Abstract
Nearly all bacterial species, including pathogens, have the ability to form biofilms. Biofilms are defined as structured ecosystems in which microbes are attached to surfaces and embedded in a matrix composed of polysaccharides, eDNA, and proteins, and their development is a multistep process. Bacterial biofilms constitute a large medical problem due to their extremely high resistance to various types of therapeutics, including conventional antibiotics. Several environmental and genetic signals control every step of biofilm development and dispersal. From among the latter, quorum sensing, cyclic diguanosine-5’-monophosphate, and small RNAs are considered as the main regulators. The present review describes the control role of these three regulators in the life cycles of biofilms built by Pseudomonas aeruginosa, Staphylococcus aureus, Salmonella enterica serovar Typhimurium, and Vibrio cholerae. The interconnections between their activities are shown. Compounds and strategies which target the activity of these regulators, mainly quorum sensing inhibitors, and their potential role in therapy are also assessed.
Keywords: Biofilm, Cyclic diguanosine-5’-monophosphate, Quorum sensing, Small RNAs
Introduction
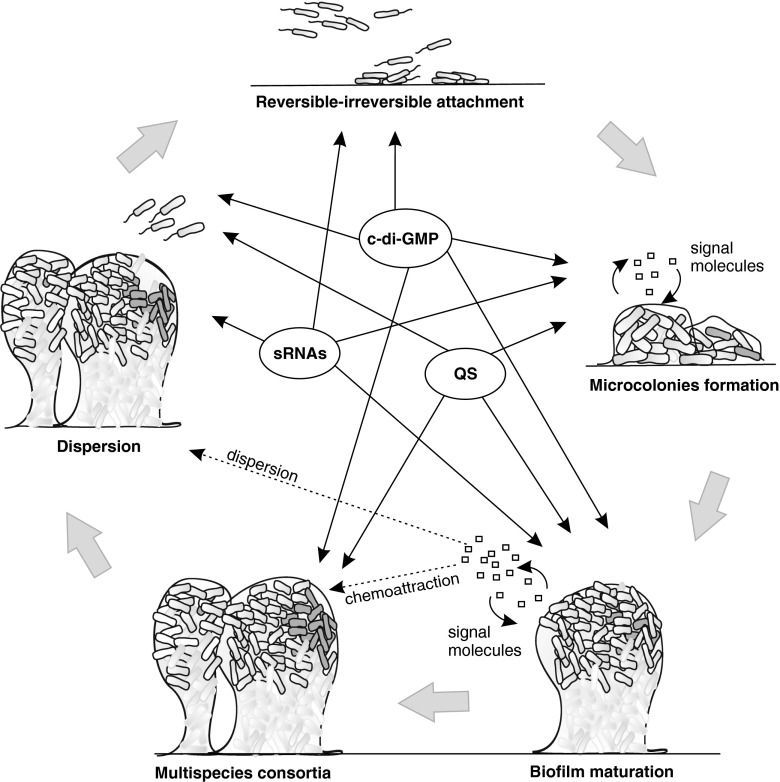
The majority of bacteria, including clinically relevant microorganisms, are able to grow in biofilms adhering to abiotic and biotic surfaces (for a review, see Donlan and Costerton 2002). A biofilm is defined as a structured microbial community whose development requires a significant change in bacterial physiology and results in increased tolerance to exogenous stress, including treatment with antibiotics and other biocides (Hall-Stoodley and Stoodley 2009). Bacterial biofilms can form a monolayer or, most frequently, multilayers in which bacteria are attached both to the surface and to neighboring bacteria by an extracellular matrix consisting of polysaccharides, proteins, and DNA (Karatan and Watnick 2009). The biofilm formation process always has several stages that include: (i) attachment to the carrier surface, (ii) reversible, followed by irreversible, binding to the surface with the participation of adhesins, (iii) development of microcolonies, and (iv) maturation of biofilm architecture (Donlan 2001). Under unfavorable conditions, the synthesis of matrix compounds decreases and the matrix is enzymatically cleaved, leading to biofilm dispersion (Gjermansen et al. 2005). A scheme of the biofilm life cycle is presented in Fig. 1.
Fig. 1.
Subsequent stages of bacterial biofilm formation/dispersal and their genetic regulation. (i) reversible, followed by irreversible, attachment to the surface, (ii) formation of microcolonies, (iii and iv) biofilm maturation leading to the formation of bacterial consortia, and (v) biofilm dispersal. The regulatory involvement of quorum sensing (QS), bis-(3’-5’)-cyclic diguanosine monophosphate (c-di-GMP), and small RNAs (sRNAs) is shown by the arrows
Biofilm formation and dispersal are highly controlled processes regulated at the genetic level and by environmental signals. Current knowledge points to quorum sensing (QS), bis-(3’-5’)-cyclic diguanosine monophosphate (c-di-GMP), and small RNAs (sRNAs) as the main regulators of bacterial biofilms, at least in several Gram-negative species (Fazli et al. 2014).
QS is considered a special “language” used for intercellular communication, which is based on small, self-generated signal molecules called autoinducers. When sufficient bacteria are present and the concentration of autoinducers reaches a threshold level, the bacteria start to sense their critical mass and answer by repressing or activating target genes (de Kievit and Iglewski 2000). QS-controlled genes can constitute around 10 % of the bacterial genome (Wagner et al. 2003). QS systems play a very important role during the development and dispersal of bacterial biofilms. Although these systems are not involved in the attachment and initial biofilm growth stages, they are required for further biofilm development and, also, are the main regulators of biofilm dispersal (Davies et al. 1998). The QS signaling pathways in Pseudomonas aeruginosa, Staphylococcus aureus, and Vibrio cholerae are shown in Fig. 2.
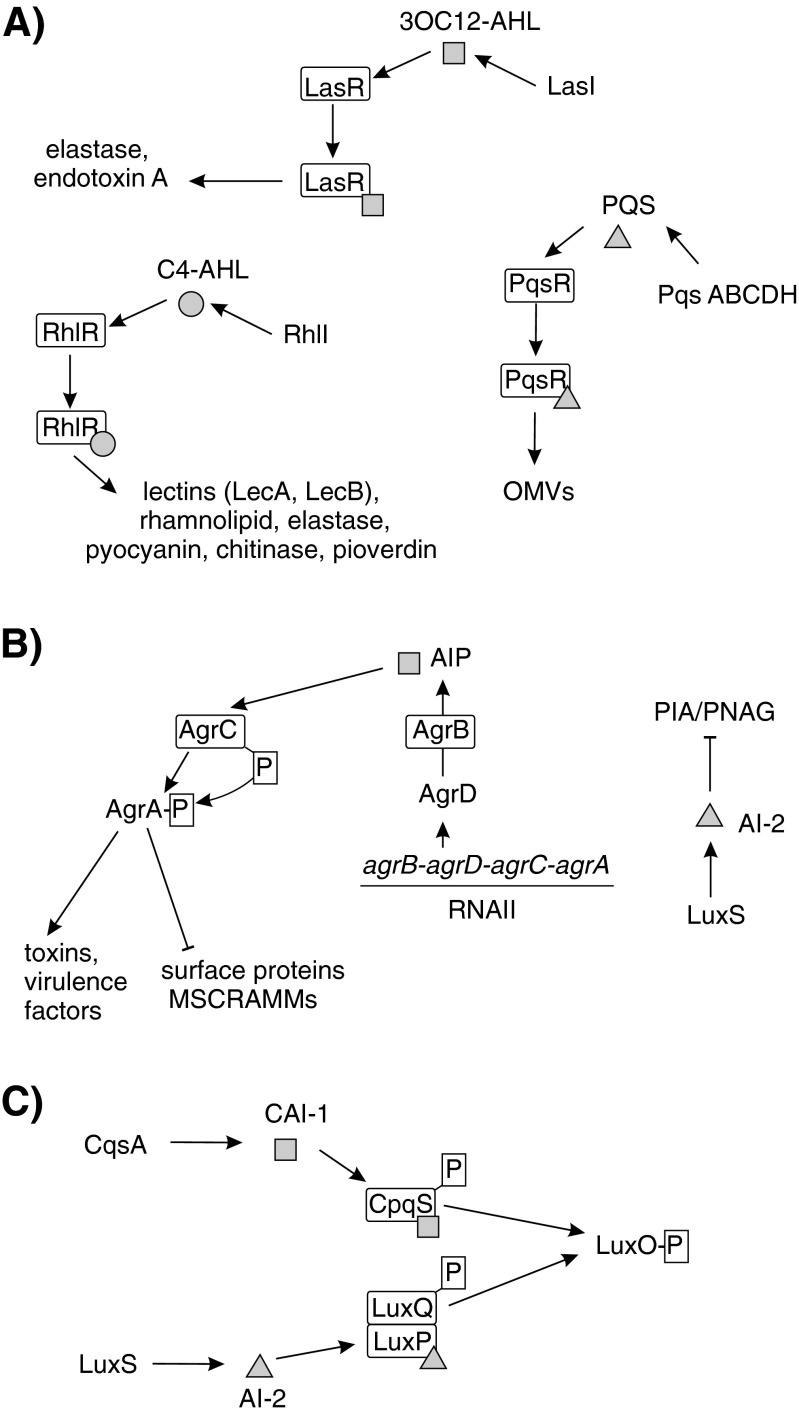
Fig. 2.
Schematic representation of QS system. a Pseudomonas aeruginosa LasI, RhlI, and PqsABCDH synthesize the QS signal molecules: N-(3-oxo-dodecanoyl)-L-homoserine lactone (3OC12-AHL), C4-AHL, and 2-heptyl-3-hydroxy-4-quinolone (PQS), respectively. The transcription factors LasR, RhlR, and PqsR detect their respective signal molecules, resulting in the regulation of target genes transcription. b Staphylococcus aureus QS peptide AIP is synthesized as a longer precursor by AgrD and is processed and secreted via AgrB. The extracellular signal is detected by the membrane-located histidine kinase AgrC and signal transduction occurs by phosphorelay to the AgrA response regulator. In the second QS system, LuxS synthesizes AI-2, which inhibits PIA/PNAG exopolysaccharide synthesis through an unknown QS cascade. c Vibrio cholerae LuxS and CqsA synthesize AI-2 and CAI-1 signal molecules, respectively. These signal molecules are detected by their corresponding receptors, the two-component histidine kinases LuxPQ and CpqS, which transfer phosphate residue to transcription activator LuxO. The regulation by LuxO–P is presented in Fig. 5
The second main biofilm regulator, the c-di-GMP signaling network, is considered the most complex secondary signaling system discovered in bacteria. However, its complexity varies significantly and this type of signaling is absent in some bacteria (Römling and Balsalobre 2012). After binding to a variety of cellular receptors, c-di-GMP controls bacterial transcription, the activity of enzymes, and even the functioning of larger cellular structures (Hengge 2009). c-di-GMP plays a crucial role in the bacterial decision between planktonic and biofilm-associated lifestyle (Jenal and Malone 2006). The factors regulated by c-di-GMP and important for three-dimensional biofilm structure development are: synthesis of exopolysaccharides, adhesive pili and adhesins, secretion of extracellular DNA (eDNA), and also control of cell death and motility. Regulatory connections between QS and c-di-GMP have been proved; it has been shown that cell density itself is one of the environmental cues sensed by the c-di-GMP network (Strivastava and Waters 2012).
Finally, small non-coding RNA molecules, sRNAs, including riboswitches, have been shown to participate in post-transcriptional gene regulation in bacteria, involving a range of metabolic processes, adaptation to stress, and microbial pathogenesis (for a review, see Michaux et al. 2014; Mandin and Guillier 2013). Therefore, sRNA regulators have become powerful tools for metabolic engineering and synthetic biology (Kang et al. 2014). However, the amount of data pointing to the role of sRNA in the biofilm life cycle is rather limited.
From among the other factors involved in biofilm formation, which are beyond the scope of this review, horizontal gene transfer (Madsen et al. 2012), alternative sigma factors (Irie et al. 2010), and toxin–antitoxin systems (Wang and Wood 2011) should be mentioned.
The control of bacterial biofilm has been studied mainly in members of the genus Pseudomonas, including the human opportunistic pathogen, P. aeruginosa, but the number of papers describing biofilm regulation in other bacterial pathogens is now growing exponentially. This knowledge may be useful in biofilm manipulation, control, and eradication. The present review describes the genetics of biofilm development in representative bacterial pathogens, both Gram-negative and Gram-positive. The strategies of biofilm eradication, mainly those exploiting QS control, are also discussed.
Pseudomonas aeruginosa biofilms
Brief characteristics
P. aeruginosa is an aerobic, non-fermenting, Gram-negative rod that has become a major opportunistic human pathogen and the leading cause of nosocomial infections in cancer, transplantation, and cystic fibrosis (CF) patients. Furthermore, due to its ability to cause chronic lung infections, this species is the primary pathogen responsible for the mortality of patients with CF (Silby et al. 2011). Many reports describe P. aeruginosa as being also one of the main species found in dermal and burn wounds (Ammons et al. 2009). In this species, exopolysaccharides appear to be the most important matrix components, in contrast to the other members of the genus Pseudomonas, P. putida and P. fluorescens, where this role is played by large surface proteins, among them the most abundant is large adhesion protein, LapA (Fazli et al. 2014). P. aeruginosa produces at least three secreted polysaccharides, Pel, Psl, and alginate. The last one produced by mucoid strains is considered as a P. aeruginosa virulence factor and, next to another compound of matrix, eDNA, seems to be of particular relevance to biofilm-mediated antibiotic resistance (Aspe et al. 2012). Three already mentioned main systems based on QS, c-di-GMP signaling, or regulatory sRNA control P. aeruginosa biofilm formation and dispersal. Their mode of action is described below. All systems follow the same scheme of signal transduction pathway, starting from sensors, followed by signal transmitters, and ending in effectors, which, in turn, execute the outcome, i.e., the production or modulation of factors involved in biofilm formation and detachment. The overview of these regulatory pathways is outlined in Fig. 3. The control of biofilm development in two other species of the genus Pseudomonas, P. fluorescens and P. putida, commonly found in soil and plant rhizosphere, and in an opportunistic pathogen, Burkholderia cenocepacia infecting CF patients, generally follow the outline described for P. aeruginosa. However, quite serious differences are observed concerning the importance of the main regulatory pathways and also the involvement of additional regulatory factors (Fazli et al. 2014).
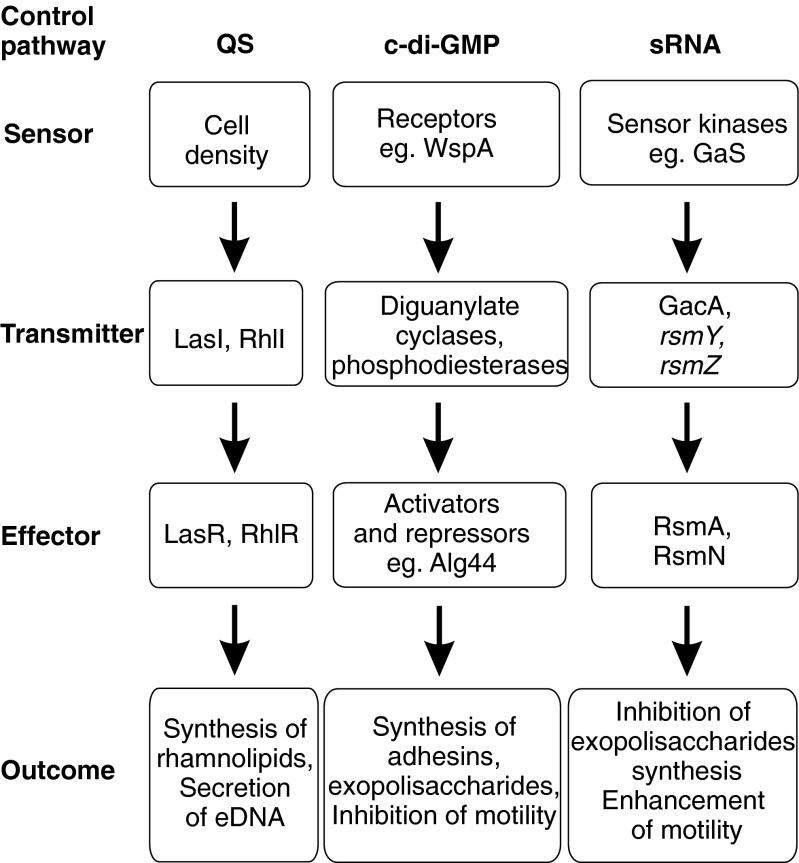
Fig. 3.
Control of biofilm formation in P. aeruginosa. Three control pathways are shown. In two AHL-based QS pathways, cell density plays a role of the environmental signal. The signal is then transferred through the transmitter proteins LasI and RhlI to the effector proteins LasR and RhlR, being transcriptional regulators. The QS system controls the synthesis of rhamnolipids and secretion of eDNA. In c-di-GMP signaling, the level of c-di-GMP is sensed by sensor proteins—receptors which govern the metabolism of this molecule through the activity of diguanylate cyclases (DCGs) and phosphodiesterases (PDEs). Upon binding to the effector proteins, which are activators or repressors acting at transcriptional or post-transcriptional levels, c-di-GMP controls the synthesis of adhesins and exopolysaccharides and inhibits the motility. The control by sRNA involves the activation of sensor kinases, e.g., GacS, which phosphorylates transmitter protein GacA, leading to the subsequent activation of small RNAs, inhibition of effector RsmA activity, and, finally, to the inhibition of exopolysaccharides synthesis and enhancement of motility
Quorum sensing (QS)
QS regulation of biofilm-related genes in P. aeruginosa in the natural environment and during persistent infections is considered as the best known example among all bacterial species (Singh et al. 2000). This bacterium has at least three QS systems: two N-acylated homoserine lactone (AHL)-based LasIR and RhlIR systems and a Pseudomonas quinolone signal (PQS)-based system. Both AHL systems contain a gene encoding AHL sensor/transcriptional regulator, lasR and rhlR, respectively, and a gene encoding an autoinducer, lasI, required for the synthesis of N-(3-oxo-dodecanoyl)-L-homoserine lactone and rhlI responsible for the synthesis of N-(butanoyl)-L-homoserine lactone (C4-AHL) (Pearson et al. 1994, 1995). In the PQS system, 2-heptyl-3-hydroxy-4-quinolone transported by outer membrane vesicles (OMVs) is a sensing molecule, and, in turn, PQS causes the biogenesis of OMVs (Mashburn-Warren et al. 2008; Kulkarni and Jagannadham 2014). All three systems are regulated hierarchically: LasR positively regulates RhlR and PQS and RhlR negatively regulates PQS (Pesci et al. 1997; Wade et al. 2005).
QS signaling controls the synthesis of rhamnolipids, which are important in the late stage of biofilm development, maintaining the channels in mushroom-shaped structures, resulting in the proper distribution of nutrient and oxygen and removal of waste products (Davey and O’Toole 2000). Rhamnolipids synthesis is induced in the center of biofilm mushroom caps, which is consistent with the control role of QS (Lequette and Greenberg 2005). The overproduction of these biosurfactants causes the biofilm detachment from the surface, leading to its dispersal (Boles et al. 2005). QS also plays a role in the release of a large amount of eDNA at the late stage of biofilm development, as a consequence of the autolysis of a bacterial subpopulation (Allesen-Holm et al. 2006). It is well established that autolysis is regulated by PQS, but the mechanism of this process is still not fully understood (Fazli et al. 2014). The QS system regulates the production of yet other compounds important for biofilm formation, i.e., LecA and LecB lectins (Tielker et al. 2005; Diggle et al. 2006) and siderophores pyoverdine and pyochelin (Banin et al. 2005). The last two exert their action through participation in iron metabolism; it was shown that too low or too high concentration of iron results in the inhibition of biofilm formation (Singh et al. 2002).
c-di-GMP signaling
In general, high c-di-GMP level induces the biosynthesis of adhesins and matrix polysaccharides and inhibits various types of motility and, therefore, stimulates biofilm formation. In contrast, low c-di-GMP level downregulates the production of adhesins and exopolysaccharides and enhances bacterial motility, leading to biofilm dispersal (for a review, see Hengge 2009). Two types of enzymes control the level of c-di-GMP in bacteria. Diguanylate cyclases (DGCs), which contain typical domain, CGDEF, produce this nucleotide from two molecules of GTP, whereas c-di-GMP is broken down into 5’-phosphoguanylyl-(3’-5’) guanosine (pGpG) by specific photodiesterases (PDEs), which activity is associated with EAL or HD-GYP specific domains. The activity of both types of enzymes is controlled by environmental cues (Römling et al. 2013).
In P. aeruginosa, the so-called “basic c-di-GMP signaling module” consists of five components: (i) sensors of environmental signals, (ii) enzymes involved in the synthesis and degradation of c-di-GMP (see above), (iii) specific effectors which can be proteins or riboswitches, both being allosterically regulated by c-di-GMP, (iv) targets, e.g., promoter DNA, enzymes, or cellular structures such as flagellar basal body or exopolysaccharide synthetic and secretion apparatus, (v) molecular output produced by effectors after their activation by c-di-GMP (Hengge 2009).
P. aeruginosa contains three signal sensors (receptors), WspA, YfiB, and RocS1, which sense the level of c-di-GMP, five diguanylate cyclases, WspR, YfiN, SadC, RoeA, and SiaD, and five phosphodiesterases, BifA, DipA, RocR, MucR, and NbdA. The best characterized chemosensor, WspA, senses growth on surfaces and then becomes activated and able to phosphorylate its cognate diguanylate cyclase, WspR (Güvener and Harwood 2007). While phosphorylated, WspR forms clusters, changes the location within the cell, and its cyclase activity is increased (Huangyutitham et al. 2013). Yet another sensor, the outer membrane protein YfiB, responds to cell membrane stress and activates YfiN cyclase (Malone et al. 2012). PDEs enzymes causing biofilm dispersal are activated by such environmental cues as starvation, low concentration of oxygen, and nitric acid, but their precise mechanism of activation has not been elucidated (An et al. 2010; Li et al. 2013). The multitude of sensors, as well as c-di-GMP synthesizing and breaking enzymes, allows the c-di-GMP to be controlled by various environmental cues.
Four P. aeruginosa effectors, Alg44, FleQ, PelD, and FimX, regulate different targets involved in biofilm development. Alg44 activated by c-di-GMP controls the synthesis of alginate, an exopolysaccharide important in the late stages of infection with P. aeruginosa mucoid strains (Merighi et al. 2007). FleQ acts as both a repressor and an activator of pel operon encoding Pel exopolysaccharide synthesis (Baraquet and Harwood 2013), regulates the synthesis and transport of outer membrane adhesin, CdrA (Borlee et al. 2010), and represses the expression of flagellum biosynthesis genes (Baraquet and Harwood 2013). Two other effectors, PelD and FimX, regulate Pel synthesis at a post-transcriptional level and control twitching motility (Lee et al. 2007; Jain et al. 2012).
Control by sRNA
Regulation by sRNAs, rsmY and rsmZ, is the best known example. In this pathway, the role of a sensor is played by three sensor kinases, RetS, LadS, but mainly GacS (Ventre et al. 2006). GacS phosphorylates GacA (Goodman et al. 2009), which, in turn, activates the transcription of rsmZ and rsmY. These two sRNAs are also controlled by the other proteins participating in the P. aeruginosa phosphorelay system (Petrova and Sauer 2010). rsmZ and rsmY reduce the activity of effector protein RsmA, being a negative post-transcriptional regulator of the biofilm matrix polysaccharide Psl (Irie et al. 2010), and also downregulate another effector, RsmN, controlling the same functions as RsmA (Marden et al. 2013). It was shown that an increased expression of rsmY and rsmZ results in enhanced initial attachment to abiotic surfaces but, on the contrary, subsequent biofilm development is hampered by the high level of these sRNAs (Chambers and Sauer 2013). Another sRNA, phrS, stimulates the P. aeruginosa PQS pathway. phrS expression requires global oxygen-responsible regulator ANR, which provides a regulatory link between oxygen availability and PQS (Sonnleitner et al. 2011).
The list of factors controlling P. aeruginosa biofilm development should be extended by the alternative sigma factor, RpoS. It was shown that, in P. aeruginosa, PAO1 biofilm rpoS expression is increased (Waite et al. 2006) and RpoS acts as a positive regulator of the expression of the psl gene (Irie et al. 2010). Finally, it was postulated that yet another, fatty, cis-2-decenoic acid-mediated, signaling may play a role in biofilm dispersal (Amari et al. 2013). However, the mechanism of its activity has not yet been resolved.
Staphylococcus aureus biofilms
Brief characteristics
S. aureus is a Gram-positive, nonmotile coccus able to form cell clusters and producing yellow pigment. This bacterium is a causative agent of acute and chronic infections. Its ecological niche in humans is the anterior nares. S. aureus biofilm persists on medical implants and catheters, constituting a significant healthcare problem (Kiedrowski and Horswill 2011). The list of S. aureus biofilm-related diseases is long and includes: osteomyelitis, indwelling medical device infections, periodontitis and peri-implantitis, chronic wound infection, chronic rhinosinusitis, endocarditis, and ocular infections (for a review, see Archer et al. 2011).
S. aureus biofilm life cycle follows the typical scheme already described for P. aeruginosa. Due to its lack of motility, the biofilm is flatter than those formed by motile genera, although mushroom forms can also be observed (Mann et al. 2009). In the biofilm, a great number of slow-growing cells and persister cells—nondividing and tolerant to antibiotics—are present (Lewis 2007). S. aureus biofilm is embedded within a glycocalyx or slime layer composed primarily of teichoic acid and staphylococcal and host proteins (Husain et al. 2013). The polysaccharide PIA (polysaccharide intercellular antigen), composed mainly of polymeric N-acetyl-glucosamine and eDNA, is also a significant biofilm constituent. The genetic control of PIA synthetic operon, icaADBC, involves many factors, among them the main IcaR repressor and the second TcaR repressor (Cramton et al. 1999; Jefferson et al. 2004). The expression of icaR gene is positively regulated by protein Spx (suppressor of clpP and clpX), a global regulator of stress response and negatively regulated by Rbf protein (Cue et al. 2009; Pamp et al. 2006). Moreover, SrrAB (staphylococcal respiratory response regulator) is responsible for PIA induction under anaerobic conditions (Ulrich et al. 2007). It was shown that biofilm formation in several S. aureus strains, including MRSA (methicillin-resistant S. aureus), does not depend on PIA production (Fitzpatrick et al. 2005), which is substituted by proteinaceous cell-to-cell adhesion with the participation of, e.g., biofilm-associated protein, Bap (Lasa and Penadés 2006). Another important component of staphylococcal biofilm, eDNA, is released as a consequence of cell lysis by holin homolog CidA and other proteins (Rice et al. 2007; Brady et al. 2006). However, massive cell lysis takes place at the late stage of biofilm development, eDNA is also released at an early stage, thus participating in cell attachment (Mann et al. 2009). In highly aggressive S. aureus isolates, a novel toxin family, phenol-soluble modulins (PSMs), contributes to biofilm development and dispersal and, therefore, also in the dissemination of biofilm-associated infections (Peschel and Otto 2013). It should be noted here that appreciable strain-dependent variations across staphylococcal biofilm composition are observed (Kiedrowski and Horswill 2011).
S. aureus biofilm development is regulated by many environmental conditions and genetic signals. From among the latter, the most important are QS and control by small molecules, including sRNA.
Control by QS, c-di-GMP, and sRNA
The accessory gene regulator (Agr) system plays a crucial role in the functioning of the S. aureus QS system. In this genus, QS positively regulates toxins and acute virulence factors and negatively regulates surface proteins named microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), thus inhibiting adhesion to human matrix proteins, e.g., fibrinogen (Clarke and Foster 2006; Pei et al. 1999).
QS also enhances cell detachment from mature biofilm. The agr locus contains agrA, agrC, agrD, and agrB genes, constituting the so-called transcript RNAII. The prepheromone AgrD is exported and modified by AgrB, which results in formation of the characteristic thiolactone-containing autoinducing peptide (AIP). AIP activates the two-component AgrC/AgrA system, which, in turn, activates the transcription of RNAII, providing an autofeedback loop (Ji et al. 1995). AIP also influences the transcription of bifunctional RNAIII regulating MSCRAMMs and encoding d-hemolysin (Fechter et al. 2014). The expression of psm genes coding for PSMs surfactants responsible for biofilm maturation and dispersal by the disruption of noncovalent interactions between biofilm cells and matrix components are also under the influence of the QS system (Periasamy et al. 2012; Otto 2014). The staphylococcal QS system upregulates the expression of peptidases and nucleases, which also increases biofilm detachment (Boles and Horswill 2008; Lauderdale et al. 2009). The data of several experiments support the inhibitory role of QS on biofilm development; for example, it was demonstrated that agr mutants form a thicker biofilm compared with wild-type strains (Vuong et al. 2003).
It was postulated that the ability to form a biofilm is closely related to the character of infection, and the determinants of acute and chronic virulence are regulated by QS in an opposite fashion. QS is important in expressing acute virulence and the formation of a differentiated biofilm with the capacity for dissemination, whereas chronic infections are concomitant with biofilm downregulation and mutation in the QS system. This postulate is supported by the observation that QS mutants are found in elevated numbers in chronic infections (Shopsin et al. 2010). However, it was recently shown that a significant fraction of S. aureus bacteremia cases are caused by agr-defective strains; thus, the role of QS in invasive staphylococcal infections can be questionable (Painter et al. 2014).
The data on c-di-GMP involvement in S. aureus biofilm formation seems to be controversial. Ishihara and coworkers (2009) suggested that this nucleotide is important for biofilm formation because a mutation in GdpS protein containing the GGDEF domain characteristic for diguanylate cyclases hampers its development. This defect was complemented by the addition of external c-di-GMP in physiological concentration. Opposite results were obtained by Holland and coworkers (2008) who demonstrated that, however, GdpS does, in fact, affect biofilm formation, but the mechanism of its activity is independent of c-di-GMP. The reason for the reported discrepancy can be due to physiological differences between the strains used. It was also shown that the treatment of S. aureus with extracellular c-di-GMP applied in high concentration, ranging between 0.02 and 0.2 μM, suppressed biofilm formation by the inhibition of intercellular adhesive interactions (Karaolis et al. 2005). These authors even suggested that c-di-GMP can be used as a novel anti-biofilm agent.
In S. aureus, 250 sRNA genes were discovered; however, functional studies are still lagging behind (Romilly et al. 2012). It was shown that the 3’ domain of the already mentioned RNAIII transcript represses, at the post-transcriptional level, the synthesis of cell-wall hydrolytic enzymes and, thus, negatively influences biofilm formation (Boisset et al. 2007). In a similar way, the 3’ untranslated domain of icaR transcript encoding a transcriptional repressor of biofilm polysaccharide synthesis interferes with the translation initiation of its own RNA (Ruiz de los Mozos et al. 2013). It was also shown that the 5’ untranslated, 196 nucleotides long, region of sarA transcript, designated teg49, induces the formation of biofilm through the positive regulation of the sar locus encoding SarA protein, promoting the initial steps of biofilm formation (Kim et al. 2014).
Two other major factors relevant for biofilm development in S. aureus are SarA (staphylococcal accessory regulator) and SigB. The sarA transcript is upregulated in biofilm when compared to planktonic cultures (Beenken et al. 2003) and its product inhibits the expression of nuclease Nuc and also proteases, thus preventing the degradation of biofilm structural components (Tsang et al. 2008). It was shown that SarA regulates the expression of cell wall-associated and certain extracellular proteins in agr-dependent and agr-independent pathways (Arya and Princy 2013). In turn, an alternative sigma factor, SigB (Kullik and Giachino 1997), is involved in the early stages of biofilm formation. It was shown that sigB-deficient S. aureus does not form a biofilm and upregulates RNAIII, which promotes the antibiofilm Agr system (Rachid et al. 2000).
The factors involved in S. aureus biofilm formation, maintenance, and detachment are presented in Fig. 4.
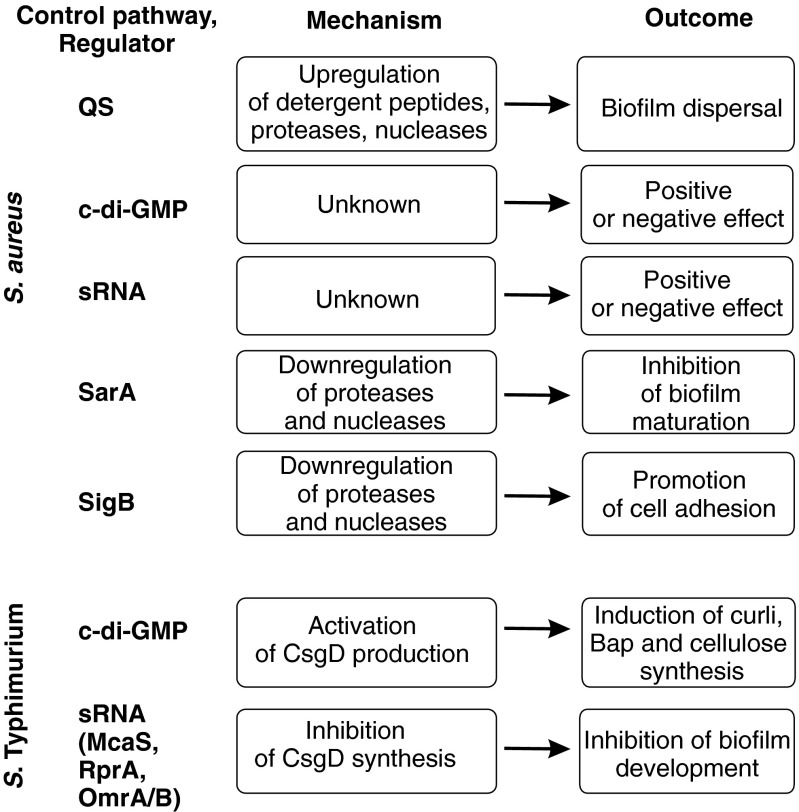
Fig. 4.
Control of biofilm formation in S. aureus and S. Typhimurium. S. aureus: biofilm control by five main regulatory factors is shown. QS positively regulates the synthesis of detergent-like peptides, proteases, and nucleases, resulting in biofilm dispersal. The involvement of c-di-GMP and sRNAs in biofilm regulation is still controversial, and the mechanism of their activity is unknown. SarA inhibits the expression of proteases and nucleases and, thus, promotes the development of immature biofilm. Alternative sigma factor SigB promotes the expression of adherence factors and, thus, positively regulates the initial steps of biofilm formation. S. Typhimurium: c-di-GMP activates the master CsgD and, subsequently, increases the synthesis of curli, Bap, and cellulose. Several sRNAs (McaS, RprA, OmrA/B, and possibly GcvB) inhibit the translation of CsgD mRNA and inhibit biofilm development
Biofilms of the enteric bacteria Salmonella Typhimurium and Vibrio cholerae
Gastrointestinal diseases usually arise upon the ingestion of food or water contaminated by enteric bacterial pathogens (Fàbrega and Vila 2013). Microorganisms frequently associated with illness are Salmonella enterica serovars, Escherichia coli pathovars, Shigella spp., Campylobacter spp., Yersinia enterocolitica, V. cholerae, and Listeria monocytogenes. The genetics of biofilm formation and dispersal are the most intensively studied in S. enterica sv. Typhimurium and V. cholerae.
Salmonella enterica serovar Typhimurium
The ability to form a biofilm is an important factor of Salmonella virulence. For example, S. enterica serovar Typhi frequently forms biofilms on gallstones, resulting in a chronic infection of the gall bladder and the development of Salmonella carrier state—a serious public health problem (Gonzalez-Escobedo et al. 2011). Striking similarities in biofilm composition and regulatory circuits between several serovars of S. enterica and E. coli have been shown. The control of various stages of biofilm development is the best studied in S. enterica sv. Typhimurium, the most common causal agents of gastrointestinal diseases.
S. Typhimurium biofilm matrix is composed of proteins and exopolysaccharides. A major protein component is curli (amyloid fimbriae), encoded by csg operons (Yaron and Römling 2014). Protein BapA constitutes another important component of the matrix (Barnhart and Chapman 2006) and major biofilm exopolysaccharides are cellulose (Zogaj et al. 2001) and colonic acid (Gibson et al. 2006). Adhesion-mediated type I fimbriae, Lpf and Pef, also contribute to the early steps of biofilm formation (Ledeboer et al. 2006).
Biofilm formation is controlled by the master regulator, CsgD protein, belonging to the LuxR family of regulators. csgD expression is positively regulated by an alternative sigma factor, σs; thus, the level of CsgD is high in the stationary phase of growth (Yaron and Römling 2014). CsgD increases curli and Bap expression and also, post-transcriptionally, indirectly activates cellulose biosynthesis (Fàbrega and Vila 2013). In the regulation of S. Typhimurium biofilm, c-di-GMP and sRNA play a crucial role. The elevated level of CsgD activates the transcription of the adrA gene encoding diguanylate cyclase synthesizing signal molecule c-di-GMP. In turn, c-di-GMP activates csgD expression in a complex way, involving at least eight GG(D/E)EF/EAL domain proteins (Ahmad et al. 2011; Anwar et al. 2014; Römling 2012). Moreover, c-di-GMP modulates cellulose biosynthesis (Latasa et al. 2005) and is responsible for the so-called rdar morphotype (red, dry, rough), characteristic for a potent biofilm producer (Ahmad et al. 2011). In addition to c-di-GMP, CsgD synthesis is also regulated at the post-transcriptional level by sRNAs. csgD mRNA is a direct target for several sRNAs, McaS, RprA, OmrA/OmrB, and possibly GcvB. All these sRNAs negatively regulate CsgD synthesis by binding to the overlapping 5’-region of the transcript, masking the ribosome binding site and, thus, inhibiting translation or inducing mRNA degradation (Mika and Hengge 2013). The principles of the regulation of S. Typhimurium biofilm are outlined in Fig. 4.
Vibrio cholerae
V. cholerae is a ubiquitous bacterium in aquatic systems but also causes cholera, a severe diarrheal disease resulting from the consumption of contaminated drinking water (Faruque et al. 1998). It has a capacity to form biofilm, in both aquatic ecosystems and within the host (Watnick and Kolter 1999). The initial stages of biofilm formation are promoted by flagella-mediated motility and three types of pili (Yildiz and Visick 2009). The biofilm matrix is composed of Vibrio polysaccharide (VPS) containing glucose and galactose and minor constituents, N-acetyl glucosamine, mannose, and xylose (Yildiz and Schoolnik 1999), and matrix proteins RbmA, RbmC (rugosity and biofilm structure modulators), and Bap1 (Berk et al. 2012). The positive regulators of VPS production are the proteins VpsR and VpsT, which promote the transcription of vps structural genes (Yildiz et al. 2004; Casper-Lindley and Yildiz 2004). Biofilm formation in V. cholerae is regulated by QS, c-di-GMP, and sRNA. Very tight regulatory connections between these three factors have been proven (Srivastava and Waters 2012).
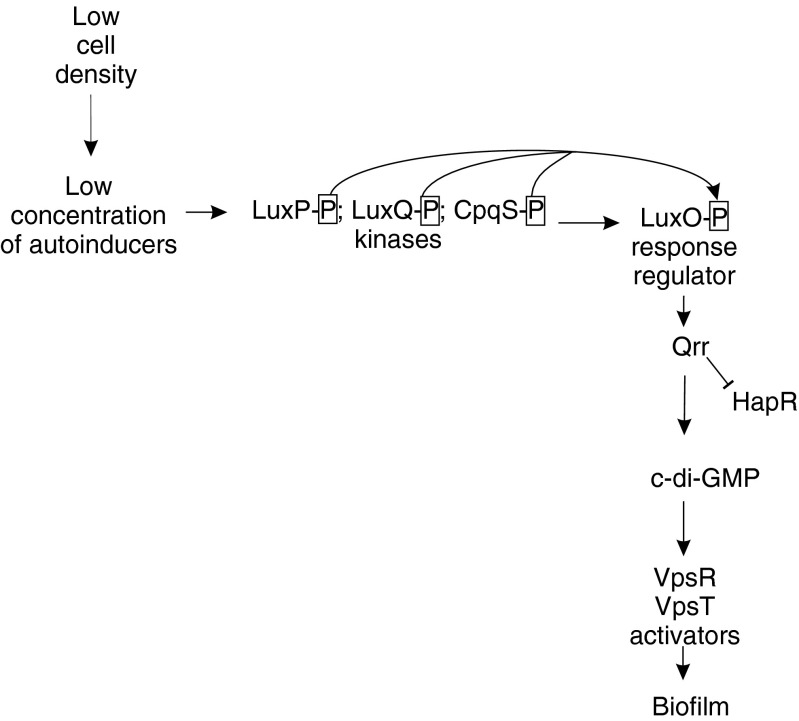
The formation of V. cholerae biofilm is induced at low cell density and repressed at high cell density (Ng and Bassler 2009). The Vibrio QS system is composed of two sensory circuits that respond to two different autoinducers: Als–AI-2 or a hydroxylated alkyl ketone, CAI-1 (Higgins et al. 2007; Tiaden et al. 2010). At low concentrations of inducers, the periplasmic receptors, respectively, histidine kinases LuxPQ and CpqS, phosphorylate the response regulator LuxO. When phosphorylated, LuxO activates the expression of four small RNAs, Qrr 1–4 (quorum-regulated RNAs). These RNAs are transcriptionally activated not only by LuxO–P but also by an alternative σ54, and their activity appears to require the RNA-binding chaperone Hfq (Bardill and Hammer 2012; Lilley and Bassler 2000). Qrrs repress the synthesis of protein HapR, a negative regulator of c-di-GMP synthesis, and, at the same time, enhance the production of c-di-GMP synthesizing enzymes. Control of the c-di-GMP level by QS and Qrr constitutes a complicated circuit, moreover in that V. cholerae contains 61 predicted enzymes involved in the synthesis/degradation of this nucleotide (Galperin 2004). Summarizing, the activity of Qrr results in high c-di-GMP level at low cell density. c-di-GMP positively controls biofilm development by binding to VpsR and VpsT, which are the direct activators of biofilm genes (Hammer and Bassler 2003; Vance et al. 2003; Srivastava and Waters 2012). The control of biofilm formation in V. cholerae is schematically presented in Fig. 5.
Fig. 5.
Control of biofilm development in Vibrio cholerae at low cell density. At low concentration of autoinducers, histidine kinases LuxP and CpqS are phosphorylated and able to phosphorylate regulator LuxO. LuxO–P activate the expression of Qrr 1–4 RNAs, which, in turn, positively influence the level of c-di-GMP. c-di-GMP activates VpsR and VpsT proteins, which positively regulate biofilm genes
At high cell density, the interaction of inducers and receptors switches their activity to phosphatases and LuxO is dephosphorylated. As a result, Qrr level decreases, HapR protein is synthesized, c-di-GMP level drops, and biofilm formation is inhibited (Tu and Bassler 2007). Repression of biofilm formation at high cell density could lead to the dispersal of mature V. cholerae biofilm (Srivastava and Waters 2012). Histone-like protein, H-NS, is another factor involved in the negative control of V. cholerae biofilm (Teschler et al. 2015).
It was also shown that, in addition to c-di-GMP, other nucleotides control V. cholerae biofilm; cAMP represses biofilm formation (Liang et al. 2007), while ppGpp positively regulates this process (He et al. 2012).
Targeting genetic determinants as a mode of biofilm modulation
A study aimed to discover new promising agents and strategies against bacterial infections, especially those associated with biofilms, is an urgent task due to biofilm resistance to already used antimicrobial agents. Innovative anti-biofilm strategies are the subject of recent reviews (Cushnie and Lamb 2011; Joo and Otto 2012; Römling and Balsalobre 2012; Markowska et al. 2013; Chung and Toh 2014; Masák et al. 2014; Tan et al. 2014).
QS inhibition, also called quorum quenching, QQ, is considered one of the promising anti-biofilm strategies. However, the recent view of the role of QS in infections, pointing to its importance in biofilm dispersal, provides an argument against the practical use of the QQ approach to cure diseases caused by biofilm (Otto 2014). Many compounds, both natural and synthetic, affect bacterial QS systems (Kalia 2013), thus influencing biofilm development. Two of them, vanillin and cinnamic acid, were even shown to stimulate the formation of biofilm due to their ability to induce AHL synthesis (Plyuta et al. 2013). Unfortunately, up to 2014, only two clinical trials on QS inhibitors have been performed (Scutera et al. 2014).
The ideal QS inhibitors should fulfill the following criteria: (i) they should be low molecular weight and stable compounds, (ii) their activity should be highly specific and not toxic for the eukaryotic hosts, (iii) they should not interfere with the basal metabolic processes that can be targets for the development of drug resistance (Bhardwaj et al. 2013).
There are several potential classes of QS inhibiting strategies. One class targets QS signal production. For example, it was demonstrated that the analog of methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAM) blocks AI-1 and AI-2 acyl-lactone-based signal molecules (Gutierrez et al. 2009), methyl anthranilate inhibits the production of PQS (Calfee et al. 2001), and eugenol, the major compound of clove extract, decreases the transcriptional activation of P. aeruginosa las and pqs systems (Zhou et al. 2013). The second group of strategies neutralizes QS signals by enzymatic or antibody-mediated inactivation (Kalia and Purohit 2011). QQ enzymes can either hydrolyze the AHL molecules, e.g., AHL-lactonase, or reduce carbonyl to hydroxyl groups by the activity of oxidoreductases (Scutera et al. 2014). Long-acyl AHLs are degraded with the participation of AHL-acylase (Huang et al. 2003). The immunological approach includes the use of monoclonal antibodies, such as AP4-24H11, against S. aureus autoinducer (Park et al. 2007) or antibodies against P. aeruginosa homoserine lactones (Palliyil et al. 2014). Another class of strategies explores compounds whose targets are autoinducer–receptor interactions and/or receptor-mediated signals. Synthetic brominated derivatives of furanone known as C-30, AHL analogs, hamamelitannin, and also the extracts of several common fruits, herbs, and spices inhibit QS receptors, LasR and RhlR (Vattem et al. 2007; Sintim et al. 2010; O’Loughlin et al. 2013). For example, clove oil inhibits Las- and Rhl-regulated virulence factors, swimming motility, and extracellular polymeric substance (EPS) production in P. aeruginosa (Husain et al. 2013). In turn, solonamides isolated from Photobacterium, due to their structural similarity to agr AI, competitively inhibit the agr system in S. aureus (Mansson et al. 2011). Yet another compound, a sesquiterpene alcohol, farnesol, produced by Candida albicans and also present in the essential oils of citrus fruits, affects biofilm formation by P. aeruginosa, C. albicans, S. aureus, and Streptococcus mutans by interfering with the transcription of QS operons (Cugini et al. 2007; Jabra-Rizk et al. 2006). A number of compounds were identified that target the response regulator LuxO and the transcriptional repressor HtpR in V. cholerae. One of the most potent is pyrrole analog precursor of cholerae autoinducer 1, CA1 (Perez et al. 2014).
Several QS inhibitors can also enhance the activity of existing antibiotics and restore immune response efficiency (Jakobsen et al. 2012). For example, it was demonstrated that the QS inhibitor, hamamelitannin, acts synergistically with vancomycin or clindamycin against S. aureus (Brackman et al. 2011). It was also shown that biofilms treated with synthetic furanone C-30 are susceptible to tobramycin and readily dispersed by detergents (Hentzer et al. 2003). Moreover, QS mutants and cells treated with QS inhibitors were found to be prone to oxidative burst and phagocytosis (Bjarnsholt et al. 2005).
In our group, the trial was performed to evaluate the inhibitory activity of nanosilver on the QS system in P. aeruginosa. Silver nanoparticles were chosen due to their strong antibacterial activity, pleiotropic effect on bacterial cell, and lack of documented resistance development (Markowska et al. 2013). Unfortunately, no inhibition of QS LasI and Rhl1 systems was determined (data not published).
Some researchers stress the advantages connected with the use of QQ compounds. Because QS inhibitors do not exert a direct bactericidal effect, it is commonly considered that there is less selection pressure and less likelihood of resistance development (Pan and Ren 2009). Unfortunately, recent evidence indicates the development of the resistance to QS inhibitors in P. aeruginosa (Kalia et al. 2014). The increased resistance exploits the efflux mechanisms, e.g., mexR and nalC mutants demonstrated the increased resistance to C-30 (Maeda et al. 2012). The potential risk connected with the use of all QQ strategies described above should also be mentioned. QS inhibitors can select more virulent strains by interfering with natural selection towards reduced virulence (Köhler et al. 2010). Moreover, the elimination of a particular pathogen can predispose patients to be infected with others.
As the prominent role of c-di-GMP is the activation of biofilm formation, the signals that downregulate its concentration in a cell can be considered potential anti-biofilm agents. There are several approaches to interfere with c-di-GMP signaling, e.g., manipulation with enzymatic activities, interference with signal perception, and direct inactivation (Römling and Balsalobre 2012). The inhibition of c-di-GMP synthesizing activity or stimulation of phosphodiesterase activity can diminish or enhance biofilm formation, respectively (Chávez de Paz et al. 2012; An et al. 2010). There are no natural compounds interfering with the synthesis or activity of c-di-GMP, but among synthetic compounds, sulfathiazole and N-(4-aminophenyl) benzamide were shown to be potent inhibitors of biofilm formation, this being achieved by suppressing diguanlyate cyclases (Antoniani et al. 2010). Several inhibitors targeting diguanylate cyclases, named DCG inhibitors, and, thus, influencing c-di-GMP metabolism in V. cholerae were identified (Sambanthamoorthy et al. 2012, 2014). In turn, azathioprine interferes with intracellular nucleotide pool availability (Antoniani et al. 2013). Other signals controlling the c-di-GMP level can also be used to regulate c-di-GMP signaling. For example, P. aeruginosa exposure to nitric oxide (NO) stimulates c-di-GMP-specific phosphodiesterase activity, thus promoting biofilm dispersal (Barraud et al. 2009), whereas the sequestration of c-di-GMP by high-affinity receptors removes the available nucleotide pool and promotes biofilm dispersal, phenotypically mimicking phosphodiesterase activity (Ma et al. 2011). c-di-GMP stimulates biofilm development only in a relatively narrow range of concentration; it was shown that extracellular c-di-GMP, when applied in high concentration, acts as an inhibitor of biofilm formation by S. aureus (Karaolis et al. 2005). It should also be mentioned that c-di-GMP and other cyclic di-nucleotides can serve as potential adjuvants, and their high efficacy to stimulate an immune response can constitute a future strategy to inhibit biofilm formation (Karaolis et al. 2007).
Targeting the regulatory sRNAs is another potential way to modulate the expression of genes important for biofilm development (Kang et al. 2014). Experiments performed with E. coli have shown that the modulation of expression of several sRNAs, OmrR, OmrB, and McaS, leading to the change in cell motility, production of curli, and export of exopolysaccharides, results in the inhibition of biofilm formation. Also, the knockout of other sRNAs, Arc2, SdsR, GadY, and MicA, affects biofilm development and motility, although their mode of action remains elusive (Mandin and Guillier 2013). Metabolic engineering and the possibility to synthesize artificial RNAs of choice (Man et al. 2011) create the opportunity for the silencing of any specific gene and, therefore, inhibit various steps of biofilm formation or enhance biofilm dispersal.
Concluding remarks
The biofilm formation and dispersal in pathogenic bacteria has been studied extensively, and a large number of literature positions dealing with these processes, including their genetic regulation, has been published. The regulators being the subject of this paper, QS, c-di-GMP, and sRNA, are necessary for biofilm biology in all four described bacterial species, but the details of their regulatory role and the importance of the particular factor vary between species.
In P. aeruginosa and S. aureus, QS systems regulate mainly biofilm dispersal, while in V. cholerae, QS is important for biofilm formation, and it was proved that this process is induced at low cell density. There are no available data on the regulation of S. Typhimurium biofilm by QS. In turn, c-di-GMP signaling is a factor regulating biofilm formation in P. aeruginosa, S. Typhimurium, and V. cholerae, but its involvement in the regulation of S. aureus biofilm is still controversial. The interactions between QS and c-di-GMP regulatory pathways exist and are especially well documented for V. cholerae. Finally, as the variety of sRNAs regulates the large spectrum of bacterial genes, it can be expected that these molecules can also be involved in biofilm biology. Indeed, sRNAs have been shown to regulate post-transcriptionally, usually negatively, biofilm formation in all four described species. The interconnections between sRNAs and other regulators are proved.
The number of attempts to target genetic determinants in order to modulate biofilms formed by bacterial pathogens is growing exponentially. In the in vitro experiments, several small-molecule therapeutics were discovered. They can be divided into four classes: QS inhibitors, disruptors of c-di-GMP signaling, inhibitors of sRNAs activity, and compounds with unknown target. The technique combining microscopic imaging with cellular viability measurements allows to identified the compound that selectively disrupt biofilm formation without affecting cell survival (Teschler et al. 2015). All compounds giving positive results during the in vitro studies should pass preclinical and clinical trials in order to be accepted in the therapy arena.
References
- Ahmad I, Lamprokostopoulou A, Le Guyon S, Streck E, Barthel M, Peters V, Hardt W-D, Römling U. Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS One. 2011;6:e28351. doi: 10.1371/journal.pone.0028351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- Amari DT, Marques CN, Davies DG. The putative enoyl-coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic acid. J Bacteriol. 2013;195:4600–4610. doi: 10.1128/JB.00707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammons MC, Ward LS, Fisher ST, Wolcott RD, James GA. In vitro susceptibility of established biofilms composed of a clinical wound isolate of Pseudomonas aeruginosa treated with lactoferrin and xylitol. Int J Antimicrob Agents. 2009;33:230–236. doi: 10.1016/j.ijantimicag.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Wu J, Zhang LH. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol. 2010;76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniani D, Bocci P, Maciąg A, Raffaelli N, Landini P. Monitoring of diguanylate cyclase activity and of cyclic-di-GMP biosynthesis by whole-cell assays suitable for high-throughput screening of biofilm inhibitors. Appl Microbiol Biotechnol. 2010;85:1095–1104. doi: 10.1007/s00253-009-2199-x. [DOI] [PubMed] [Google Scholar]
- Antoniani D, Rossi E, Rinaldo S, Bocci P, Lolicato M, Paiardini A, Raffaelli N, Cutruzzolà F, Landini P. The immunosuppressive drug azathioprine inhibits biosynthesis of the bacterial signal molecule cyclic-di-GMP by interfering with intracellular nucleotide pool availability. Appl Microbiol Biotechnol. 2013;97:7325–7336. doi: 10.1007/s00253-013-4875-0. [DOI] [PubMed] [Google Scholar]
- Anwar N, Rouf SF, Römling U, Rhen M. Modulation of biofilm-formation in Salmonella enterica serovar Typhimurium by the periplasmic DsbA/DsbB oxidoreductase system requires the GGDEF-EAL domain protein STM3615. PLoS One. 2014;9:e106095. doi: 10.1371/journal.pone.0106095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, Princy SA. An insight into pleiotropic regulators Agr and Sar: molecular probes paving the new way for antivirulent therapy. Future Microbiol. 2013;8:1339–1353. doi: 10.2217/fmb.13.92. [DOI] [PubMed] [Google Scholar]
- Aspe M, Jensen L, Melegrito J, Sun M. The role of alginate and extracellular DNA in biofilm-mediated Pseudomonas aeruginosa gentamicin resistance. J Exp Microbiol Immunol. 2012;16:42–48. [Google Scholar]
- Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A. 2005;102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraquet C, Harwood CS. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci U S A. 2013;110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardill JP, Hammer BK. Non-coding sRNAs regulate virulence in the bacterial pathogen Vibrio cholerae. RNA Biol. 2012;9:392–401. doi: 10.4161/rna.19975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol. 2009;191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj AK, Vinothkumar K, Rajpara N. Bacterial quorum sensing inhibitors: attractive alternatives for control of infectious pathogens showing multiple drug resistance. Recent Pat Antiinfect Drug Discov. 2013;8:68–83. doi: 10.2174/1574891X11308010012. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PØ, Burmølle M, Hentzer M, Haagensen JA, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Høiby N, Givskov M. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol. 2005;57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob Agents Chemother. 2011;55:2655–2661. doi: 10.1128/AAC.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady RA, Leid JG, Camper AK, Costerton JW, Shirtliff ME. Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect Immun. 2006;74:3415–3426. doi: 10.1128/IAI.00392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfee MW, Coleman JP, Pesci EC. Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2001;98:11633–11637. doi: 10.1073/pnas.201328498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol. 2004;186:1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JR, Sauer K. Small RNAs and their role in biofilm formation. Trends Microbiol. 2013;21:39–49. doi: 10.1016/j.tim.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez de Paz LE, Lemos JA, Wickström C, Sedgley CM. Role of (p)ppGpp in biofilm formation by Enterococcus faecalis. Appl Environ Microbiol. 2012;78:1627–1630. doi: 10.1128/AEM.07036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung PY, Toh YS. Anti-biofilm agents: recent breakthrough against multi-drug resistant Staphylococcus aureus. Pathog Dis. 2014;70:231–239. doi: 10.1111/2049-632X.12141. [DOI] [PubMed] [Google Scholar]
- Clarke SR, Foster SJ. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol. 2006;51:187–224. doi: 10.1016/S0065-2911(06)51004-5. [DOI] [PubMed] [Google Scholar]
- Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D, Lei MG, Luong TT, Kuechenmeister L, Dunman PM, O’Donnell S, Rowe S, O’Gara JP, Lee CY. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol. 2009;191:6363–6373. doi: 10.1128/JB.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugini C, Calfee MW, Farrow JM, 3rd, Morales DK, Pesci EC, Hogan DA. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:896–906. doi: 10.1111/j.1365-2958.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- Cushnie TP, Lamb AJ. Recent advances in understanding the antibacterial properties of flavonoids. Int J Antimicrob Agents. 2011;38:99–107. doi: 10.1016/j.ijantimicag.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- de Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relationships. Infect Immun. 2000;68:4839–4849. doi: 10.1128/IAI.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Stacey RE, Dodd C, Cámara M, Williams P, Winzer K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol. 2006;8:1095–1104. doi: 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- Donlan RM. Biofilm formation: a clinically relevant microbiological process. Clin Infect Dis. 2001;33:1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbrega A, Vila J. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev. 2013;26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol. 2014;16:1961–1981. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- Fechter P, Caldelari I, Lioliou E, Romby P. Novel aspects of RNA regulation in Staphylococcus aureus. FEBS Lett. 2014;588:2523–2529. doi: 10.1016/j.febslet.2014.05.037. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick F, Humphreys H, O’Gara JP. The genetics of staphylococcal biofilm formation—will a greater understanding of pathogenesis lead to better management of device-related infection? Clin Microbiol Infect. 2005;11:967–973. doi: 10.1111/j.1469-0691.2005.01274.x. [DOI] [PubMed] [Google Scholar]
- Galperin MY. Bacterial signal transduction network in a genomic perspective. Environ Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DL, White AP, Snyder SD, Martin S, Heiss C, Azadi P, Surette M, Kay WW. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J Bacteriol. 2006;188:7722–7730. doi: 10.1128/JB.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ Microbiol. 2005;7:894–906. doi: 10.1111/j.1462-2920.2005.00775.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escobedo G, Marshall JM, Gunn JS. Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat Rev Microbiol. 2011;9:9–14. doi: 10.1038/nrmicro2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez JA, Crowder T, Rinaldo-Matthis A, Ho MC, Almo SC, Schramm VL. Transition state analogs of 5’-methylthioadenosine nucleosidase disrupt quorum sensing. Nat Chem Biol. 2009;5:251–257. doi: 10.1038/nchembio.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- He H, Cooper JN, Mishra A, Raskin DM. Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol. 2012;194:2962–2972. doi: 10.1128/JB.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Høiby N, Givskov M. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- Holland LM, O’Donnell ST, Ryjenkov DA, Gomelsky L, Slater SR, Fey PD, Gomelsky M, O’Gara JP. A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J Bacteriol. 2008;190:5178–5189. doi: 10.1128/JB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JJ, Han JI, Zhang LH, Leadbetter JR. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2003;69:5941–5949. doi: 10.1128/AEM.69.10.5941-5949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangyutitham V, Güvener ZT, Harwood CS. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. MBio. 2013;4 doi: 10.1128/mBio.00242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain FM, Ahmad I, Asif M, Tahseen Q. Influence of clove oil on certain quorum-sensing-regulated functions and biofilm of Pseudomonas aeruginosa and Aeromonas hydrophila. J Biosci. 2013;38:835–844. doi: 10.1007/s12038-013-9385-9. [DOI] [PubMed] [Google Scholar]
- Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara Y, Hyodo M, Hayakawa Y, Kamegaya T, Yamada K, Okamoto A, Hasegawa T, Ohta M. Effect of cyclic bis(3’-5’)diguanylic acid and its analogs on bacterial biofilm formation. FEMS Microbiol Lett. 2009;301:193–200. doi: 10.1111/j.1574-6968.2009.01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabra-Rizk MA, Meiller TF, James CE, Shirtliff ME. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob Agents Chemother. 2006;50:1463–1469. doi: 10.1128/AAC.50.4.1463-1469.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Behrens AJ, Kaever V, Kazmierczak BI. Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of cyclic di-GMP concentrations. J Bacteriol. 2012;194:4285–4294. doi: 10.1128/JB.00803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen TH, van Gennip M, Phipps RK, Shanmugham MS, Christensen LD, Alhede M, Skindersoe ME, Rasmussen TB, Friedrich K, Uthe F, Jensen PØ, Moser C, Nielsen KF, Eberl L, Larsen TO, Tanner D, Høiby N, Bjarnsholt T, Givskov M. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother. 2012;56:2314–2325. doi: 10.1128/AAC.05919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson KK, Pier DB, Goldmann DA, Pier GB. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol. 2004;186:2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci U S A. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H-S, Otto M. Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem Biol. 2012;19:1503–1513. doi: 10.1016/j.chembiol.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Kalia VC, Purohit HJ. Quenching the quorum sensing system: potential antibacterial drug targets. Crit Rev Microbiol. 2011;37:121–140. doi: 10.3109/1040841X.2010.532479. [DOI] [PubMed] [Google Scholar]
- Kalia VC, Wood TK, Kumar P. Evolution of resistance to quorum-sensing inhibitors. Microb Ecol. 2014;68:13–23. doi: 10.1007/s00248-013-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Z, Zhang C, Zhang J, Jin P, Zhang J, Du G, Chen J. Small RNA regulators in bacteria: powerful tools for metabolic engineering and synthetic biology. Appl Microbiol Biotechnol. 2014;98:3413–3424. doi: 10.1007/s00253-014-5569-y. [DOI] [PubMed] [Google Scholar]
- Karaolis DK, Rashid MH, Chythanya R, Luo W, Hyodo M, Hayakawa Y. c-di-GMP (3’-5’-cyclic diguanylic acid) inhibits Staphylococcus aureus cell–cell interactions and biofilm formation. Antimicrob Agents Chemother. 2005;49:1029–1038. doi: 10.1128/AAC.49.3.1029-1038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, Philpott D, Schroeder JT, Hyodo M, Hayakawa Y, Talbot BG, Brouillette E, Malouin F. Bacterial c-di-GMP is an immunostimulatory molecule. J Immunol. 2007;178:2171–2181. doi: 10.4049/jimmunol.178.4.2171. [DOI] [PubMed] [Google Scholar]
- Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski MR, Horswill AR. New approaches for treating staphylococcal biofilm infections. Ann N Y Acad Sci. 2011;1241:104–121. doi: 10.1111/j.1749-6632.2011.06281.x. [DOI] [PubMed] [Google Scholar]
- Kim S, Reyes D, Beaume M, Francois P, Cheung A. Contribution of teg49 small RNA in the 5’ upstream transcriptional region of sarA to virulence in Staphylococcus aureus. Infect Immun. 2014;82:4369–4379. doi: 10.1128/IAI.02002-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T, Perron GG, Buckling A, van Delden C. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog. 2010;6:e1000883. doi: 10.1371/journal.ppat.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni HM, Jagannadham MV. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology. 2014;160:2109–2121. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- Kullik I, Giachino P. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- Lasa I, Penadés JR. Bap: a family of surface proteins involved in biofilm formation. Res Microbiol. 2006;157:99–107. doi: 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penadés JR, Lasa I. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol. 2005;58:1322–1339. doi: 10.1111/j.1365-2958.2005.04907.x. [DOI] [PubMed] [Google Scholar]
- Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledeboer NA, Frye JG, McClelland M, Jones BD. Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect Immun. 2006;74:3156–3169. doi: 10.1128/IAI.01428-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequette Y, Greenberg EP. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187:37–44. doi: 10.1128/JB.187.1.37-44.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Natl Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J Bacteriol. 2013;195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Silva AJ, Benitez JA. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl Environ Microbiol. 2007;73:7482–7487. doi: 10.1128/AEM.01564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Bassler BL. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol. 2000;36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- Ma Q, Zhang G, Wood TK. Escherichia coli BdcA controls biofilm dispersal in Pseudomonas aeruginosa and Rhizobium meliloti. BMC Res Notes. 2011;4:447. doi: 10.1186/1756-0500-4-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- Maeda T, García-Contreras R, Pu M, Sheng L, Garcia LR, Tomás M, Wood TK. Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 2012;6:493–501. doi: 10.1038/ismej.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone JG, Jaeger T, Manfredi P, Dötsch A, Blanka A, Bos R, Cornelis GR, Häussler S, Jenal U. The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog. 2012;8:e1002760. doi: 10.1371/journal.ppat.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S, Cheng R, Miao C, Gong Q, Gu Y, Lu X, Han F, Yu W. Artificial trans-encoded small non-coding RNAs specifically silence the selected gene expression in bacteria. Nucleic Acids Res. 2011;39:e50. doi: 10.1093/nar/gkr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Guillier M. Expanding control in bacteria: interplay between small RNAs and transcriptional regulators to control gene expression. Curr Opin Microbiol. 2013;16:125–132. doi: 10.1016/j.mib.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson M, Nielsen A, Kjærulff L, Gotfredsen CH, Wietz M, Ingmer H, Gram L, Larsen TO. Inhibition of virulence gene expression in Staphylococcus aureus by novel depsipeptides from a marine photobacterium. Mar Drugs. 2011;9:2537–2552. doi: 10.3390/md9122537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden JN, Diaz MR, Walton WG, Gode CJ, Betts L, Urbanowski ML, Redinbo MR, Yahr TL, Wolfgang MC. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2013;110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowska K, Grudniak AM, Wolska KI. Silver nanoparticles as an alternative strategy against bacterial biofilms. Acta Biochim Pol. 2013;60:523–530. [PubMed] [Google Scholar]
- Masák J, Čejková A, Schreiberová O, Rezanka T. Pseudomonas biofilms: possibilities of their control. FEMS Microbiol Ecol. 2014;89:1–14. doi: 10.1111/1574-6941.12344. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, Brandenburg K, Whiteley M. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol. 2008;69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3’-5’)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- Michaux C, Verneuil N, Hartke A, Giard J-C. Physiological roles of small RNA molecules. Microbiology. 2014;160:1007–1019. doi: 10.1099/mic.0.076208-0. [DOI] [PubMed] [Google Scholar]
- Mika F, Hengge R. Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int J Mol Sci. 2013;14:4560–4579. doi: 10.3390/ijms14034560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci U S A. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Phenol-soluble modulins. Int J Med Microbiol. 2014;304:164–169. doi: 10.1016/j.ijmm.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter KL, Krishna A, Wigneshweraraj S, Edwards AM. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol. 2014;22:676–685. doi: 10.1016/j.tim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Palliyil S, Downham C, Broadbent I, Charlton K, Porter AJ. High-sensitivity monoclonal antibodies specific for homoserine lactones protect mice from lethal Pseudomonas aeruginosa infections. Appl Environ Microbiol. 2014;80:462–469. doi: 10.1128/AEM.02912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol. 2006;188:4861–4870. doi: 10.1128/JB.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Ren D. Quorum sensing inhibitors: a patent overview. Expert Opin Ther Pat. 2009;19:1581–1601. doi: 10.1517/13543770903222293. [DOI] [PubMed] [Google Scholar]
- Park J, Jagasia R, Kaufmann GF, Mathison JC, Ruiz DI, Moss JA, Meijler MM, Ulevitch RJ, Janda KD. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem Biol. 2007;14:1119–1127. doi: 10.1016/j.chembiol.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci U S A. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Palma M, Nilsson M, Guss B, Flock JI. Functional studies of a fibrinogen binding protein from Staphylococcus epidermidis. Infect Immun. 1999;67:4525–4530. doi: 10.1128/iai.67.9.4525-4530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez LJ, Karagounis TK, Hurley A, Bassler BL, Semmelhack MF. Highly potent, chemically stable quorum sensing agonists for Vibrio cholerae. Chem Sci. 2014;5:151–155. doi: 10.1039/C3SC52572D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A. 2012;109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A, Otto M. Phenol-soluble modulins and staphylococcal infection. Nat Rev Microbiol. 2013;11:667–673. doi: 10.1038/nrmicro3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyuta V, Zaitseva J, Lobakova E, Zagoskina N, Kuznetsov A, Khmel I. Effect of plant phenolic compounds on biofilm formation by Pseudomonas aeruginosa. APMIS. 2013;121:1073–1081. doi: 10.1111/apm.12083. [DOI] [PubMed] [Google Scholar]
- Rachid S, Ohlsen K, Wallner U, Hacker J, Hecker M, Ziebuhr W. Alternative transcription factor sigma(B) is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J Bacteriol. 2000;182:6824–6826. doi: 10.1128/JB.182.23.6824-6826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romilly C, Caldelari I, Parmentier D, Lioliou E, Romby P, Fechter P. Current knowledge on regulatory RNAs and their machineries in Staphylococcus aureus. RNA Biol. 2012;9:402–413. doi: 10.4161/rna.20103. [DOI] [PubMed] [Google Scholar]
- Römling U. Cyclic di-GMP, an established secondary messenger still speeding up. Environ Microbiol. 2012;14:1817–1829. doi: 10.1111/j.1462-2920.2011.02617.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;272:541–561. doi: 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de los Mozos I, Vergara-Irigaray M, Segura V, Villanueva M, Bitarte N, Saramago M, Domingues S, Arraiano CM, Fechter P, Romby P, Valle J, Solano C, Lasa I, Toledo-Arana A. Base pairing interaction between 5’- and 3’-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 2013;9:e1004001. doi: 10.1371/journal.pgen.1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambanthamoorthy K, Sloup RE, Parashar V, Smith JM, Kim EE, Semmelhack MF, Neiditch MB, Waters CM. Identification of small molecules that antagonize diguanylate cyclase enzymes to inhibit biofilm formation. Antimicrob Agents Chemother. 2012;56:5202–5211. doi: 10.1128/AAC.01396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambanthamoorthy K, Luo C, Pattabiraman N, Feng X, Koestler B, Waters CM, Palys TJ. Identification of small molecules inhibiting diguanylate cyclases to control bacterial biofilm development. Biofouling. 2014;30:17–28. doi: 10.1080/08927014.2013.832224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutera S, Zucca M, Savoia D. Novel approaches for the design and discovery of quorum-sensing inhibitors. Expert Opin Drug Discov. 2014;9:353–366. doi: 10.1517/17460441.2014.894974. [DOI] [PubMed] [Google Scholar]
- Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, Agolory S, Altman DR, Holzman RS, Kreiswirth BN, Novick RP. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2010;202:1593–1599. doi: 10.1086/656915. [DOI] [PubMed] [Google Scholar]
- Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev. 2011;35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- Sintim HO, Smith JA, Wang J, Nakayama S, Yan L. Paradigm shift in discovering next-generation anti-infective agents: targeting quorum sensing, c-di-GMP signaling and biofilm formation in bacteria with small molecules. Future Med Chem. 2010;2:1005–1035. doi: 10.4155/fmc.10.185. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Hüttenhofer A, Haas D, Bläsi U. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol. 2011;80:868–885. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Waters CM. A tangled web: regulatory connections between quorum sensing and cyclic Di-GMP. J Bacteriol. 2012;194:4485–4493. doi: 10.1128/JB.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SY, Chew SC, Tan SY, Givskov M, Yang L. Emerging frontiers in detection and control of bacterial biofilms. Curr Opin Biotechnol. 2014;26:1–6. doi: 10.1016/j.copbio.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Teschler JK, Zamorano-Sánchez D, Utada AS, Warner CJA, Wong GCL, Linington RG, Yildiz FH. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol. 2015;13:255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden A, Spirig T, Hilbi H. Bacterial gene regulation by alpha-hydroxyketone signaling. Trends Microbiol. 2010;18:288–297. doi: 10.1016/j.tim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Tielker D, Hacker S, Loris R, Strathmann M, Wingender J, Wilhelm S, Rosenau F, Jaeger KE. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology. 2005;151:1313–1323. doi: 10.1099/mic.0.27701-0. [DOI] [PubMed] [Google Scholar]
- Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One. 2008;3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–223. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich M, Bastian M, Cramton SE, Ziegler K, Pragman AA, Bragonzi A, Memmi G, Wolz C, Schlievert PM, Cheung A, Döring G. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol. 2007;65:1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x. [DOI] [PubMed] [Google Scholar]
- Vance RE, Zhu J, Mekalanos JJ. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect Immun. 2003;71:2571–2576. doi: 10.1128/IAI.71.5.2571-2576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem DA, Mihalik K, Crixell SH, McLean RJ. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia. 2007;78:302–310. doi: 10.1016/j.fitote.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188:706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- Wade DS, Calfee MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005;187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst JM, Saqi M, Littler E, Curtis MA. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics. 2006;7:162. doi: 10.1186/1471-2164-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wood TK. Toxin–antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron S, Römling U. Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb Biotechnol. 2014;7:496–516. doi: 10.1111/1751-7915.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- Zhou L, Zheng H, Tang Y, Yu W, Gong Q. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotechnol Lett. 2013;35:631–637. doi: 10.1007/s10529-012-1126-x. [DOI] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella Typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]