfiler pipeline employed to detect and measure nuclei from H&E images is available at http://dna-discovery.stanford.edu/projects/completed-projects/pan-cancer-ith.html
AUTHOR CONTRIBUTIONS
N.A. developed analytic methods, analyzed the data and wrote the manuscript. T.A.G. developed analytic methods, gave technical support and conceptual advice and wrote the manuscript. M.J. analyzed the histopathology images and advised on data visualization and interpretation. L.C.X. advised on the choice of statistical methods and design of statistical analysis. C.A.A. gave technical support and conceptual advice. C.C.M. developed analytic methods, wrote the manuscript and supervised the project. H.P.J. wrote the manuscript and supervised the project. C.P. supervised the project. All authors edited the manuscript.
References
- 1.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 5.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGranahan N, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7:283ra54–283ra54. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almendro V, et al. Inference of Tumor Evolution during Chemotherapy by Computational Modeling and In Situ Analysis of Genetic and Phenotypic Cellular Diversity. Cell Rep. 2014;6:514–527. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerlinger M, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landau DA, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mroz EA, Tward AM, Hammon RJ, Ren Y, Rocco JW. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oesper L, Satas G, Raphael BJ. Quantifying tumor heterogeneity in whole-genome and whole-exome sequencing data. Bioinforma Oxf Engl. 2014;30:3532–3540. doi: 10.1093/bioinformatics/btu651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Li JZ. A general framework for analyzing tumor subclonality using SNP array and DNA sequencing data. Genome Biol. 2014;15:473. doi: 10.1186/s13059-014-0473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth A, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andor N, Harness JV, Müller S, Mewes HW, Petritsch C. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinforma Oxf Engl. 2014;30:50–60. doi: 10.1093/bioinformatics/btt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha G, et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res. 2014;24:1881–1893. doi: 10.1101/gr.180281.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sathirapongsasuti JF, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinforma Oxf Engl. 2011;27:2648–2654. doi: 10.1093/bioinformatics/btr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barber LJ, Davies MN, Gerlinger M. Dissecting cancer evolution at the macro- heterogeneity and micro-heterogeneity scale. Curr Opin Genet Dev. 2015;30:1–6. doi: 10.1016/j.gde.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yadav VK, De S. An assessment of computational methods for estimating purity and clonality using genomic data derived from heterogeneous tumor tissue samples. Brief Bioinform. 2014 doi: 10.1093/bib/bbu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4 doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tajiri R, et al. Intratumoral heterogeneous amplification of ERBB2 and subclonal genetic diversity in gastric cancers revealed by multiple ligation-dependent probe amplification and fluorescence in situ hybridization. Hum Pathol. 2014;45:725–734. doi: 10.1016/j.humpath.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Sakurada A, Lara-Guerra H, Liu N, Shepherd FA, Tsao MS. Tissue heterogeneity of EGFR mutation in lung adenocarcinoma. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2008;3:527–529. doi: 10.1097/JTO.0b013e318168be93. [DOI] [PubMed] [Google Scholar]
- 30.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitale M. Intratumor BRAFV600E heterogeneity and kinase inhibitors in the treatment of thyroid cancer: a call for participation. Thyroid Off J Am Thyroid Assoc. 2013;23:517–519. doi: 10.1089/thy.2012.0614. [DOI] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, Ozolek JA, Rohde GK. Detection and Classification of Thyroid Follicular Lesions Based on Nuclear Structure from Histopathology Images. Cytom Part J Int Soc Anal Cytol. 2010;77:485–494. doi: 10.1002/cyto.a.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartwell KA, et al. Niche-based screening identifies small-molecule inhibitors of leukemia stem cells. Nat Chem Biol. 2013;9:840–848. doi: 10.1038/nchembio.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto S, et al. Clinical relevance of Ki67 gene expression analysis using formalin- fixed paraffin-embedded breast cancer specimens. Breast Cancer Tokyo Jpn. 2013;20:262–270. doi: 10.1007/s12282-012-0332-7. [DOI] [PubMed] [Google Scholar]
- 38.Cazier J-B, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun. 2014;5 doi: 10.1038/ncomms4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostrow SL, Barshir R, DeGregori J, Yeger-Lotem E, Hershberg R. Cancer evolution is associated with pervasive positive selection on globally expressed genes. PLoS Genet. 2014;10:e1004239. doi: 10.1371/journal.pgen.1004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgess DJ. Evolution: Cancer drivers everywhere? Nat Rev Genet. 2014 doi: 10.1038/nrg3718. advance online publication. [DOI] [PubMed] [Google Scholar]
- 41.Heim D, et al. Cancer beyond organ and tissue specificity: next-generation-sequencing gene mutation data reveal complex genetic similarities across major cancers. Int J Cancer J Int Cancer. 2014 doi: 10.1002/ijc.28882. [DOI] [PubMed] [Google Scholar]
- 42.Yuan Y, et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat Biotechnol. 2014;32:644–652. doi: 10.1038/nbt.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanton C. Cancer Evolution Constrained by Mutation Order. N Engl J Med. 2015;372:661– 663. doi: 10.1056/NEJMe1414288. [DOI] [PubMed] [Google Scholar]
- 44.Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015 doi: 10.1056/NEJMoa1402121. 0, null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birkbak NJ, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roylance R, et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2011;20:2183–2194. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biebricher CK, Eigen M. The error threshold. Virus Res. 2005;107:117–127. doi: 10.1016/j.virusres.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Szathmáry E, Smith JM. The major evolutionary transitions. Nature. 1995;374:227–232. doi: 10.1038/374227a0. [DOI] [PubMed] [Google Scholar]
- 49.Merlo LMF, et al. A comprehensive survey of clonal diversity measures in Barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res Phila Pa. 2010;3:1388–1397. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maley CC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 51.Bochtler T, et al. Clonal heterogeneity as detected by metaphase karyotyping is an indicator of poor prognosis in acute myeloid leukemia. J Clin Oncol Off J Am Soc Clin Oncol. 2013;31:3898–3905. doi: 10.1200/JCO.2013.50.7921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.