Abstract
The goal of this study was to compare significant SNP selection approaches in the context of complex traits based on SNP estimates obtained by models: a model fitting a single SNP (M1), a model fitting a single SNP and a random polygenic effect (M2), the nonparametric CAR score (M3), a SNP-BLUP model with random effects of all SNPs fitted simultaneously (M4). There were 46,267 SNPs tested in a population of 2601 Holstein Friesian bulls, four traits (milk and fat yields, somatic cell score, non-return rate for heifers) were considered. The numbers of SNPs selected as significant differed among models. M1 selected a very large number of SNPs, except for a NRH in which no SNPs were significant. M2 and M3 both selected similar and low number of SNPs for each trait. M4 selected more SNPs than M2 and M3. Considering linkage disequilibrium between SNPs, for MY M2 and M3 selected SNPs more highly correlated with each other than in the case of M4, while for FY M3 selection contained more correlated SNPs than M2 and M4. In conclusion, if the research interest is to identify SNPs not only with strong, but also with moderate effects on a complex trait a multiple–SNP model is recommended. Such models are capable of accounting for at least a part of linkage disequilibrium between SNPs through the design matrix of SNP effects. Functional annotation of SNPs significant in M4 reveals good correspondence between selected polymorphisms and functional information as well as with QTL mapping results.
Keywords: Complex traits, GWAS, Mixed model, Significance testing
Introduction
For many years genome-wide association studies (GWAS) have been a useful tool for detecting genetic variants associated with traits in human genetics (Visscher et al. 2012). With the advancement of genotyping technology high-density single nucleotide polymorphism (SNP) platforms have also been developed for other species including livestock. A whole bunch of statistical models have been applied to perform GWAS based on SNP chips. These vary from models where each SNP is considered individually to models with effects of all the available SNPs fitted simultaneously. However, the major drawback of interpreting GWAS results is the optimal selection of polymorphisms which are to be claimed as significantly associated with the analyzed trait and poor repeatability of results across methods and data sets. This problem is especially important when analyzing traits with a complex mode of inheritance (summarized by Visscher et al. 2012).
The variety of GWAS models applied to complex traits covers linear regression, penalized regression approaches with various shrinkage priors for SNP effects, like the least absolute shrinkage and selection operator regression (LASSO) introduced by Tibshirani (1996) (e.g., applied by Wu et al. 2009), the elastic net introduced by Zou and Hastie (2005) (e.g., applied by Do et al. 2011), ridge regression introduced by Hoerl and Kennard (1970) (e.g., applied and extended by Zhan and Xu 2012), normal exponential gamma distribution proposed by Hoggart et al. (2008), or by incorporating the functional information into SNP selection through biological pathways (Braun and Buetow 2011).
Our study was focused on the comparison of significant SNP selection approaches in the context of complex traits. Significance of SNPs selected based on estimates obtained from a mixed model routinely used in genomic evaluation of dairy cattle in Poland was compared to three single SNP models. In particular, the genomic selection model, which considers all polymorphisms simultaneously by applying shrinkage on SNP estimates through fitting a normal distribution with a predefined variance, is compared with (i) a linear model with a fixed effect of a single SNP which accounts neither for SNP intercorrelation, nor for family relationships (ii) a linear mixed model with a fixed, single SNP effect and a random additive polygenic effect for capturing the family relationships, and (iii) the CAR score regression proposed by Zuber and Strimmer (2011), which represents a fully nonparametric approach. Generally, we were interested in quantifying the differences between models in defining significant SNPs. Our particular focus though was on assessing the validity of the genomic selection model for GWAS purposes, since such a model is routinely evaluated on large and very informative data sets in many countries and, besides selection purposes, could be a potential source of information on single gene effects on traits undergoing selection.
Materials and methods
Data set
The material of our study consists of 2601 Polish Holstein-Friesian bulls genotyped with the Illumina BovineSNP50 BeadChip, which consists of 54,001 SNPs (version 1) and 54,609 SNPs (version 2). The applied SNP selection criteria comprised polymorphism, expressed by the minor allele frequency (MAF), with the minimum MAF of 0.01, and technical quality of a SNP expressed by the minimum call rate of 90 % within the analyzed sample of bulls. After quality control 46,267 SNPs were selected for further analysis.
Four traits undergoing a complex mode of inheritance were considered in the study: somatic cell score (SCS) representing a trait with “pure” polygenic mode of inheritance, milk (MY) and fat (FY) yields representing traits with a polygenic mode of inheritance enhanced by single genes with large effects, and non-return rate for heifers (NRH) as a trait with a very strong environmental component expressed by heritability of 0.02. Deregressed proofs of bulls were used as pseudophenotypes. Deregression, which was performed in order to remove ancestral information from the conventional breeding values of bulls, was based on a method proposed by Jairath et al. (1998). The corresponding conventional breeding values were estimated based on a random regression test day model for MY, FY, and SCS (Strabel and Jamrozik 2006) and based on a lactation model for NRH (Jagusiak and Żarnecki 2006) using phenotypic information corresponding to the routing national evaluation from April 2012. For each trait 2588 (SCS), 2601 (MY and FY), and 2524 (NRH) records were available.
Models for SNP effect estimation
The SNP effects were estimated by four different models. Single SNP models comprising (M1) y = μ + Xβ + ε and (M2) y = μ + Xβ + Z1α + ε were solved using the ASReml3 software (Gilmour et al. 2009). In the above models y represents a vector of deregressed breeding values for MY, FY, SCS or NRH; β denotes a fixed SNP effect with a design matrix X ∈ {−1, 0, 1} for a homozygous, a heterozygous, and an alternative homozygous SNP genotype respectively; α denotes a random polygenic effect of a bull with an incidence matrix Z1, where α is distributed as , with A being a relationship matrix for bulls and representing the estimate of total additive genetic variance of a given trait calculated elsewhere for the whole active population of Polish Holstein-Friesian dairy cattle. The residual effects vector ε is distributed as N(0, Dσ2ε) where D is a diagonal matrix weighted by the effective daughter contribution for each bull and σ2ε denotes a residual variance; μ denotes an overall mean. The CAR score regression (M3) proposed by Zuber and Strimmer (2011) was selected as the third model for its simplicity and computational efficiency of variable ranking in linear regression based on the Mahalanobis-decorrelation of the explanatory variables representing a nonparametric approach. According to Zuber and Strimmer (2011) this approach is very effective computationally and yields prediction errors as well as true and false positive rates that compare favorably with other regression techniques such as elastic net and boosting. The CAR scores ωi, which were considered as the SNP selection criterion, are defined as: , where Pβy is the marginal correlation vector between deregressed breeding values and SNPs, P denotes shrinkage estimator given by: λId + (1 − λ)Rempirical, where is a shrinkage intensity and Rempirical is the empirical non-regularized correlation matrix among SNP genotypes. The CAR criterion was computed using an R package CARE. The genomic selection model (M4) y = μ + Z2g + ε, equivalent to the so called SNP-BLUP model, is routinely used for the prediction of direct genomic breeding values for the Polish Holstein-Friesian population (Szyda et al. 2011). Here Z2 is a design matrix for SNP genotypes, which is parameterized as −1, 0, or 1 for a homozygous, a heterozygous, and an alternative homozygous genotype respectively and g is a vector of random additive SNP effects distributed defined as: , with I being an identity matrix.
The SNP effects in M1 were estimated by weighted least squares with the effective number of daughters corresponding to teach observation y used as a weighting variable. To estimate β in M2 the objective function log fy(y|α, β, G) + log fα(α, G), where , was used. Differentiating it with respect to β and α leads to the mixed model equations (Henderson 1984). M3 is a model free procedure in which CAR scores are functions of empirical SNP-pseudophenotype and SNP-SNP correlations. The estimation of parameters of M4 was based on solving the corresponding mixed model equations using the iteration on data technique applying the Gauss-Seidel algorithm with residuals update (Legarra and Misztal 2008).
Significant SNP selection
In case of M1, M2, and M4 the Wald test was used to obtain the nominal type I error corresponding to a standard normal distribution. For single SNP models M1 and M2 the resulting P values were subjected to multiple testing correction for the number of SNP tested (N = 46,267) via Bonferroni’s approach, while for a multiple SNP model M4 a nominal P value was used as a selection criterion. In M3 the null distribution of the function of CAR scores was used for obtaining type I error rates, which follows the Student t distribution with (κ − 1) degrees of freedom estimated by the R package fdrtool (Strimmer 2008). For all models SNPs were selected as significant when P values associated with their estimates did not exceed the 0.001 threshold.
Genomic annotation of SNPs was performed using SNPchiMp (Nicolazzi et al. 2014) for the identification of SNP positions corresponding to the UMD3.1 bovine genome assembly and Variant Effect Predictor (McLaren et al. 2010) for the identification of genomic positions of SNPs.
Results
The number of significant SNPs selected
The numbers of SNPs selected as significant by different models are presented in Table 1. For MY, FY, and SCS the simplest model M1 always selected a very large number of SNPs ranging between 2242 (SCS) and 3398 (MY), widely exceeding the number of SNPs selected by M2-M4. Although models 2 and 3 markedly differ in modeling and hypothesis testing assumptions, they both select a very similar and low number of SNPs for each trait. The genomic selection model M4 is intermediate in terms of the number of SNPs selected.
Table 1.
The numbers of SNPs selected as significant
| Trait | M1 | M2 | M3 | M4 |
|---|---|---|---|---|
| Fat yield | 2435 | 72 | 48 | 182 |
| Milk yield | 3398 | 66 | 78 | 153 |
| Somatic cell score | 2242 | 4 | 0 | 163 |
| Non-return rate for heifers | 0 | 0 | 0 | 125 |
Except for M4, traits with different putative inheritance modes resulted in different numbers of significant SNPs. For MY and FY the largest numbers of SNPs were selected at BTA14 — a chromosome harboring DGAT1 gene of high effect on milk production traits. For SCS, a trait with a pure polygenic mode of inheritance, a lower number of SNPs was indicated as significant, with only four SNPs selected by M2. For NRH, a trait with a very weak genetic component (as compared to the environmental based variation) no SNPs were identified by M1-M3. A very different pattern was observed for M4. Since all polymorphisms’ effects are estimated simultaneously with the underlying normal distribution shrinkage, the SNP selection procedure based on the 0.001 threshold chooses 0.1 % of the most significant SNPs regardless of a trait. As a consequence a very similar number of polymorphisms varying between 125 (NRH) and 182 (FY) for each trait was selected. Since, based on the very large number of SNPs selected, M1 does not seem to be a valid model for GWAS on complex traits, in further result description and discussion we confine ourselves to M2-M4.
Influence of SNP informativeness on SNP selection
No marked differences between models were observed regarding the information content, expressed by MAF, between SNPs selected as significant by M2–M4. The average MAF of polymorphisms selected for MY by M2 amounted to 0.39, by M3 to 0.30 and M4 to 0.36 and was even more similar for FY with 0.37 (M2), 0.38 (M3) and 0.49 (M4). The average MAF in group of SNPs common between all considering models is 0.40 for MY and 0.38 for FY. On the other hand, considering MY, on average M2 and M3 selected SNPs which were more highly correlated with each other than was the case for M4 since the average LD expressed by pairwise correlation between significant SNPs was equal to 0.18 and 0.17 for M2 and M3, but only 0.08 for the genomic selection model. For FY, the average LD of 0.11 among SNPs selected by M4 was also lower from the corresponding values form M2 and M3 which were equal to 0.17 and 0.21 respectively. The highest average pairwise correlation was observed between significant SNPs which were common for models M2 and M3.
Comparison of significant SNP sets between models
Figure 1 presents the percentage of SNPs significant between M2 and M4. For production traits a relatively large proportion of significant SNPs was common among all three models making 24 SNPs common for MY and 41 SNPs common for FY, while no common for all models polymorphisms were identified for SCS and NRH. Focusing on the genomic selection model M4 as the reference it is evident from Fig. 2 that more mutual SNPs existed between M4 and M2 than between M4 and M3. In particular, in addition to the polymorphisms common to all three models, the genomic selection model had 12 and 11 additional SNPs in common with M2 for MY and FY respectively, but only one SNP for MY mutual with M3. For MY there are four polymorphisms common for models M2 and M3.
Fig. 1.
The percentage of SNPs significant between models
Fig. 2.
Genomic location of significant SNPs common between models
Genomic SNP annotation
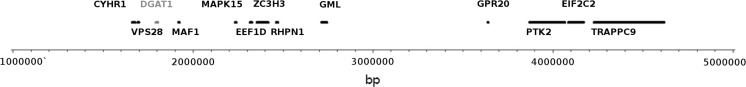
Genomic annotation was carried out for SNPs which were selected as significant by all three modles, i.e., M2, M3, and M3 (Table 2). Since M3 indicated no significant polymorphisms for SCS and NRH for those traits no annotation was considered. For FY 22 genes were marked by significant SNPs common between models, while for MY a lower number of eight genes was identified with six of them also significant for FY. All genes are located on BTA14 with varying distances from DGAT1 — a well known candidate gene for milk production traits (Grisart et al. 2002), ranging between 104,646 and 3,768,523 bp (calculated as a distance between corresponding significant SNP positions) (Fig. 3). Most of the SNPs are located within genomic range of the gene, mainly within intronic regions, but rs110323635 and rs41256919 lie in exons of MAPK15 and MAF1 genes respectively.
Table 2.
Annotation of SNPs significant in M2, M3, and M4. Genes significant for both MY and FY are marked in bold. For significant genes all SNPs located within intron/exon regions are listed
| Gene acronym | Gene name | Ensemble ID | BTA | SNP | SNP annotation | SNP position |
|---|---|---|---|---|---|---|
| Fat yield | ||||||
| FOXH1 | Forkhead box H1 | ENSBTAG00000004761 | 14 | rs109146371 | 3390 bp upstream | 1,651,311 |
| CYHR1 | Cysteine/histidine-rich 1 | ENSBTAG00000035254 | 14 | rs109968515 | intron | 1,675,278 |
| VPS28 | Homologous to vacuolar protein sorting 28 | ENSBTAG00000026320 | 14 | rs17870736 | intron | 1,696,470 |
| DGAT1 | Diacylglycerol O-acyltransferase 1 | ENSBTAG00000026356 | 14 | rs109421300 | intron | 1,801,116 |
| MAF1 | Homologous to RNA polymerase III-inhibiting protein | ENSBTAG00000012242 | 14 | rs41256919 | exon – missense variant | 1,923,292 |
| SPATC1 | Spermatogenesis and centriole associated 1 | ENSBTAG00000026350 | 14 | rs41629750 | 4472 bp upstream | 2,002,873 |
| PLEC | Plectin | ENSBTAG00000011922 | 14 | rs109350371 | 460 bo upstream | 2,054,457 |
| MAPK15 | Mitogen-activated protein kinase 15 | ENSBTAG00000019864 | 14 | rs110323635 | exon – missense variant | 2,239,085 |
| EEF1D | Elongation factor 1-delta | ENSBTAG00000014643 | 14 | rs109661298 | intron | 2,319,504 |
| ZC3H3 | Zinc finger CCCH-type containing 3 | ENSBTAG00000021472 | 14 | rs109617015 | intron | 2,386,688 |
| RHPN1 | Rhophilin, Rho GTPase binding protein 1 | ENSBTAG00000002104 | 14 | rs109529219 | intron | 2,468,020 |
| Novel gene | ENSBTAG00000003606 | 14 | rs110199901 | intron | 2,524,432 | |
| LY6K | Lymphocyte antigen 6 complex, locus K | ENSBTAG00000000158 | 14 | rs110174651 | 297 bp downstream | 2,754,909 |
| LY6D | Lymphocyte antigen 6 complex, locus D | ENSBTAG00000034498 | 14 | rs110237430 | 978 bp downstream | 2,803,998 |
| LYPD2 | LY6/PLAUR domain containing 2 | ENSBTAG00000016210 | 14 | rs109476486 | 2653 bp upstream | 2,826,632 |
| BAI1 | Brain-specific angiogenesis inhibitor 1 | ENSBTAG00000006385 | 14 | rs109545018 | intron | 3,006,509 |
| GPR20 | G protein-coupled receptor 20 | ENSBTAG00000015985 | 14 | rs110411273 | 71 bp downstream | 3,640,788 |
| TSNARE1 | t-SNARE domain containing 1 | ENSBTAG00000009974 | 14 | rs109875744 rs110888717 rs109295487 |
intron intron intron |
3,078,843 3,117,493 3,137,184 |
| PTK2 | PTK2 protein tyrosine kinase 2 | ENSBTAG00000009578 | 14 | rs109670279 rs41624797 rs109131748 rs110185345 |
intron intron intron intron |
3,885,798 3,956,956 4,017,201 4,043,743 |
| EIF2C2 | Argonaute RISC catalytic component 2 | ENSBTAG00000001579 | 14 | rs109948273 rs41576704 rs108980964 |
intron intron intron |
4,103,850 4,127,413 4,149,375 |
| TRAPPC9 | Trafficking protein particle complex 9 | ENSBTAG00000013955 | 14 | rs109807697 rs109248069 rs111018678 rs110017379 rs55617160 rs110805364 |
intron intron intron intron intron intron |
4,240,287 4,267,053 4,336,714 4,364,952 4,468,478 4,583,344 |
| COL22A1 | Collagen, type XXII, alpha 1 | ENSBTAG00000015374 | 14 | rs110444021 rs110351374 |
intron intron |
5,225,467 5,428,037 |
| Milk yield | ||||||
| C8orf33 | Chromosome 14 open reading frame 33 | ENSBTAG00000000879 | 14 | rs109752439 | 87 bp downstream | 1,489,496 |
| FAM135B | Family with sequence similarity 135, member B | ENSBTAG00000018218 | 14 | rs110622450 rs109118650 rs110501942 rs109402117 |
intron intron intron intron |
5,428,037 5,462,752 5,494,654 5,569,639 |
Fig. 3.
Position of genes on BTA14 marked by significant SNPs common for models 2–4
Although some of the genes may represent spurious associations arising through high LD to DGAT1, those which are located in more distant regions of BTA14 are potential candidate genes. Special attention is to be focused on EEF1D which according to a recent study of Xie et al. (2014) in dairy cattle exhibits significantly higher expression in mammary gland as in other tissues.
Discussion
We observed that for complex traits approaches which model the genetic component by a single SNP — here represented by M1, which are very common and successful in GWAS for disease traits, have methodological problems. Since such models do not account for the polygenic background differentiating between SNPs in high LD, especially in the neighborhood of genes with strong effects is problematic and the multiple testing correction is not sufficient to remove the SNP effect upward bias due to correlated SNPs. An intensive post-processing of SNP estimates and/or corresponding P values regarding LD is then required.
A possible alternative, which is non-intensive computationally, is to use a single SNP model with the genetic background described by an additive polygenic effect (like M2) or a multiple–SNP without any assumptions regarding the inheritance mode (like M3). In our study M2 and M3 for MY and for FY, above and beyond the M2-M3, also the genomic model (M4) selected a similar number of SNPs as the number 55 resulting from the estimator proposed by Hayes and Goddard (2001) of the total number of genes influencing the variance of production traits (M): , where p = 1/2Ne. Note, that here we used the estimated number of heterozygous QTL segregating for production traits as N = 10.73 (Hayes and Goddard 2001) and the effective population size for dairy cattle as Ne= 103 (Qanbari et al. 2010).
However, if the research interest is to identify SNPs not only with strong, but also with moderate effects on a complex trait a multiple–SNP model – here represented by M4, is recommended. Such models are capable of accounting for at least a part of LD between SNPs through the design matrix of SNP effects. A fine tuning of SNP selection procedure is needed based on the expected number of SNPs influencing the variability of a given trait, e.g., based on heritability estimates.
Recently, Wang et al. (2012) applied a genomic selection model (single step GBLUP) for GWAS on complex traits based on simulated data and found a good accuracy of prediction of QTL effects through SNPs. As indicated in that study it is important to realize that the selected SNPs do not necessarily represent underlying genes and that a chromosomal region in high LD (which is usually, but not always, equivalent to a region in physical neighborhood of the SNP) should be considered in search for causal mutations. Comparing the average LD of SNPs selected by single gene methods with the set selected by the genomic selection model it is evident that the latter is able to better deal with correlations between particular SNPs which occur through LD. Moreover, as pointed out by Dekkers (2012) a genomic selection model directly accounts for the population structure, not only on an averaged genome-wide level (through the additive polygenic covariance, as incorporated into M2), but also at a particular SNP sites (through the design matrix of SNP genotypes, as incorporated into M4). Another very important, practical advantage of using the genomic selection model for GWAS is that in most countries the model and underlying SNP effects are anyway evaluated on large, informative data sets.
Acknowledgments
The study was financed by the Polish State Committee for Scientific Research, grant no. N N311 609639. The data set was generated within the framework of the MASinBULL project (currently Genomika Polska).
References
- Braun R, Buetow K. Pathways of distinction analysis: a new technique for multi–SNP analysis of GWAS data. PLoS Genet. 2011;7:6. doi: 10.1371/journal.pgen.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers JCM. Application of genomics tools to animal breeding. Curr Genomics. 2012;13:207–212. doi: 10.2174/138920212800543057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, Tung JY, Dorfman E, Kiefer AK, Drabant EM, Francke U, Mountain JL, Goldman SM, Tanner CM, Langston JW, Wojcicki A, Eriksson N. Web-based genome-wide association study identifies two Novel Loci and a substantial genetic component for Parkinson’s Disease. PLoS Genet. 2011;7:6. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2009) ASReml user guide release 3.0. VSN International Ltd, Hemel Hempstead
- Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, Cambisano N, Mni M, Reid S, Simon P, Spelman R, Georges M, Snell R. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002;12:222–231. doi: 10.1101/gr.224202. [DOI] [PubMed] [Google Scholar]
- Hayes B, Goddard ME. The distribution of the effects of genes affecting quantitative traits in livestock. Genet Sel Evol. 2001;33:209–229. doi: 10.1186/1297-9686-33-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CR (1984) Applications of linear models in animal breeding. University of Guelph
- Hoerl AE, Kennard R (1970) Ridge regression: biased estimation for nonorthogonal problems. Technometrics 12:55–67
- Hoggart CJ, Whittaker JC, De Iorio M, Balding DJ (2008) Simultaneous analysis of All SNPs in genome-wide and re-sequencing association studies. PLoS Genet 4:7. doi:10.1371/journal.pgen.1000130 [DOI] [PMC free article] [PubMed]
- Jagusiak W, Żarnecki A. Genetic evaluation for fertility traits in Polish Holsteins. Interbull Bulletin. 2006;35:37–41. [Google Scholar]
- Jairath L, Dekkers JCM, Schaeffer LR, Liu Z, Burnside EB, Kolstad B (1998) Genetic evaluation for herd life in Canada. J Dairy Sci 81:550–562 [DOI] [PubMed]
- Legarra A, Misztal I. Technical note: computing strategies in genome-wide selection. J Dairy Sci. 2008;91:360–366. doi: 10.3168/jds.2007-0403. [DOI] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolazzi EL, Picciolini M, Strozzi F, Schnabel RD, Lawley C, Pirani A, Brew F, Stella A. SNPchiMp: a database to disentangle the SNPchip jungle in bovine livestock. BMC Genomics. 2014;15:123. doi: 10.1186/1471-2164-15-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanbari S, Pimentel ECG, Tetens J, Thaller G, Lichtner P, Sharifi AR, Simianer H. The pattern of linkage disequilibrium in German Holstein cattle. Anim Genet. 2010;41:346–356. doi: 10.1111/j.1365-2052.2009.02011.x. [DOI] [PubMed] [Google Scholar]
- Strabel T, Jamrozik J (2006) Genetic analysis of milk production traits of Polish black and white cattle using large-scale random regression test-day models. J Dairy Sci 89:3152–3163 [DOI] [PubMed]
- Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- Szyda J, Żarnecki A, Suchocki T, Kamiński S. Fitting and validating the genomic evaluation model to Polish Holstein-Friesian cattle. J Appl Genet. 2011;52:363–366. doi: 10.1007/s13353-011-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selction via the lasso. J R Statist Soc B. 1996;58:267–288. [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Misztal I, Aguilar I, Legarra A, Muir WM. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet Res. 2012;94:73–83. doi: 10.1017/S0016672312000274. [DOI] [PubMed] [Google Scholar]
- Wu TT, Chen YF, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25:714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Yang S, Cui X, Jiang L, Zhang S, Zhang Q, Zhang Y, Sun D. Identification and expression pattern of two novel alternative splicing variants of EEF1D gene of dairy cattle. Gene. 2014;534:189–196. doi: 10.1016/j.gene.2013.10.061. [DOI] [PubMed] [Google Scholar]
- Zhan H, Xu S. Adaptive ridge regression for rare variant detection. PLoS ONE. 2012;7(8):e44173. doi: 10.1371/journal.pone.0044173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Statist Soc B. 2005;67:301–320. doi: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- Zuber V, Strimmer K. High-dimensional regression and variable selection using CAR scores. Statist Appl Genet Mol Biol. 2011;10:34. [Google Scholar]