Abstract
Development of colorectal cancer (CRC) associates with accumulation of genetic mutations include the epidermal growth factor receptor (EGFR) signaling pathway. However, whether mutations in KRAS together with downstream factors BRAF, PIK3CA and NRAS impact prognosis is still unclear for stage II-III colon cancer. In the present study a total of 228 stage II-III colon cancer samples were retrospectively collected, KRAS (codons 12, 13 and 61), BRAF (exon 11 and exon 15), PIK3CA (exon 9 and exon 20) and NRAS (codons 12, 13 and 61) status was detected by Sanger sequencing, 37.89% (86/227) tumors harbored a KRAS mutation, 7.02% (16/228) harbored a BRAF mutation, 13.18% (29/220) harbored a PIK3CA mutation and 0.89% (2/224) harbored a NRAS mutation. NRAS mutations existed only in stage II colon cancer. Older groups harbored a higher KRAS and BRAF mutation (P < 0.05), PIK3CA (exon9) mutations appeared more common in worse differentiation tumors (P = 0.032). Moreover, PIK3CA (E545K) mutation was significantly associated with tumor recurrence (P = 0.031) and acted independently prognostic for poor OS (P = 0.044), while only in stage III colon cancer. KRAS, BRAF and NRAS mutations do not have major prognostic value in stage II and III colon cancer, subtypes of gene mutations should be further investigated for a better understanding in CRC.
Colorectal cancer (CRC) is ranking the third most common cancer worldwide and the fourth leading cause of cancer-related death, which makes CRC a heavy burden in many countries and districts, including Asia1,2. Since the precision medicine initiated this year, complex diseases such as cancer and diabetes seem to start a new Odyssey3,4. Targeted therapy has already achieved great progress in many kinds of cancers included CRC, the monoclonal antibody (MoAb) cetuximab or panitumumab which targeted on epidermal growth factor receptor (EGFR) prolonged survival of metastatic CRC (mCRC) patients5, although only selected groups may benefit from these agents. However, patients with stage II or III colon cancer experienced obstacles with targeted therapy, no matter anti-EGFR or anti-VEGF MoAbs, which both did not appear effective when added to chemotherapy6,7,8.
Although KRAS mutation status test is recommended before targeted therapy selection9, the prognostic value of KRAS in early stage of colon cancer remains controversial. Previous studies are inconsistent in the impact of KRAS mutation on recurrence and death for patients with chemotherapy in early stage colon cancer10,11,12,13. BRAF gene consists one of the RAF gene family, which encodes a serine/threonine protein kinase belonging to the RAS-RAF-MEK pathway regulated by activated KRAS14,15. Generally, KRAS and BRAF mutations are mutually exclusive, moreover, BRAF indicates poorer prognosis for mCRC patients treated with anti-EGFR therapy16, while the role of BRAF in other cancer stage is still unclear, reported studies did not sustain accordance about this issue17,18,19,20. Since a myriad of PIK3CA-AKT pathway aberrations have been observed in human cancers, gain-of-function mutations of this gene appeared in many cancer types included CRC21,22. As the main effector in the downstream of EGFR intracellular signaling pathway, PIK3CA mutation has appeared a negative effect in MoAbs treatment in mCRC patients16. However, there exists limited data on mutant PIK3CA associated with prognostic value for stage II or III cancers. NRAS is closely to KRAS, which also a member of RAS gene23, although NRAS gene mutation is rare in CRC, approximately 2.2–4.19%16,22,24, it appears a valuable prognostic biomarker in anti-EGFR MoAbs therapy for mCRC patients5,25, then NRAS status for indicating prognosis should be further confirmed in early stage cancers.
The tumorigenesis and development of CRC is a multistep process with different genetic mutation accumulation26, driver mutations included somatic changes in KRAS, BRAF, PIK3CA and NRAS represent the main aspect in genetic alternations for CRC27. To date, major of the presented data were derived from western populations and few data was available for the Chinese. Herein we design this study to investigate the mutation spectrum in stage II/III colon cancer and the potential correlations with clinicopathological characteristics, furthermore, survival data of patients treated with 5-fluorouracil (5-FU)/oxaliplatin-based chemotherapy was collected, which may provide an appropriate insight between gene mutation and survival status in Chinese populations.
Results
Frequency and association with tumor mutations
In the cohort of 228 colon cancer patients, 124 stage II and 104 stage III tumor specimens were collected for gene mutation analysis. KRAS (codons 12, 13 and 61) mutants were found in 37.89% (86/227) cases, 26.87% (61/227) in codon 12, 6.61% (15/227) in codon 13 and 3.54% (8/226) in codon 61. The corresponding order of KRAS codon 12 mutation was G12D, G12V, G12C, G12S and G12A. For codon 13, 93.3% (14/15) mutations were G13D, and followed by G13S. Codon 13 mutations were more frequent in stage II colon cancers, with a significant association compared to stage III tumors (10.5% vs. 1.9%, P = 0.013). Q61H was the most frequent mutant subtype in codon 61, and Q61L together with Q61R were also observed in this study.
The status of BRAF (exon 11 and exon 15) mutations was detected in all samples, 7.02% (16/228) tumors harbored a BRAF mutation, 1.75% (4/228) in exon 11 and 5.26% (12/228) in exon 15. V600E was the major subtype in BRAF mutation (43.75%, 7/16), which appeared only in KRAS wild type (P = 0.046), while BRAF mutation was not mutually exclusive with KRAS mutation, for exon 11 or exon 15 (V600M) could be found in KRAS mutant tumors.
PIK3CA (exon 9 and exon 20) mutations could not be assigned to 3.5% (8/228) samples, 13.18% (29/220) harbored a PIK3CA mutation, 10.45% (23/220) in exon 9 and 2.7% (6/221) in exon 20. E545K mutation in exon 9 was more frequent than other subtypes, followed by E542K and E545G, we also found E545D, Q546R and Q546K mutation in these samples. The H1047R consisted 83.3% (5/6) mutations in exon 20, and only one specimen harbored a H1047Y mutation. PIK3CA mutation appeared a strong association with KRAS mutation, 21.69% (18/83) in mutants versus 8.09% (11/136) in wild types (P = 0.005). However, only exon 9 mutation had the association (P = 0.022) and exon 20 did not share this (P = 0.202).
NRAS (codons 12, 13 and 61) mutations existed only in stage II colon cancer, 0.89% (2/224) tumors harbored a NRAS mutation, of which were G12D in codon 12 and Q61R in codon 61. The rare mutants in NRAS did not yield any significant association with other mutants. Besides, we also found that NRAS and other gene mutations were mutually exclusive in this cohort.
Clinicopathological characteristics of KRAS/BRAF/PIK3CA/NRAS gene mutations
In the univariate analysis, both KRAS and BRAF mutations were more frequent in older patients (P < 0.05), and female patients shared higher KRAS mutations (codon 12 and codon 61) frequency, meanwhile patients with tumor family history showed more KRAS (G12D) mutations (30.4% vs. 11.8%, P = 0.023) and stage II tumors harbored a higher KRAS codon 13 mutations (10.5% vs. 1.9%, P = 0.013). Moreover, with N stage accelerated, KRAS codon 13 mutations declined (P < 0.05). We found BRAF mutants only existed in non-smokers (P = 0.046) and no other significant differences were discovered when compared to wild type. As for PIK3CA mutations, no significant association appeared between gene mutants and the characteristics, while subgroup mutants such asPIK3CA (exon9) mutations appeared more common in worse differentiation tumors (P = 0.032), and exon 20 mutations did not share this (P = 0.593). Besides, exon 9 (E545K) mutation was significantly associated with tumor recurrence (10.7% vs. 0.0%, P = 0.031) in stage III patients, but not for stage II or combined together. NRAS mutations were too rare to attain significant associations with any clinicopathological characteristics in this study (Table 1).
Table 1. Characteristics of 228 colon cancer patients and association of gene mutations with clinicopathological parameters.
| Characteristics | Number (%) |
KRAS |
BRAF |
PI3KCA |
NRAS |
||||
|---|---|---|---|---|---|---|---|---|---|
| Mutations (%) | P | Mutations (%) | P | Mutations (%) | P | Mutations (%) | P | ||
| Sex | |||||||||
| Male | 137 (60.1) | 49 (36.0) | 0.489 | 8 (5.8) | 0.434 | 18 (13.5) | 0.849 | 0 (0) | 0.157 |
| Female | 91 (39.9) | 37 (40.7) | 8 (8.8) | 11 (12.6) | 2 (2.2) | ||||
| Age | |||||||||
| ≤60 | 111 (48.7) | 34 (30.9) | 0.041 | 4 (3.6) | 0.049 | 13 (12.1) | 0.694 | 1 (0.9) | 0.96 |
| >60 | 117 (51.3) | 52 (44.4) | 12 (10.3) | 16 (14.2) | 1 (0.9) | ||||
| Mean | 60 ± 12 | ||||||||
| Range | 23-84 | ||||||||
| Tumor family history | |||||||||
| Yes | 23 (10.2) | 9 (40.9) | 0.82 | 0 (0) | 0.382 | 6 (26.1) | 0.09 | 0 (0) | 1.0 |
| No | 203 (89.0) | 77 (37.9) | 16 (7.9) | 22 (11.3) | 2 (1.0) | ||||
| Missing | 2 (0.8) | ||||||||
| Tumor location* | |||||||||
| Right | 107 (46.9) | 43 (40.2) | 0.584 | 7 (6.5) | 0.792 | 18 (17.1) | 0.097 | 1 (0.9) | 1.0 |
| Left | 121 (53.1) | 43 (35.8) | 9 (7.4) | 11 (9.6) | 1 (0.8) | ||||
| Tumor differentiation | |||||||||
| Well /Moderate | 191 (84.1) | 69 (36.3) | 0.356 | 13 (6.8) | 0.724 | 21 (11.5) | 0.104 | 2 (1.1) | 1.0 |
| Poor | 36 (15.5) | 16 (44.4) | 3 (8.3) | 8 (22.2) | 0 (0) | ||||
| Missing data | 1 (0.4) | ||||||||
| Tumor stage↑ | |||||||||
| II | 124 (54.4) | 48 (38.7) | 0.779 | 8 (6.5) | 0.715 | 14 (11.8) | 0.5 | 2 (1.7) | 0.5 |
| III | 104 (45.6) | 38 (36.9) | 8 (7.7) | 15 (14.9) | 0 (0) | ||||
| Depth of invasion↑ | |||||||||
| T2 | 7 (3.1) | 3 (42.9) | 1.0 | 0 (0) | 1.0 | 0 (9.7) | 0.122 | 0 (0.0) | 1.0 |
| T3 | 207 (91.2) | 78 (37.9) | 15 (7.2) | 25 (12.6) | 2 (1.0) | ||||
| T4 | 13 (5.3) | 5 (38.5) | 1 (7.7) | 4 (30.8 | 0 (0.0) | ||||
| Missing data | 1 (0.4) | ||||||||
| Nodal stage↑ | |||||||||
| N0 | 124 (54.4) | 49 (39.5) | 0.62 | 8 (6.5) | 0.94 | 14 (11.8) | 0.768 | 2 (1.7) | 1.0 |
| N1 | 58 (25.5) | 19 (32.8) | 5 (8.6) | 8 (14.5) | 0 (0.0) | ||||
| N2 | 45 (19.7) | 18 (40.9) | 3 (6.7) | 7 (15.6) | 0 (0.0) | ||||
| Missing data | 1 (0.4) | ||||||||
| Relapse | |||||||||
| Yes | 35 (15.3) | 14 (41.2) | 0.699 | 3 (8.6) | 0.725 | 4 (11.4) | 1.0 | 0 (0.0) | 1.0 |
| No | 170 (74.6) | 64 (37.6) | 12 (7.1) | 22 (13.6) | 2 (1.2) | ||||
| Missing data | 23 (10.1) | ||||||||
Tumor location*: Right is defined as right colon and transverse colon; left is defined as left colon and sigmoid colon.
↑: Seventh edition of the AJCC/UICC TNM staging systems.
Univariate analysis of outcomes predictors
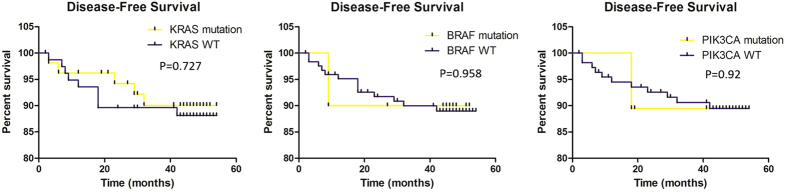
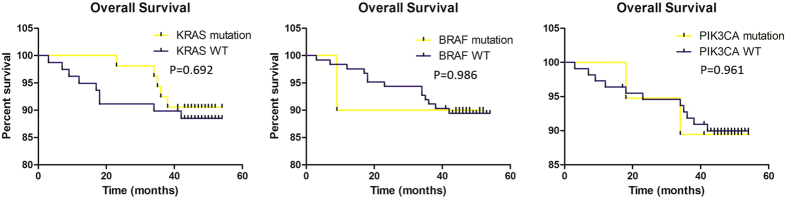
The median follow-up for outcomes analysis in this cohort was 46.0 months, 55.9% (66/118) stage II and 84.9% (79/93) stage III patients had received 5-FU/ oxaliplatin-based standard chemotherapy after surgery, consequent treatment of the other 17 patients were unclear. Only patients who received adjuvant chemotherapy after surgery were recruited in the final analysis and none of them had accepted radiotherapy or anti-EGFR/VEGF MoAbs targeted therapy. No significant difference in DFS was observed between patients with KRAS wild type and mutants (P = 0.727), furthermore subgroups such as codon 12/13 and mutation types (G12D/G12V/G12C/G12S/G13D) did not appear as prognostic factors for DFS, stratified by tumor stage or analyzed together, we also failed to discover significant associations between BRAF/PIK3CA/NRAS mutations and DFS (Fig. 1). However, KRAS codon 61 and PIK3CA (E545K) mutations were prognostic for DFS in stage III alone, but not in stage II or the whole population. KRAS mutation status was not prognostic for OS, no matter analyzed together or separately. Other gene mutations also did not obtain significance with OS (Fig. 2), while similarly KRAS codon 61 and PIK3CA (E545K) mutants showed prognostic for OS (Table 2). We also discovered that tumor differentiation had significant relationship with DFS and OS, in patients for stage III and combined together (P < 0.05).
Figure 1. Associations between gene mutations and disease-free survival (DFS).
No significant associations existed. P value for KRAS/BRAF/PIK3CA gene were 0.727, 0.958 and 0.92, respectively. WT: wild type.
Figure 2. Associations between gene mutations and over-all survival (OS).
No significant associations existed. P value for KRAS/BRAF/PIK3CA gene were 0.692, 0.986 and 0.961, respectively. WT: wild type.
Table 2. Univariate analysis of outcome predictors.
| DFS |
OS |
|||||||
|---|---|---|---|---|---|---|---|---|
| Stage/Gene | Mutant Number | P | Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | |
| KRAS | ||||||||
| Stage II | 28 | 0.389 | 2.749 | 0.249 to 30.324 | 0.400 | 2.802 | 0.254 to 30.911 | |
| Stage III | 25 | 0.412 | 0.574 | 0.152 to 2.163 | 0.404 | 0.568 | 0.151 to 2.144 | |
| Stage II and III | 53 | 0.727 | 0.824 | 0.276 to 2.458 | 0.692 | 0.802 | 0.269 to 2.393 | |
| BRAF | ||||||||
| Stage II | 6* | – | – | – | – | – | – | |
| Stage III | 4 | 0.578 | 1.792 | 0.229 to 14.030 | 0.563 | 1.833 | 0.234 to 14.336 | |
| Stage II and III | 10 | 0.958 | 0.946 | 0.124 to 7.235 | 0.986 | 0.982 | 0.128 to 7.510 | |
| PIK3CA | ||||||||
| Stage II | 8* | – | – | – | – | – | – | |
| Stage III | 11 | 0.945 | 1.056 | 0.228 to 4.891 | 0.904 | 1.099 | 0.237 to 5.086 | |
| Stage II and III | 19 | 0.920 | 0.926 | 0.207 to 4.140 | 0.961 | 0.964 | 0.216 to 4.306 | |
*No death events in this group.
Multivariate analysis of outcome predictors
In the multivariate analysis, we selected sex, age, tumor differentiation, nodal stage, KRAS, BRAF and PIK3CA gene mutations in relation to DFS and OS by stage and in both stages combined together. The KRAS codon 61 mutation still achieved significance after adjustment, which acted as an independent prognostic factor for DFS and OS in stage III alone (P = 0.026 and 0.018 respectively). However, PIK3CA (E545K) mutant status was prognostic for OS (P = 0.044), while a significant trend for DFS only in stage III (P = 0.051). Other gene mutations still failed to obtain significance for DFS or OS in the multivariate analysis (mutant subtypes and tumor stage involved). The interaction emerged for DFS and OS with tumor differentiation still exited in patients for stage III and combined together (P < 0.05) (Table 3).
Table 3. Multivariate analysis of outcome predictors.
| Prognostic marker | Stage II (n = 66) |
Stage III (n = 79) |
Stage II/III (n = 145) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| DFS | P | Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI |
| Sex | 0.978 | 278790.681 | 0.0001 to ∞ | 0.464 | 1.701 | 0.411 to7.0 | 0.352 | 1.869 | 0.501 to 9.0 |
| Age | 0.419 | 2.729 | 0.239 to 31.2 | 0.152 | 3.199 | 0.652 to 15.7 | 0.069 | 3.305 | 0.91 to 12.0 |
| Tumor differentiation | 0.217 | 5.049 | 0.386 to 66.1 | 0.022 | 3.879 | 1.215 to 12.4 | 0.007 | 4.264 | 1.486 to 12.2 |
| Nodal stage | – | – | – | 0.895 | 1.086 | 0.317 to 3.7 | 0.142 | 1.675 | 0.841 to 3.3 |
| KRAS mutation | 0.542 | 2.234 | 0.169 to 29.5 | 0.372 | 0.543 | 0.142 to 2.1 | 0.702 | 0.807 | 0.269 to 2.4 |
| BRAF mutation | 0.990 | <0.0001 | 0.0001 to ∞ | 0.927 | 1.103 | 0.136 to 8.9 | 0.762 | 0.729 | 0.094 to 5.7 |
| PIK3CA mutation | 0.964 | <0.0001 | 0.0001 to ∞ | 0.383 | 0.461 | 0.081 to 2.6 | 0.368 | 0.48 | 0.097 to 2.4 |
| OS | |||||||||
| Sex | 0.978 | 285332.546 | 0.0001 to ∞ | 0.462 | 1.697 | 0.415 to 6.9 | 0.339 | 1.904 | 0.508 to 7.1 |
| Age | 0.454 | 2.537 | 0.222 to 28.9 | 0.112 | 3.623 | 0.742 to 17.7 | 0.050 | 3.59 | 0.98 to 12.9 |
| Tumor differentiation | 0.190 | 5.486 | 0.429 to 70.1 | 0.029 | 3.642 | 1.14 to 11.6 | 0.006 | 4.358 | 1.526 to 12.4 |
| Nodal stage | – | – | – | 0.934 | 1.054 | 0.305 to 3.6 | 0.170 | 1.618 | 0.813 to 3.2 |
| KRAS mutation | 0.540 | 2.234 | 0.171 to 29.2 | 0.334 | 0.514 | 0.133 to 2.0 | 0.567 | 0.725 | 0.24 to 2.2 |
| BRAF mutation | 0.990 | <0.0001 | 0.0001 to ∞ | 0.992 | 0.989 | 0.121 to 8.1 | 0.652 | 0.622 | 0.079 to 4.9 |
| PIK3CA mutation | 0.963 | <0.0001 | 0.0001 to ∞ | 0.430 | 0.496 | 0.087 to 2.8 | 0.427 | 0.523 | 0.105 to 2.6 |
Discussion
In this retrospective study, we evaluated common hot spot gene mutations located downstream of EGFR signaling pathway, the potential prognostic value for KRAS, BRAF, PIK3CA and NRAS was assessed in 228 stage II-III colon cancer. Since anti-EGFR MoAbs improved survival in mCRC patients based on specific RAS gene mutations5,28, biomarkers such as gene mutations or amplifications has sparked thriving researches for outcomes prognosis. As for stage II-III colon cancer, KRAS and BRAF gene mutations were evaluated frequently in different studies, however, no consistence was observed10,11,12,29. Gene mutation profiling alters for different ethnics was confirmed in previous studies22,30,31, then potential prognostic value of gene mutations should be reconsidered in Chinese populations.
Our data presented here demonstrated that PIK3CA (E545K) mutation was significantly associated with tumor recurrence in stage III, besides, E545K also played as an independent prognostic gene biomarker for adverse outcomes in stage III colon cancer alone. Although KRAS codon 61 appeared significant association with DFS and OS for stage III cancer, while only one patient died with mutant KRAS codon 61 tumor, considering the rare death event in this group, the magnitude of KRAS codon 61 status for prognosis was confused, which should be investigated further in the future. We did not discover any other gene mutant had impact for DFS or OS on stage II-III colon cancer patients treated with 5-FU/oxaliplatin-based standard chemotherapy, KRAS tumor mutation status has no major prognostic value for survival was consistent with a previous analysis in a large cohort, though BRAF mutations achieved significance for OS in their study11. Associations between KRAS or BRAF mutation and worse outcomes were reported in North Central Cancer Treatment Group N0147 trial, in which prognostic impact of KRAS and BRAF mutants were involved with stage III colon cancer patients received FOLFOX with or without cetuximab32. The adverse effects of KRAS or BRAF mutation in stage II/III colon cancer was not observed in this study, which may be due to the small sample size and consequent lower mutant tumors. However, considering the different treatment strategy in these studies (i.e. FU/FA alone or combined with irinotecan11), comparison of the prognostic value of these biomarkers should be more careful. Previous studies which involved Eastern Asia populations showed KRAS mutation was associated with worse prognosis in stage III or high-risk stage II colon cancer patients10, while lack of significant association between BRAF mutation and DFS was in accordance with our results and other clinical trial researches based on western populations11,12,33. Different populations and treatment regime may be responsible for the final outcomes, we should also notice that diverse therapies were performed after tumor relapse, the over-all survival of patients would be influenced in more aspects. To what extent the KRAS/BRAF gene mutations impact on survival in stage II/III colon patients should be further confirmed.
We found PIK3CA (E545K) was prognostic for poor OS independently in stage III colon cancers, the adverse effect of PIK3CA in early stage colon cancers were previously reported34,35, although other studies presented no associations between PIK3CA mutation and survival36. Reasons for the differences may include different target populations, covariates studied in the Cox model, diverse drugs usage and the methodologies for mutations detection. PIK3CA mutation (exon9) significantly associated with KRAS mutant tumors, which was consistent with reported results16,22,37, the exon9 mutations were highly dependent on RAS-GTP binding, especially in E542K and E545K38, which may suggest that PIK3CA mutations occur after KRAS mutations, in a later step during the tumorigenesis of CRC39. E545K also related to the tumor recurrence in stage III cancer patients, all the three E545K mutations appeared only in relapse tumors (10.7% vs. 0.0%, P = 0.031), while E545K was rare in this cohort because of the small sample size, and which consisted a relative small proportion (23.1%, 3/13) in PIK3CA exon9 mutants, then precise conclusions on the prognostic value of E545K should be investigated more in the future. NRAS mutations were only 0.89% (2/224) in this cohort, which was obviously lower than reported results (2.9–4.0%)13,36, the different detection method and study population may explain the distinct NRAS mutant frequency. We did not find significant associations between NRAS mutation and patients’ survival, which was consistent with previous studies12,13,36. In multivariate analysis, tumor differentiation appeared independently prognostic value in colon cancer patients for stage III and combined together, poor differentiation indicated more malignant tumors, and the results were in accordance with a recent study40, though the tumor differentiation acted as prognostic for survival only in univariate analysis in their study.
The frequencies of mutations in KRAS, BRAF, PIK3CA and NRAS were compared with previous researches, the KRAS and BRAF mutation frequencies were close to early studies11,12,18,41, however, data based on another population presented lower KRAS and BRAFmutants10, more sigmoid and rectum tumors involved in their cohort may contribute to the lower mutation frequency. Older patients harbored a higher gene mutation showed aberration accumulation with aging12. The PIK3CA mutation (13.18%) was lower than some previous data, which ranged from 12.2 to 20.18%12,35,36,42,43,44, ethnic population, various detection methods and whether rectum cancer involved may explain the difference, the PIK3CA mutation did not significantly associated with clinicopathological characteristics, which was consistent with reported results35,43,44. However, subtype of PIK3CA mutation may impact the tumor recurrence and survival, because few E545K mutants in our cohort, the relationship involved in this analysis may be not robust, then further studies or pool analysis of reported researches may focus on this. NRAS mutations were only 0.89% in present study, while another study based on high sensitive detection method showed a 2.9% mutation frequency12, and both studies did not discover any correlation between NRAS mutation and clinicopathological characteristics or patients’ survival. Considering rare data provided for NRAS mutations in stage II/III colon cancer, large population cohort investigation for this gene mutation detection should be performed, as NRAS closely related to KRAS and NRAS mutation was prognostic for EGFR MoAbs therapy inmCRC28. Several limitations were included in present study such as relatively small sample size and limited information of rare gene mutants to support confirmed conclusions. Though KRAS codon 61 appeared as prognostic biomarker in stage III cancer, the rare event may indicate indeterminacy at present, considering the low codon 61 mutation frequency in CRC22,27, confirmation of the prognostic value required further investigation with a larger sample size. Additionally, other potentially biomarkers for prognosis were not evaluated, which contained DNA mismatch repair (MMR) and microsatellite instability (MSI) status, or the CpG island methylator phenotype (CIMP). Reported studies provided the relationship between survival and these biomarkers, though the conclusions were still controversial10,12,18,32,35. We should strive to obtain more precise and simpler genome aberrations for prognosis, even in cancers marked as a complicated multistep disease, more investigations should be implemented to bring us a step closer to personalized medicine in the future.
In conclusion, we conducted this study to describe the gene mutation profiling in stage II/III colon cancer in Eastern Asia populations, PIK3CA (E545K) mutation was independently prognostic for OS in stage III and different subtypes of gene mutants should be further investigated in colon cancers, additional analysis with these molecular prognostic factors may provide a better insight and help us understand the mutation profiling in the evolution of this disease.
Methods
Patients
We totally recruited 124 stage II and 104 stage III colon cancer patients who received complete resection of tumor at the Cancer Hospital of the Chinese Academy of Medical Sciences (Beijing, China) between September 2010 and October 2011. None of them was treated with chemotherapy or radiotherapy previously and each of them was contacted to provide formalin-fixed, paraffin-embedded (FFPE) tumor tissues. Disease-free survival (DFS) was defined as time from operation to the first confirmed relapse, or alive without recurrence at last contact, and time to death or last follow-up from surgery was defined as overall survival (OS). The patients’ demographic and clinicopathological data are presented in Table 1.
Ethics statement
The experimental protocol in this study was approved by the Institutional Review Board (IRB) in Cancer Hospital of Chinese Academy of Medical Sciences and Peking Union Medical College, and informed consent was obtained from all patients. All experiments were carried out in accordance with relevant guidelines and regulations.
DNA isolation and mutation assessment
Hematoxylin and eosin (HE) staining was implemented to guarantee specimens tested contained >80% cancer cells, and areas enriched malignant cells were identified before DNA extraction by two independent pathologists. DNA was extracted by the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and stored at −80 °C until use.
Mutation hotspots in KRAS (codons 12 and 13), BRAF (exon15), PIK3CA (exon 9 and exon 20) and NRAS (codon 61) were detected, moreover, rare types of mutants for KRAS (codon 61 in exon 3), BRAF (exon 11) and NRAS (codons 12 and 13) were also included. We performed the polymerase chain reaction (PCR) amplification as followed for KRAS, BRAF, NRAS and PIK3CA exon 20: 1 min of initial denaturation at 95 °C, 35 cycles of amplification consisting of 30 s at 94 °C, 40 s at 57 °C, and 30 s at 72 °C, with a final additional elongation at 72 °C for 7 min. PIK3CA exon 9 amplification was carried out with a touchdown PCR program: 94 °C (2 min); 3 cycles of 94 °C (30 sec), 64 °C (30 sec), 70 °C (30 sec); 3 cycles of 94 °C (30 sec), 61 °C (30 sec), 70 °C (30 sec); 3 cycles of 94 °C (30 sec), 58 °C (30 sec), 70 °C (30 sec); 32 cycles of 94 °C for (30 sec), 57 °C (30 sec), 70 °C (40 sec); 1 cycle of 70 °C (5 min). Non-template control was included in each batch during the experiment, sequencing was applied using ABI 3500xL Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA).
Statistical analysis
Univariate relationships between gene mutation status and clinicopathological characteristics were performed by the Chi-square (χ2) test. Multivariate relationships were evaluated by fitting logistic regression analysis, using a backward stepwise (likelihood ratio) method and variables which showed statistically significant association with gene mutations were subjected to final regression analysis. The clinical database was last updated in March 2015. DFS and OS were done with the Kaplan-Meier survival function with the method of log-rank test, the hazard ratios were calculated with Cox proportional hazard regression models. All statistical tests were two-sided and the significance level was set at P < 0.05. Data analysis was performed by the SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA).
Additional Information
How to cite this article: Shen, Y.C. et al. Prognostic impact of mutation profiling in patients with stage II and III colon cancer. Sci. Rep. 6, 24310; doi: 10.1038/srep24310 (2016).
Acknowledgments
This study was supported in part by grants from the International Science & Technology Cooperation Program of China (2013DFE33110), Chinese National High Technology Research and Development Program of China (863 Program, 2011AA02A110), Chinese National Major Project for New Drug Innovation (2012ZX09303012; 2013ZX09101002).
Footnotes
Author Contributions Y.K.S., S.-H.L. and X.H.H. conceived and designed the research. Y.C.S. and J.F.W. performed the experiments. Y.K.S., S.-H.L., X.H.H., Y.C.S., J.F.W. and S.W. contributed to analyzing the results. H.Y.Y. provided samples, pathological diagnosis and analyzed the results. Y.K.S., Y.C.S., X.H.H. and S.-H.L. wrote the manuscript. All authors have reviewed and approved the final manuscript.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69 (2011). [DOI] [PubMed] [Google Scholar]
- Chen W., Zheng R., Zeng H., Zhang S. & He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 27, 2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. S. & Varmus H. A new initiative on precision medicine. N Engl J Med 372, 793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin M. A. Health: Make precision medicine work for cancer care. NATURE 520, 290 (2015). [DOI] [PubMed] [Google Scholar]
- Douillard J. Y. et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 369, 1023 (2013). [DOI] [PubMed] [Google Scholar]
- de Gramont A. et al. Bevacizumab plus oxaliplatin-based chemotherapy as adjuvant treatment for colon cancer (AVANT): a phase 3 randomised controlled trial. LANCET ONCOL 13, 1225 (2012). [DOI] [PubMed] [Google Scholar]
- Alberts S. R. et al. Effect of oxaliplatin, fluorouracil, and leucovorin with or without cetuximab on survival among patients with resected stage III colon cancer: a randomized trial. JAMA 307, 1383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegra C. J. et al. Phase III trial assessing bevacizumab in stages II and III carcinoma of the colon: results of NSABP protocol C-08. J CLIN ONCOL 29, 11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson A. R. et al. Metastatic colon cancer, version 3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw 11 141, 152 (2013). [DOI] [PubMed] [Google Scholar]
- Lee D. W. et al. KRAS mutation is associated with worse prognosis in stage III or high-risk stage II colon cancer patients treated with adjuvant FOLFOX. ANN SURG ONCOL 22, 187 (2015). [DOI] [PubMed] [Google Scholar]
- Roth A. D. et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J CLIN ONCOL 28, 466 (2010). [DOI] [PubMed] [Google Scholar]
- Gavin P. G. et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. CLIN CANCER RES 18, 6531 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov D. et al. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. AM J GASTROENTEROL 108, 1785 (2013). [DOI] [PubMed] [Google Scholar]
- Rajagopalan H. et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. NATURE 418, 934 (2002). [DOI] [PubMed] [Google Scholar]
- Davies H. et al. Mutations of the BRAF gene in human cancer. NATURE 417, 949 (2002). [DOI] [PubMed] [Google Scholar]
- De Roock W. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. LANCET ONCOL 11, 753 (2010). [DOI] [PubMed] [Google Scholar]
- Ogino S. et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. CLIN CANCER RES 18, 890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins G. et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J CLIN ONCOL 29, 1261 (2011). [DOI] [PubMed] [Google Scholar]
- Ogino S. et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. GUT 58, 90 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin P. G. et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. CLIN CANCER RES 18, 6531 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney K. D., Corcoran R. B. & Engelman J. A. The PI3K pathway as drug target in human cancer. J CLIN ONCOL 28, 1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. et al. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLOS ONE 8, e81628 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. NAT REV CANCER 3, 11 (2003). [DOI] [PubMed] [Google Scholar]
- Irahara N. et al. NRAS mutations are rare in colorectal cancer. DIAGN MOL PATHOL 19, 157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe A. et al. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC CANCER 15, 258 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz S. D. & Bertagnolli M. M. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 361, 2449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roock W., De Vriendt V., Normanno N., Ciardiello F. & Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. LANCET ONCOL 12, 594 (2011). [DOI] [PubMed] [Google Scholar]
- Al-Shamsi H. O., Alhazzani W. & Wolff R. A. Extended RAS testing in metastatic colorectal cancer-Refining the predictive molecular biomarkers. J Gastrointest Oncol 6, 314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins G. et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J CLIN ONCOL 29, 1261 (2011). [DOI] [PubMed] [Google Scholar]
- Kumar K. et al. Distinct BRAF (V600E) and KRAS mutations in high microsatellite instability sporadic colorectal cancer in African Americans. CLIN CANCER RES 15, 1155 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J THORAC ONCOL 9, 154 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinicrope F. A. et al. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX-based adjuvant chemotherapy. J CLIN ONCOL 31, 3664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S. et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. CLIN CANCER RES 18, 890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S. et al. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. INT J CANCER 121, 1771 (2007). [DOI] [PubMed] [Google Scholar]
- Ogino S. et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J CLIN ONCOL 27, 1477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin P. G. et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. CLIN CANCER RES 18, 6531 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. et al. Coexistent mutations of KRAS and PIK3CA affect the efficacy of NVP-BEZ235, a dual PI3K/MTOR inhibitor, in regulating the PI3K/MTOR pathway in colorectal cancer. INT J CANCER 133, 984 (2013). [DOI] [PubMed] [Google Scholar]
- Zhao L. & Vogt P. K. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA 105, 2652 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz S. D. & Bertagnolli M. M. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 361, 2449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. A prognostic analysis of 895 cases of stage III colon cancer in different colon subsites. INT J COLORECTAL DIS 30, 1173 (2015). [DOI] [PubMed] [Google Scholar]
- Ogino S. et al. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. CLIN CANCER RES 15, 7322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barault L. et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. INT J CANCER 122, 2255 (2008). [DOI] [PubMed] [Google Scholar]
- Benvenuti S. et al. PIK3CA cancer mutations display gender and tissue specificity patterns. HUM MUTAT 29, 284 (2008). [DOI] [PubMed] [Google Scholar]
- Abubaker J. et al. Clinicopathological analysis of colorectal cancers with PIK3CA mutations in Middle Eastern population. ONCOGENE 27, 3539 (2008). [DOI] [PubMed] [Google Scholar]