Abstract
c-MYC (MYC) overexpression or hyperactivation is one of the most common drivers of human cancer. Despite intensive study, the MYC oncogene remains recalcitrant to therapeutic inhibition. MYC is a transcription factor, and many of its pro-tumorigenic functions have been attributed to its ability to regulate gene expression programs1–3. Notably, oncogenic MYC activation has also been shown to increase total RNA and protein production in many tissue and disease contexts4–7. While such increases in RNA and protein production may endow cancer cells with pro-tumor hallmarks, this elevation in synthesis may also generate new or heightened burden on MYC-driven cancer cells to properly process these macromolecules8. Herein, we discover the spliceosome as a new target of oncogenic stress in MYC-driven cancers. We identify BUD31 as a MYC-synthetic lethal gene, and demonstrate that BUD31 is a component of the core spliceosome required for its assembly and catalytic activity. Core spliceosomal factors (SF3B1, U2AF1, and others) associated with BUD31 are also required to tolerate oncogenic MYC. Notably, MYC hyperactivation induces an increase in total pre-mRNA synthesis, suggesting an increased burden on the core spliceosome to process pre-mRNA. In contrast to normal cells, partial inhibition of the spliceosome in MYC-hyperactivated cells leads to global intron retention, widespread defects in pre-mRNA maturation, and deregulation of many essential cell processes. Importantly, genetic or pharmacologic inhibition of the spliceosome in vivo impairs survival, tumorigenicity, and metastatic proclivity of MYC-dependent breast cancers. Collectively, these data suggest that oncogenic MYC confers a collateral stress on splicing and that components of the spliceosome may be therapeutic entry points for aggressive MYC-driven cancers.
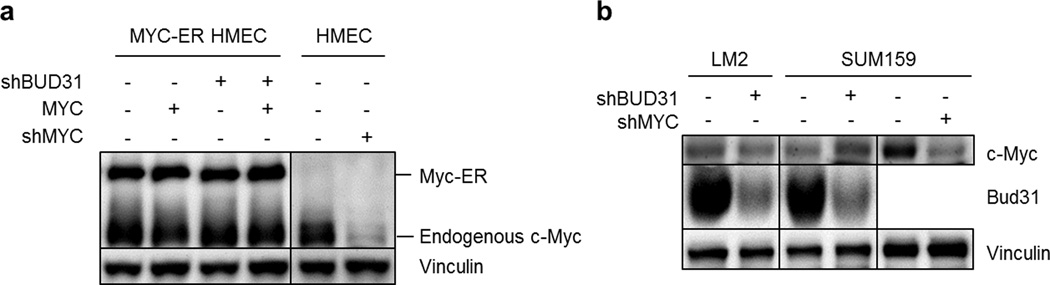
To discover genes and cellular processes required to tolerate oncogenic MYC, we previously performed a genome-wide MYC-synthetic lethal screen in human mammary epithelial cells engineered with inducible-MYC (MYC-ER HMECs) for candidates affecting cell viability in a MYC-selective manner9. This screen nominated BUD31 as a candidate MYC-synthetic lethal gene (Fig. 1a), wherein barcoded BUD31-shRNAs consistently dropped out of the population in MYC-hyperactivated cells relative to cells without MYC induction (Fig. 1b). In validation experiments, BUD31 depletion restrained clonogenic growth and activated apoptosis in MYC-induced cells, as compared to MYC-normal cells (Extended Data Fig. 1a–c). Expression of shRNA-resistant BUD31 rescued the MYC-synthetic lethal phenotype of BUD31-shRNA (Fig. 1c, Extended Data Fig. 1d), indicating the phenotype is an RNAi on-target effect.
Figure 1. The spliceosome is required for cells to tolerate oncogenic MYC hyperactivation.
a, BUD31 is a MYC-synthetic lethal gene. b, shBUD31 barcode abundances +/−MYC-ER hyperactivation (mean±s.e.m., n=3 biological replicates). c, Relative number of MYC-ER HMECs with dox-inducible shBUD31-UTR and constitutive Flag-GFP or -BUD31 expression (mean±s.e.m., n=4 technical replicates). d, Flag-BUD31 co-immunoprecipitation for core spliceosomal factors. e, Interaction between BUD31 and spliceosomal proteins assessed by BiFC (mean±s.e.m., n=3 technical replicates). f, GFP+ MYC-dependent cells with inducible shBUD31-UTR and constitutive WT, mutant BUD31, or negative control cDNA expression were mixed with GFP− cells and passaged (mean±s.e.m, n=8 technical replicates, two-tailed Student’s t-test). g, Change in MYC-ER HMEC clonogenicity after SD6 treatment (mean±s.e.m., n=4 technical replicates, two-tailed Student’s t-test). h–k, Relative number of MYC-ER HMECs after partial depletion of core spliceosomal proteins (mean±s.e.m., n=4 technical replicates, one-way ANOVA). **P<0.01, ***P<0.001.
BUD31 has been linked to the spliceosome in yeast10, but its function in mammalian systems has not been elucidated. To uncover the molecular function(s) of BUD31, we identified BUD31-interacting proteins via Flag-BUD31 immunoprecipitation from cells with or without RNase A (which eliminates protein-protein interactions mediated by RNA tethering), and mass spectrometry. Remarkably, 79 of 134 core spliceosomal components were associated with BUD31 (Extended Data Fig. 2a), suggesting a strong association between BUD31 and the spliceosome in human cells.
The spliceosome is a dynamic molecular machine consisting of several nuclear protein complexes that cycle on and off of pre-mRNA during intronic splicing11. Coimmuniprecipitation experiments confirmed that BUD31 associates with multiple subcomplexes of the spliceosome including the Prp19/CDC5L subcomplex (PRPF19), the U2 snRNP (SF3B1, SF3A1), U2-related factors (U2AF1), the U5 snRNP (EFTUD2), and Sm proteins (SNRPF) (Fig. 1d and Extended Data Fig. 2c), but interaction with non-spliceosomal proteins was not detected (Extended Data Fig. 2d–e). To test more broadly the association of BUD31 with subcomplexes of the spliceosome, we performed bimolecular fluorescence complementation (BiFC) between BUD31 and proteins from each major spliceosomal subcomplex. BiFC analysis indicated that BUD31 associates with components of the major snRNPs (U1, U2, U4/U6, and U5) as well as Sm proteins (Fig. 1e and Extended Data Fig. 2b), indicating that BUD31 is present at several stages of spliceosomal assembly.
To examine more directly whether BUD31 plays a role in pre-mRNA splicing, we tested in vitro splicing efficiency using nuclear extracts with or without BUD31 knockdown. BUD31 loss significantly inhibited pre-mRNA splicing (Extended Data Fig. 2f–i). In addition, knockdown of BUD31 led to defects in early spliceosome assembly, as indicated by impaired formation of complex A (Extended Data Fig. 2h–i). Collectively, these data indicate that HMECs require a core spliceosomal protein (BUD31) to tolerate dysregulated MYC.
We hypothesized that cells with oncogenic MYC required BUD31 for cell survival because of its role in the spliceosome. To test this hypothesis, we generated a BUD31 mutant deficient in binding core spliceosomal proteins by mutating a highly conserved region spanning a C2-C2 zinc finger. Mutation of this region abrogated BUD31 interaction with spliceosomal proteins (Extended Data Fig. 2j). To determine whether this region is also necessary for cells to tolerate MYC hyperactivation, we performed an in vitro competition assay. GFP-expressing MYC-driven breast cancer cells encoding inducible shBUD31 were transduced with shRNA-resistant wild-type or mutant BUD31 cDNA, and these cells were mixed with non-transduced, GFP-negative cells. BUD31 knockdown significantly inhibited proliferation of MYC-driven cancer cells. Proliferation was fully rescued by wild-type BUD31 cDNA but not a BUD31 mutant deficient in spliceosomal binding (Fig. 1f), suggesting BUD31 association with the spliceosome is required to support survival of MYC-hyperactivated cells. More broadly, these results indicate that oncogenic MYC may increase cellular dependency on spliceosome function. In contrast, ectopic expression of oncogenes HER2 and EGFR did not enhance the effects of BUD31 depletion (Extended Data Fig. 3a–b), suggesting the stress imposed by MYC on spliceosomal function is not a universal feature of the oncogenic state.
To test whether one or more subcomplexes of the spliceosome are required to tolerate aberrant MYC activity, we examined additional components of spliceosome assembly and catalysis including SF3B1 (U2 snRNP), U2AF1 (U2-related splicing factor), EFTUD2 (U5 snRNP), and SNRPF (core Sm protein found in every snRNP complex). Notably, partial depletion of each spliceosomal component led to loss of cell viability (Fig. 1h–k, Extended Data Fig. 4a–d) and increased apoptosis (Extended Data Fig. 4e–h) in MYC-hyperactivated cells. This suggests that (a) multiple subcomplexes of the core spliceosome are required for cells to tolerate oncogenic MYC, and (b) MYC-hyperactivated cells are sensitive to modest perturbations in spliceosome function.
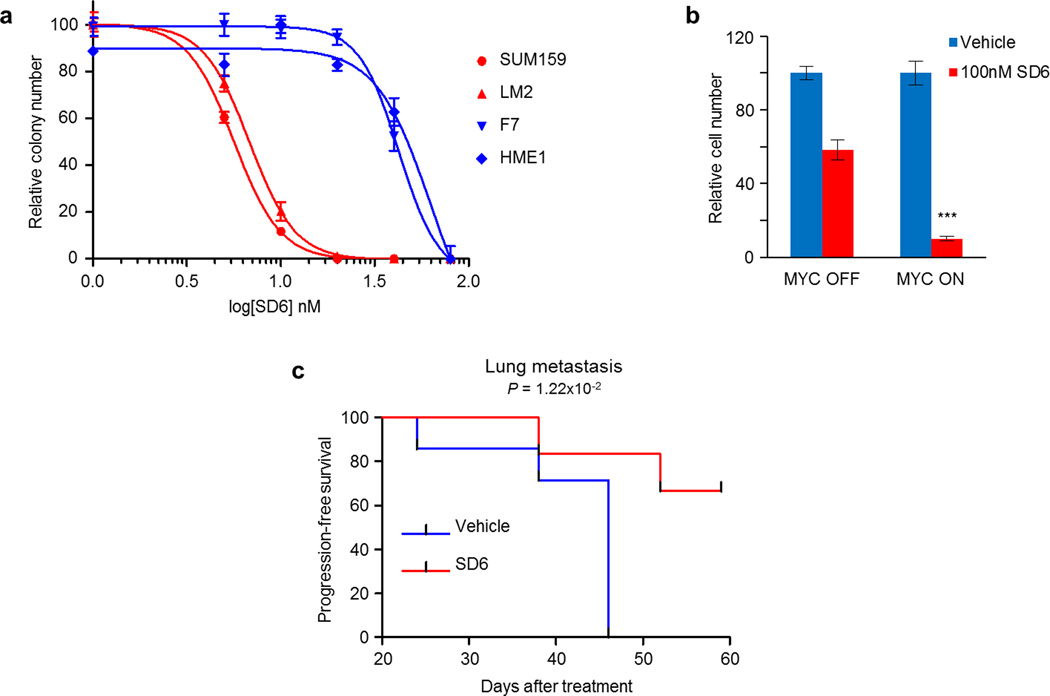
Next we investigated whether pharmacologic inhibition of the spliceosome is also synthetic lethal with MYC. Several pharmacologic agents (e.g., FR901464, pladienolides, and their derivatives) have been characterized to bind the core SF3b complex components and inhibit spliceosome function12. However, most of these inhibitors are not amenable for in vivo delivery. We developed a new small molecule inhibitor of SF3B1, called SD6, that impairs spliceosome function and is bioavailable in mammals13. Consistent with our genetic data, low concentrations of SD6 significantly suppressed colony formation (Fig. 1g) and induced apoptosis (Extended Data Fig. 4i) in a MYC-selective manner. The synthetic lethal interaction between MYC hyperactivation and core spliceosome perturbation suggests that pre-mRNA splicing is necessary to tolerate oncogenic MYC.
In many different cell lineages and experimental systems, oncogenic activation of MYC has been shown to amplify the synthesis of cellular mRNA through direct or indirect mechanisms4,5,14,15. In agreement, MYC hyperactivation in HMECs increased total cellular mRNA synthesis and mRNA steady-state levels (Fig. 2a) without an increase in cellular growth rate (Extended Data Fig. 3c). In contrast to a recent report in B-cell compartments16, MYC hyperactivation did not affect the levels of spliceosome proteins in HMECs (data not shown), suggesting that increased pre-mRNA dosage is not compensated for by higher spliceosome levels. Thus, we hypothesized that the MYC-induced increase in global mRNA synthesis confers increased pressure on the spliceosome to process pre-mRNAs, and partial perturbation of the spliceosome would lead to widespread defects in splicing of pre-mRNA introns in the MYC-hyperactive state. To test this hypothesis, we compared intron retention (IR) after BUD31 knockdown in MYC-normal or MYC-hyperactivated cells. We performed RNAseq from cells in each state (normal, BUD31 knockdown, MYC-hyperactive, and MYC-hyperactive with BUD31 knockdown) and determined pre-mRNA splicing efficiency by calculating IR at junctions across the genome (Fig. 2b). Because analysis of intronic reads may be influenced by the presence of stable RNAs within introns and/or spliced lariats, we restricted the analysis to reads directly spanning exon-intron or exon-exon junction sequences (75,623 junctions in 6,861 genes) (see Supplementary Information).
Figure 2. In MYC-hyperactivated cells, perturbation of the spliceosome leads to global intron retention.
a, (Left) total poly(A)+ RNA per cell (10−4 ng) and (right) newly synthesized 4-sU-labeled poly(A)+ RNA per cell (10−5 ng) (mean±s.e.m., n=4 technical replicates for both assays, two-tailed Student’s t-test). b, Schematic of intron retention (IR) analysis. c–d, Empirical cumulative distribution of IR coefficients for (c) 75,623 exon-intron junctions or (d) 6,861 genes. Curves represent IR differences after BUD31 depletion in MYC-normal and MYC-hyperactive states. A rightward shift in the MYC-hyperactive curve indicates increased IR (Kolmogorov-Smirnov test). e–g, Log2-fold changes in junction IR relative to untreated by RNAseq of representative genes (mean±s.e.m., n=3 biological replicates, two-tailed Student’s t-test). h–j, qRT-PCR validation showing fold change in junction IR relative to untreated (mean±s.d., n=3 biological replicates, two-tailed Student’s t-test). *P<0.05, **P<0.01, ***P<0.001
To examine the effects of spliceosome perturbation in the normal and oncogenic MYC states, we compared the effect of BUD31 knockdown on junction IR coefficients in wild-type and MYC-hyperactivated cells. Notably, BUD31 depletion caused significantly more IR in the MYC-hyperactive state relative to the MYC-normal state (Fig 2c, P<10−324). Similar results were observed when junction coefficients were computed on a gene level (Fig. 2d, P<10−189). The increase in intron retention conferred by aberrant MYC activation and BUD31 shRNA was validated on individual exon-intron junctions via qRT-PCR (examples in Fig. 2e–j). IR was not limited to a few discrete genes. Instead, BUD31 knockdown in the MYC-hyperactive state led to significantly increased IR in 42% of genes analyzed (2,848 of 6,861, P<0.05). These data indicate that the combination of oncogenic MYC activation and partial spliceosome inhibition leads to a widespread increase in IR. This is consistent with the hypothesis that MYC-induced increase in pre-mRNA synthesis enhances cellular dependency on optimal spliceosome function by raising the level of pre-mRNA substrates for spliceosomal processing.
Intron-retaining pre-mRNAs often fail to complete mRNA maturation and are commonly degraded via quality control mechanisms17. Because the combination of MYC hyperactivation and spliceosome inhibition led to a global increase in intron retention (Fig. 2c–d), we hypothesized that these cells may harbor widespread defects in pre-mRNA maturation and stability (Fig. 3a). To test this hypothesis, we measured levels of cellular poly(A)+ RNA in each of four states (± MYC-hyperactivation,±BUD31-shRNA) before and after treatment with the transcriptional inhibitor actinomycin D. After actinomycin D treatment, cellular poly(A)+ RNA decreased by comparable levels (~16–19%) in control cells with or without BUD31 knockdown (Fig. 3b). Intriguingly, MYC-hyperactivated cells exhibited enhanced mRNA stability, perhaps resulting from enhanced polysomal loading of mRNA during MYC-induced translation18. In contrast, cells harboring MYC-hyperactivation and BUD31-depletion exhibited a substantially greater loss (38%) of poly(A)+ RNA after actinomycin D treatment, suggesting a defect in pre-mRNA maturation and/or stability in the combined MYC-hyperactivated/BUD31-shRNA state. Similarly, fluorescence in situ hybridization (FISH) measurements of poly(A)+ RNA revealed that the combination of MYC hyperactivation and BUD31 knockdown led to a substantially greater decrease (60%) in poly(A)+ RNA after actinomycin D treatment (Extended Data Fig. 5a). Similar trends were observed in nuclear RNA pools, consistent with defects in nuclear pre-mRNA maturation (Extended Data Fig. 5b). Consistent with this decrease in pre-mRNA maturation and stability, cells harboring oncogenic MYC and BUD31 knockdown exhibited significantly lower (54%) steady-state levels of poly(A)+ RNA (Fig. 3c). Collectively, these results indicate that MYC-hyperactivation increases cellular pre-mRNA synthesis, and inhibition of the spliceosome reduces the cellular capacity to process this pre-mRNA burden. The result of this MYC-hyperactivated and spliceosome-hypomorphic state is enhanced intron retention, decreased mRNA maturation and stability, and a significant loss of steady-state cellular mRNA.
Figure 3. Combined spliceosomal perturbation and MYC-hyperactivation inhibits pre-mRNA maturation.
a, Model of MYC-spliceosome synthetic lethality. b, Difference in cellular poly(A)+ RNA in HMECs after actinomycin D (A.D.) treatment (mean±s.e.m., n=3 biological replicates, two-tailed Student’s t-test). c, Steady-state poly(A)+ RNA levels per cell (10−4 ng) (mean±s.e.m., n=4 biological replicates, two-tailed Student’s t-test). d, Gene ontology (GO) enrichment of intron-retained genes in the MYC-hyperactive and BUD31-depleted state. Dashed line indicates P=0.05. e–f, In MYC-hyperactive BUD31-shRNA cells, representative genes display (e) increased intron retention (mean±s.e.m.) and (f) decreased steady-state RNA levels (mean±s.e.m.) after BUD31 knockdown in MYC-hyperactivated cells. Bar colors represent GO terms, see legend in Extended Data Figure 6. **P<0.01, ***P<0.001
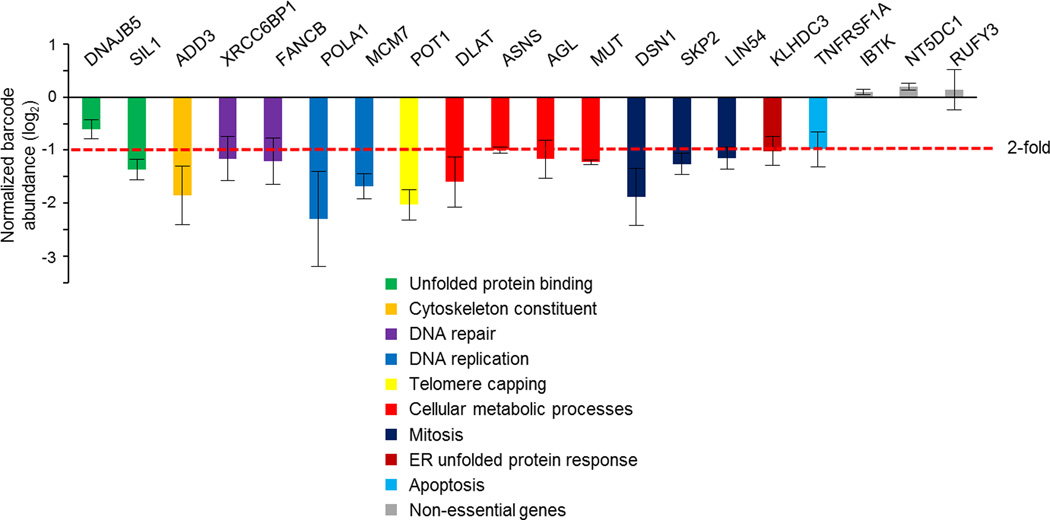
GO analysis of genes with the most significant intron retention in the combined MYC-hyperactive/BUD31-depleted state (2,848 of 6,816 genes analyzed for IR) suggests many essential processes and subcellular structures were impacted, including gene expression, DNA replication and repair, the mitotic spindle, unfolded protein response, and RNA splicing (Fig. 3d). Many genes participating in these essential cell processes exhibited increased IR in the combined MYC-hyperactive/BUD31-knockdown state (representative genes in Fig. 3e) and a concomitant decrease in RNA levels, consistent with a defect in maturation and stability of IR-containing transcripts (Fig. 3f). Consistent with their role in critical cellular processes, knockdown of these genes reduced cell number by 0.7–4.2 fold (as quantified by barcode-tag abundance, Extended Data Fig. 6). Together, these data are consistent with the hypothesis that the combination of oncogenic MYC and spliceosome inhibition leads to widespread loss of mRNA integrity, resulting in the deregulation of many essential genes and processes instead of a single pathway.
Because oncogenic MYC significantly increases the sensitivity of HMECs to inhibition of the spliceosome, we hypothesized that MYC-driven cancers may be hyper-dependent on core spliceosomal function to support their survival. We queried whether MYC-driven breast cancer cell lines exhibit increased sensitivity to knockdown of core spliceosomal genes. Recently, we conducted genome-wide RNAi screens in a panel of 72 breast cancer and immortalized cell lines for genes affecting cell viability (Fig. 4a) (R.M., A.S., B.N., in prep). From this dataset, we tested for a correlation between MYC-dependency (as indicated by sensitivity to MYC-shRNAs) and dependency on (a) the spliceosome (as indicated by sensitivity to shRNAs targeting spliceosome components in the shRNA library) or (b) 100,000 randomly drawn gene sets. Notably, MYC-dependent breast cancer cell lines were significantly more sensitive to shRNAs targeting the core spliceosome (Fig. 4b, P=0.005). The correlation between MYC-dependency and spliceosome-dependency was significantly pronounced in the basal breast cancer lines (Fig. 4c, P<0.00001), an aggressive molecular subtype of breast cancer frequently driven by MYC.
Figure 4. In vivo perturbation of spliceosomal activity impairs MYC-dependent breast tumors and metastases.
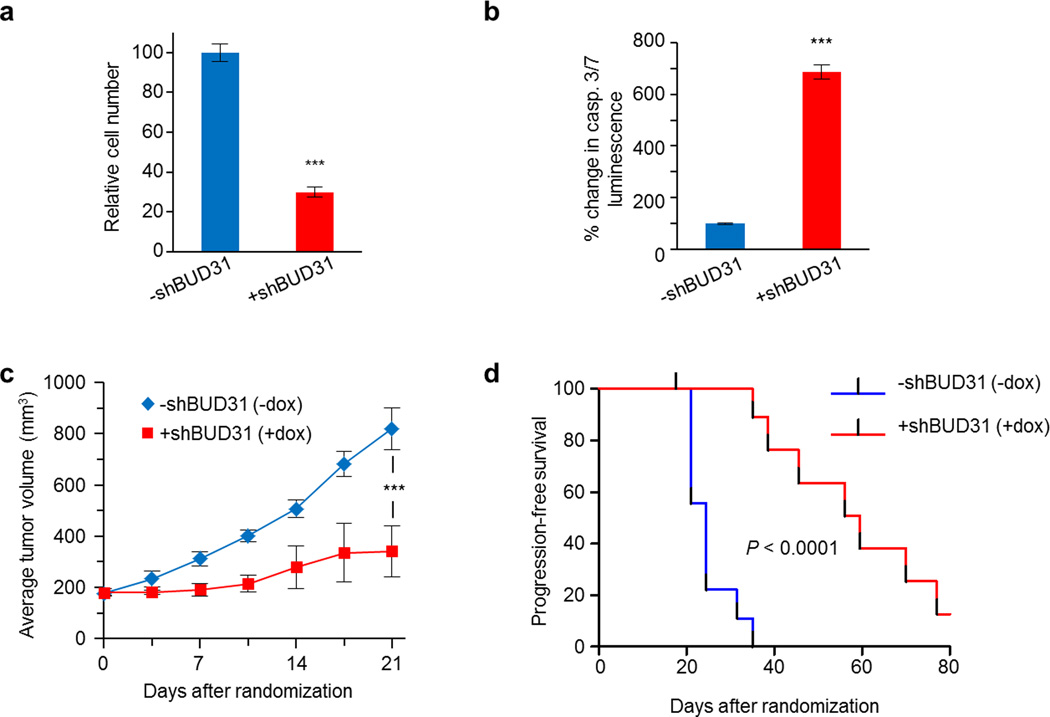
a, Schematic for identifying genetic co-dependencies in breast cancer lines. b–c, MYC-siMEM score, which represents the correlation between cell line sensitivity to MYC-shRNAs and sensitivity to shRNAs targeting random gene sets (n=100,000), is plotted against frequency of gene sets. Increasing MYC-siMEM values denote higher correlation with MYC-dependency. Red arrows indicate MYC-siMEM scores for spliceosome-dependency in (b) all breast cancer lines (n=72) and (c) the basal breast cancer subset (n=32, P-value by bootstrap analysis for both). d–e, MDA-MB-231-LM2 cells with shBUD31 display (d, bottom) diminished BUD31 protein levels, (d, top) decreased cell numbers (mean±s.e.m., n=8 technical replicates, two-tailed Student’s t-test) and (e) increased caspase-3 cleavage (bottom) and caspase-3/7 luminescence (top; mean±s.e.m., n=3 technical replicates, two-tailed Student’s t-test). f–g, Barcode-shRNA abundance of LM2 cells within (f) primary tumors or (g) pulmonary metastases. Mean barcode abundance in each tumor or lung is normalized to the injected cell population (n=3 technical replicates, two-tailed Student’s t-test). h, Change in LM2 tumor growth after 2 weeks of vehicle (n=13) or SD6 (n=10) infusion. Bars indicate mean values (two-tailed Student’s t-test). i, Pulmonary LM2 bioluminescence after 10-day infusion with vehicle (n=7) or SD6 (n=6). Bars indicate median values (Mann-Whitney test). *P<0.05, **P<0.01, ***P<0.001.
Triple-negative breast cancers (TNBCs) are commonly driven by MYC, and exhibit an aggressive, highly metastatic clinical course. To determine whether MYC-driven TNBCs are dependent on spliceosomal integrity for their tumorigenic and metastatic proclivity, we tested the effects of genetic and pharmacologic inhibition of the spliceosome on MYC-dependent and metastatic TNBC models. Inducible BUD31-shRNA reduced cell viability and increased apoptosis in MYC-dependent TNBC cells in vitro (Fig. 4d–e, and Extended Data Fig. 7a–b). Similar to MYC-ER HMECs, MYC protein levels remained unchanged during BUD31 depletion in these MYC-dependent cancer cell lines (Extended Data Fig. 8a, and 8b for HMECs), suggesting the apoptotic response was not due to loss of the driver oncogene (MYC). To assess the impact of spliceosomal perturbation on tumor growth, we established a pooled competition assay that utilizes shRNA-associated barcodes to detect changes in tumor cell fitness (Extended Data Fig. 9). In the metastatic TNBC cell line MDA-MB-231-LM2 (LM2)19, inducible-MYC-shRNA-expressing cells dropped out of the tumor population, confirming the MYC dependency of this TNBC model (Fig. 4f). Similarly, tumor cells harboring BUD31- or SF3B1-shRNA dropped out of the tumor population (Fig. 4f). Tumorigenicity of another MYC-dependent TNBC model (SUM159) was similarly impaired by BUD31 depletion (Extended Data Fig. 7c–d). These data suggest that loss of BUD31 or other core spliceosomal factors inhibits MYC-dependent breast cancer growth in vivo.
Because MYC-driven breast cancers are prone to metastasize to visceral organs including the lungs20, we tested whether perturbation of spliceosome function affected metastatic expansion of MYC-dependent LM2 cells. As shown in figure 4g, BUD31 depletion resulted in substantial drop out of this metastatic cell population (>133.5-fold change), with the majority of +dox tumors containing shBUD31-barcodes below the level of detection. This data suggests that BUD31 and the spliceosome are essential for MYC-dependent breast tumorigenicity and metastatic expansion in vivo.
Next, we tested whether pharmacologic inhibition of the spliceosome also impaired tumorigenic and metastatic potential of MYC-dependent TNBC cells. Compared to MYC-normal cell lines (IC50 ~53 nM), MYC-driven cancer cells were significantly more sensitive (IC50 ~4 nM) to the spliceosome inhibitor SD6 in vitro (Extended Data Fig. 10a). Similarly, SD6 suppressed proliferation of a MYC-driven B-cell model4 (Extended Data Fig. 10b), suggesting oncogenic MYC may confer hyper-dependency on the spliceosome in many epigenetic backgrounds and cancer types. In primary LM2 tumor xenografts, SD6 potently restrained tumor growth with no toxicities in any organ system examined, suggesting splicing is essential for the tumorigenicity of these MYC-dependent breast cancer cells (Fig. 4h). Similarly, SD6 impaired lung metastatic expansion in experimental metastasis assays (Fig. 4i), and extended progression-free survival (Extended Data Fig. 10c). Collectively, this data suggests MYC-driven breast cancers depend on spliceosomal integrity for their tumorigenic and metastatic progression.
Altogether, these data suggest MYC-driven breast cancers harbor an enhanced dependency on the core spliceosome. Recent studies have shown MYC regulates splicing of select genes via induction of alternative splicing factors or components of the core spliceosome.16,21 This study suggests MYC may induce a much broader stress on splicing via its ability to elevate global pre-mRNA synthesis. Recently, there has been considerable investigation into how MYC elicits a widespread increase in mRNA synthesis across the transcriptome4,5,22,23. Importantly, either direct or indirect mechanisms of elevated pre-mRNA synthesis elicited by MYC could lead to an enhanced dependency on the spliceosome, and thus make MYC-driven cancers candidates for spliceosome-based therapies. These observations provoke the important question of whether MYC-induced amplification of mRNA synthesis may also generate vulnerabilities in other aspects of RNA processing (such as mRNA capping, polyadenylation, or mRNA export) and downstream protein biosynthesis24 in MYC-driven cancers. Intriguingly, the spliceosome may be a target of both oncogene addiction and oncogenic stress. Components of the U2 snRNP, such as SF3B1 and U2AF1, harbor frequent and recurrent somatic mutations that cluster in an evolutionarily conserved domain, suggestive of oncogenic function.25,26 Based on such putative oncogenic functions, the spliceosome has been proposed as a target for classical oncogene addiction, in which spliceosome mutant tumors may be addicted to the oncogenic functions of spliceosome mutants and thus sensitive to spliceosome inhibitors. However, this study and others27,28 have shown that inhibition of spliceosome components is deleterious in cancer cell line models that lack spliceosome mutations, suggesting that other drivers of cancer (like MYC) are determinants of sensitivity to splicing inhibitors. Because oncogenic MYC is known to drive several pro-tumorigenic programs that include rewiring of biosynthetic pathways29,30, our model provokes the important hypothesis that cellular processes (like splicing) that enable cancer cells to tolerate such widespread shifts in macromolecular synthesis may provide entry points for anti-cancer therapies.
METHODS
Vectors and virus production
Commercially available pGIPZ shRNAs targeting BUD31 (V2LHS_47771 and V2LHS_47770), EFTUD2 (V2LHS_28167), SF3B1 (V3LHS_397872), SNRPF (V2LHS_276933), and U2AF1 (V2LHS_84677) were obtained from Open Biosystems. shRNAs targeting the 3’UTR region of BUD31 were designed using the BiopredSI and RNAi Codex algorithms (shRNA sequence TGCTGTTGACAGTGAGCGCCGCTGTCTATCAGCTGTGATTTAGTGAAGCCACAGATGTAAATCACAGCTGATAGACAGCGATGCCTACTGCCTCGGA). For inducible RNAi experiments, shRNAs were subcloned into the pINDUCER doxycycline-inducible lentiviral expression system31. Lentiviruses and retroviruses were produced by transiently transfecting shRNA or cDNA constructs using Mirus Bio TransIT transfection protocols into 293T cells and harvesting viral supernatants 48 hours after transfection.
Cell culture
HMECs expressing hTERT and inducible MYC-ER (MYC-ER HMECs), F7s, and HME1s were cultured in mammary epithelial growth medium (MEGM, Lonza). 293T cells, Hela cells, and MDA-MB-231 LM2 human breast cancer cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS). SUM159 human breast cancer cells were cultured in F12 (Gibco) media supplemented with 5% FBS, 10 mM HEPES (Gibco), 5 ug/mL insulin (Invitrogen), and 1 ug/mL hydrocortisone. P493-6 human B-cell lymphoma cell line was cultured in RPMI-1640 supplemented with 10% FBS (Clonotech) and 1% GlutaMAX (Invitrogen). All cell lines were incubated at 37°C and 5% CO2. Cell lines were obtained from ATCC, and all cell lines are tested yearly for mycoplasma contamination. Stable cell lines expressing shRNAs or cDNAs were generated by lentiviral or retroviral transduction in the presence of 8 µg/mL polybrene followed by selection with appropriate antibiotic resistance markers.
Cell proliferation assays
MYC-ER HMECs were infected with pINDUCER-shRNA viruses at an M.O.I. of 1.3–1.5, and transduced cells were seeded at a density of 3,000 onto 96-well black plates (Corning). MYC-ER HMECs with pINDUCER-shBUD31-3’UTR were treated with 300 nM 4-OHT (tam) to induce MYC hyperactivation and 32 ng/ml doxycycline (dox, Sigma) to induce shBUD31 expression. SUM159 and MDA-MB-231-LM2 (LM2) were infected with pINDUCER-shBUD31 (targeting the 3’UTR and coding region, respectively) virus at an M.O.I. of 1.5, and seeded at a density of 1,000 and 2,000, respectively. Expression of shBUD31 in LM2 and SUM159 cells was induced with 1 µg/ml dox. HMECs and breast cancer cells were re-fed every 3–4 days until cells reached confluence. At confluence, cells were fixed in 4% paraformaldehyde, and nuclei were stained with Hoeschst3321 (1:1000, Life Technologies). Nuclei were imaged and counted using the Celígo Imaging Cell Cytometer (Brooks).
For clonogenic assays, breast cancer or immortalized epithelial cells were seeded at low density (between 500–2,000 cells per plate depending on the cell line) into 6cm plates, four replicates per treatment group. MYC-ER HMECs with pINDUCER-shBUD31-3’UTR were treated with 8 ng/ml dox and 300 nM tam, and MYC-ER HMECs treated with 10 or 20 nM SD6 were also cultured with 200 nM tam. Cells were re-fed every 4 days until colonies were macroscopic. The colonies were stained using Coomassie brilliant blue. Macroscopic colonies were quantified and normalized to vehicle-treated cells for each cell line.
For P493-6 cell line with pmyc-tet construct32, MYC was reduced by treating cells with 0.1 µg/ml tetracycline (Sigma) for 72 hours. MYC was induced by washing P493-6 cells with 1× PBS twice, then culturing cells in RPMI-1640 medium with 10% Tet System Approved FBS (Clontech) and 1% GlutaMAX). P493-6 cells were treated with or without 100 nM SD6 and with or without 01. µg/ml tet for 4 days.
Immunoprecipitation and mass spectrometry
Hela cells transduced with lentivirus encoding BUD31 cDNA and non-transduced Hela cells were harvested, and nuclear extracts as well as whole cell lysates were collected as described previously33. Lysates were treated with RNase A (500 ug/mL) for 1 hour on ice. For IPs, nuclear and whole cell extracts were ultracentrifuged at 100,000 rcf, and incubated with 25 ug M2 Flag antibody (Sigma) for 1 hour, followed by ultracentrifugation and incubation with Sepharaose-CL4B Protein A beads (GE Healthcare). Beads were washed with NTN (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% NP-40), and immunocomplexes were resuspended in 1× Laemmli buffer and resolved on pre-cast 4–20% Novex Tris-Glycine gels (Life Technologies). Gels were minimally stained with Coomassie brilliant blue, cut into 8 molecular weight ranges, and digested with trypsin. Immunocomplexes were identified on a Thermo Fisher LTQ mass spectrometer, and data processing was performed as previously described33.
Enrichment analysis
Human GO annotation file (gene_association.goa_human.gz) was downloaded from geneontology.org/GO.downloads.annotations.shtml containing a GOC Validation date of 09/02/2013. Enrichment analysis was performed to consider the content of (a) BUD31-associated proteins, or (b) genes with enhanced IR. Gene symbols annotated to BUD31-associated proteins were cross tabulated against all gene ontology annotations. Genes with enhanced IR were cross tabulated against the subset of gene ontology annotations for genes considered in this analysis. We used Fisher’s exact test to determine p-values for the proportion of genes overlapping each annotation set.
Bimolecular fluorescence complementation (BiFC)
BUD31 was cloned into the pQCXIN-N-YFP fusion vector, in which the BUD31 N-terminus was fused to the N-terminal domain (residues 1–155) of Venus YFP. Human splicing factor cDNAs were individually recombined into retroviral vectors with C-terminal Venus YFP (residues 156–239) tags at the N-terminal ends. SUM159 breast cancer cells were transduced with these bait and prey BiFC retroviruses, and cellular fluorescence was analyzed by flow cytometry in triplicate.
BUD31 mutagenesis
Wild-type and mutant BUD31 cDNAs were generated by gene synthesis (IDT DNA) and recombined into the pQCXIN-N-YFP fusion vector. Mutant BUD31 consisted of substituting human BUD31 amino acid residues 105–114 with an equivalent number of glycine residues (codon GGA).
In vitro competition assay
MYC-dependent SUM159 breast cancer cells with pINDUCER-shBUD31-3’UTR were transduced with viruses containing wild-type or mutant BUD31 or negative control cDNA recombined into pQCXIN-N-YFP vectors. Infected, GFP+ cells are mixed at an 80:20 ratio with non-transduced, GFP− parental cells and seeded into 96-well plates and treated either – or + dox (1 µg/ml). At confluence, cells were passaged 1:10 and processed for flow cytometry. The in vitro competition assay was continued for two passages.
Immunoblotting
Cells were lysed in 1× SDS sample buffer (62.5mM Tris-HCl, pH6.8, 10% glycerol, 2% SDS, 2.5% β-mercaptoethanol) and heated at 95°C for 12 minutes. The following antibodies were used for Western blotting: Flag (Sigma, A8592), BUD31 (ProteinTech, 11798-1-AP), SF3B1 (Bethyl, A300-996A), Prp19 (Bethyl, A300-101A), U2AF1 (Bethyl, A302-079A), SF3A1 (Bethyl, A301-603A), EFTUD2 (Bethyl, A300-957A), SNRPF (Abcam, 154870), HER2 (Millipore, 06-562), EGFR (Cell Signaling, 2232), cleaved caspase-3 (Cell Signaling, 9664), RPS8 (Assay Biotechnology, R12-3466), EIF2S1 (Abgent, AP13469s), eIF3I (p36) (Biolegend, 646701), and c-Myc (D84C12) (Cell Signaling, 5605). Vinculin (Sigma, V9131) and ran (BD Biosciences, 610340) was used as loading control.
In vitro transcription
Uniformly 32P-UTP radiolabeled MINX pre-mRNA was in vitro transcribed from a BamHI-digested plasmid34, DNaseI (Ambion) treated and gel-isolated on a 8 M urea 6% polyacrylamide gel.
In vitro splicing
HeLa nuclear extracts used for in vitro splicing assays were made as described previously35 from HeLa cells transduced with an inducible BUD31-targeting shRNA and grown in the presence or absence of 1 ug/ml dox. Splicing reactions of 15 µl contained: 8 nM RNA substrate, 0.8 mM DTT, 1.7 mM MgOAc, 1.7 mM ATP, 17 mM phospho-creatine, 20mM glycine, 1U/µL RNasin Plus (Promega), 3.7% PVA and 50 ug of HeLa nuclear extracts. Splicing reactions were incubated for indicated time points at 30°C and stopped by digestion with proteinase K (Ambion) for 30 minutes at 45°C followed by RNA purification. RNA purified from splicing reactions was electrophoresed on 8 M urea 8% polyacrylamide gels, then exposed to a phosphorimager screen (Typhoon Trio phosphorimager, GE Healthcare). Alternatively, RNA purified from in vitro splicing reactions was added to RT-PCR reactions as previously described36 with primers in exons 1 and 2 of MINX (forward: 5'-CGGAATTCGAGCTCGCCC-3’ and reverse: 5'-GGATCCCCACTGGAAAGA-3’). PCR products were run on 6% non-denaturing polyacrylamide gels and visualized after staining with ethidium bromide.
Spliceosome complex formation assay
In vitro splicing reactions were carried out as described above, placed on ice, and heparin was added to a final concentration of 2 µg/µL. Reactions were incubated in the presence of heparin at 30°C for 5 minutes and immediately loaded onto 0.75 mm non-denaturing 4% acrylamide – 0.4% agarose composite gels. Gels were run at 250V at room temperature in 1× tris-glycine running buffer for 3 hours, then placed on Whatman paper and exposed to a phosphorimager cassette.
RNA isolation and qRT-PCR
RNA isolation was performed with the RNeasy Mini kit (Qiagen). Reverse transcription was performed using the High Capacity RNA-to-cDNA Master Mix (Applied Biosystems), and qPCR was performed using SYBR Green Master Mix (Applied Biosystems). The following primers were used:
| BUD31 | F: 5’-ACCAACTTCGGGACGAACTG-3’ |
| R: 5’-CGGCCCACTTCCAGCTT-3’ | |
| EFTUD2 | F: 5’-CCTTCGTGTTGTCAGAGAGTGTCT-3’ |
| R: 5’-TGGGTTGGAGGTTGGTGAGT-3’ | |
| SF3B1 | F: 5’-GTGGACAAAATGGCGAAGAT-3’ |
| R: 5’-GAGCTTCATCAAGAGCTGCC-3’ | |
| SNRPF: | F: 5’-GGGAATGGAGTACAAGGGCT-3’ |
| R: 5’-CCCAGATGTCCAGACAAAGC-3’ | |
| U2AF1: | F: 5’-ACGTTTAGCCAGACCATTGC-3’ |
| R: 5’-TGTTCCTGCATCTCCACATC-3’ | |
| GAPDH: | F: 5’-CCTCCCGCTTCGCTCTCT-3’ |
| R: 5’-TGGCGACGCAAAAGAAGAT-3’ |
RNA sequencing (RNAseq)
pINDUCER11-shBUD31-3’UTR infected MYC-ER HMECs were cultured for 72 hours −/+ 16 ng/mL dox and 48 hours±300 nM tamoxifen in triplicates. Total RNA was isolated using the RNeasy kit (Qiagen). RNA samples were rRNA depleted, and NGS libraries were constructed and sequenced as 75 bp paired-end reads by Illumina HiSeq 2000.
Quality assessment of RNAseq
RNA-seq NGS reads quality was evaluated using FastQC application (bioinformatics.babraham.ac.uk/projects/fastqc/).
Alignment of RNAseq data
RNA-seq NGS reads were mapped using STAR RNASeq aligner (version 2.3.1). In order to improve mapping accuracy, splice junctions’ database file (gencode.v.14.annotation.gtf.sjdb) was supplied at the genome index generation step with command line option --sjdbOverhang 74 together with hg19_Gencode14.overhang75/hg19_chrOnly.fa and default parameters. Duplicate reads were marked with the MarkDuplicates function of the Picard-tools software package (picard.sourceforge.net; version 1.107) using default settings.
Intron-exon junction definition
Rationale
To prevent confounding effects in our analysis of intron retention (IR) within HMECs, we confined our analyses to exons in non-overlapping genes that are included within all isoforms of a given gene (75,623 junctions in 6,861 genes).
Method
Intron-exon junctions were obtained using the University of California Santa Cruz Genome Browser “knownGene” table (downloaded 06/04/2014). Constitutive junctions were defined as junctions that (1) appear in each transcript annotated to a given gene symbol, (2) did not overlap with any transcript annotated to a different gene symbol, and (3) fail to mark the start or stop of a transcript.
Junction intron retention calculation
Rationale
Because analysis of intronic reads may be influenced by the presence of stable RNAs within introns and/or spliced lariats, we calculated junction IR as the ratio of exon-intron reads to exon-exon reads, restricting the analysis to reads directly spanning exon-intron or exon-exon junction sequences. Method: We used R together with the Rsamtools package to calculate intron retention (IR). Briefly, for each intron-exon junction, we extracted all non-duplicate reads overlapping this junction. Next, we assigned these reads into two categories: (1) “intronic” if the read mapped to at least the first base of the intron, (2) “exonic” if none of the bases of the read mapped to the first base of the intron and at least one base mapped to a subsequent exon. We counted the total number of reads assigned into each category for each junction. IR was calculated as:
where IRj,i represents the IR score for junction j in sample i and Ij,i and Ej,i refer to the count of reads classified as “intronic” and “exonic” for junction j in sample i, respectively. To avoid ratios with 0 in the denominator we added 1 to each of these counts. The scripts used to conduct this calculation are available upon request. We restricted all following analyses to intron-exon junctions with an average of at least 25 total (“intronic” and “exonic”) reads in the control and MYC-hyperactivated samples.
Gene intron retention calculation
For the cumulative distribution analyses, the mean IR score for all junctions in a gene were averaged.
Gene annotation
A custom gene annotation file was generated to correspond to the set of intron-exon junctions considered in the IR analysis. Briefly, exons were defined as: 1) an exon flanked by two junctions annotated to the same symbol, 2) an exonic region flanked by one junction and conserved across all transcripts annotated to the same symbol.
Statistical analysis of RNAseq
Statistical analyses were performed using the open source statistical programming environment ‘R’. Empirical cumulative distributions of IR scores were compared using two-sided Kolmogorov-Smirnov test and Wilcoxon test.
Permutation-based test of significance
The significance of the difference of empirical cumulative distributions of junction-level IR scores was evaluated using a permutation-based approach. The null hypothesis was that splicing perturbations had no effect on intron retention changes in the MYC-normal and MYC-hyperactivated states. In order to model this null hypothesis, the treatment information was blinded to the assignments of MYC activity. We generated a third Control sample by randomly selecting half of the junctions from samples 1-Control and 2-Control. Control and Myc samples were grouped as 6 “normal” samples, and LowBUD31 and Myc_LowBUD31 samples were grouped as “splicing perturbed” samples. Next, we comprehensively generated all possible “normal” and “splicing perturbed” contrasts by subtracting the average of each junction IR score of 3 “normal” samples from that of the “splicing perturbed” group. The empirical distribution of all possible double differences was generated and used to assign significance to the original observations. An analogous approach was used to evaluate the difference of empirical cumulative distributions of gene-level IR scores.
qPCR intron retention validation assay
Amplification reactions were prepared using SYBR Select Master Mix (Applied Biosystems) according to the manufacturer’s instructions with final primer concentration of 300 nM. Reactions were performed using a StepOnePlus Real Time PCR System (Applied Biosystems) with an initial incubation at 95°C for 10 minutes followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. Primers were designed using Primer3 (available at http://bioinfo.ut.ee/primer3/) and assessed for quality using Beacon Designer (http://www.premierbiosoft.com/qpcr/) and UNAFold (https://www.idtdna.com/UNAFold). Primers used for each reaction were:
| HTRA1_IE | F: 5′-GCGTTCATTTTAAGGTGCTACAGG-3′ |
| R: 5′-TGGGCATTTGTCACGATCAGT-3’ | |
| HTRA1_EE | F: 5′-GACGTGGTGGAGAAGATCGC-3’ |
| R: 5′-AAACCCAGACCCACTAGCCA-3′ | |
| PRPF19_IE | F: 5′-TCCCCTTGTGTGACCTTCTCT-3′ |
| R: 5′-AGAATCTCCGTCCATTGTTTGC-3′ | |
| PRPF19_EE | F: 5′-AGAACTTTAAGACTTTGCAGCTGG-3′ |
| R: 5′-TCTCCGTCCATTGTTTGCAGA-3′. | |
| UBALD2_IE | F: 5′-GCTGCGTTTCCTGACTCCG-3′ |
| R: 5′-GTGGTGGCTGTTGGGAATGT-3′ | |
| UBALD2_EE | F: 5′-CAGTTGCTGCAGGCGGCC-3′ |
| R: 5′-TGGAAGAACGTGCTCAGCGC-3′ |
Ultramer oligonucleotides (Integrated DNA Technologies) were synthesized to match the predicted amplicon of each primer pair, and standard curves were generated for each reaction using 3-fold serial dilutions of these control templates ranging in concentration from 4.0 × 10−12 M to 1.6 × 10−14 M. The sequence AAGAA was added to both the 5′ and 3′ ends of each template to facilitate primer binding. Control template sequences were as follows:
| HTRA1_IE: | 5′-AAGAAGCGTTCATTTTAAGGTGCTACAGGCTTAAGTGTGTACTC- CTTTGGATTTTAGGCTTCCGTTTTCTAAACGAGAGGTGCCGGTGGC- TAGTGGGTCTGGGTTTATTGTGTCGGAAGATGGACTGATCGTGAC- AAATGCCCAAAGAA-3′ |
| HTRA1_EE: | 5′-AAGAAGACGTGGTGGAGAAGATCGCCCCTGCCGTGGTTCATAT- CGAATTGTTTCGCAAGCTTCCGTTTTCTAAACGAGAGGTGCCGGTG- GCTAGTGGGTCTGGGTTTAAGAA-3′ |
| PRPF19_IE: | 5′-AAGAATCCCCTTGTGTGACCTTCTCTCTTTCTATTTCTGGCAGGT- AAAGTCACTGATCTTTGACCAGAGTGGTACCTACCTGGCTCTTGGG- GGCACGGATGTCCAGATCTACATCTGCAAACAATGGACGGAGATT- CTAAGAA-3′ |
| PRPF19_EE: | 5′-AAGAAAGAACTTTAAGACTTTGCAGCTGGATAACAACTTTGAG- GTAAAGTCACTGATCTTTGACCAGAGTGGTACCTACCTGGCTCTTG- GGGGCACGGATGTCCAGATCTACATCTGCAAACAATGGACGGAGA- AAGAA-3′ |
| UBALD2_IE: | 5′-AAGAAGCTGCGTTTCCTGACTCCGCCTGGCCCGCCGTGTCACTG- CCCTGTTTGTCCGCAGACCGCGCTGAGCACGTTCTTCCAAGAAAC- CAACATTCCCAACAGCCACCACAAGAA-3′ |
| UBALD2_EE: | 5′-AAGAACAGTTGCTGCAGGCGGCCCACTGGCAGTTCGAGACCGC- GCTGAGCACGTTCTTCCAAA-GAA |
CT values from each reaction were interpolated on the standard curve generated using the corresponding control template to approximate the concentration of cDNA template in each experimental sample. These values were then reported as the ratio of IE to total (IE + EE) cDNA template in each sample.
Transcription pulse assay
MYC-ER HMECs with pINDUCER11-shBUD31-3’UTR were cultured −/+ 16 ng/mL dox and/or −/+ 300 nM 4-OHT tamoxifen. Cells were pulsed with 500 uM 4-thiouridine (4-SU, Sigma) for 2 hours, and harvested for total RNA using RNeasy mini kit (Qiagen). 4-SU-labeled RNA was purified from 20 ug total RNA. Isolation of newly transcribed RNA was performed as described37 using 100 µL streptavidin beads (Miltenyi Biotec).
Poly(A)+ RNA isolation
Dynabeads Oligo(dT)25 (Life Technologies) were equilibrated with 50 µL Lysis/Binding Buffer, and total RNA was heat denatured (70°C for 2 minutes) prior to binding poly(A)+ RNA to Dynabeads. Isolation of mRNA was performed according to manufacturer’s instructions. Poly(A)+ RNA concentrations were measured with a fluorescence plate reader (Molecular Devices) using Quant-iT RiboGreen reagent (Life Technologies).
Poly(A)+ RNA LNA FISH
pINDUCER11-shBUD31-3’UTR transduced MYC-ER HMECs were seeded onto collagen-coated 8-well glass chamber slides and cultured −/+ 16 ng/mL dox and −/+ 300 nM tamoxifen. Cells were treated −/+ 2 ug/mL actinomycin D (Gibco) or DMSO for 5 hours prior to fixation in 4% formaldehyde and 5% acetic acid in PBS for 15 minutes at RT. Fixed cells were washed with PBS, permeabilized with proteinase K (5 ug/mL, Life Technologies), and treated −/+ RNase A (100 ug/mL, Sigma) for 30 minutes at 37°C in PBS. Dehydration of the cells was performed with 70%, 95% and 100% ethanol solutions. FITC-labeled oligo(dT)25 locked nucleic acid (LNA) probes were heated to 90°C for 4 minutes, then cooled to hybridization temperature (55°C). Dehydrated and dried cells were incubated in 40 nM of LNA probes in hybridization buffer [50% formamide, 2× SSC, 50 mM NaPi (pH 7.0), 10% dextran sulfate] overnight at 55°C. Chamber slides were washed with 5× SSC, 1× SSC, 0.2× SSC, and PBS, and dehydrated prior to counterstaining with DAPI and mounting with Fluoromount-G (Southern Biotech). Cells were imaged using a Nikon Ti-E inverted microscope with 40× air objective and Andor Zyla 4.2 sCMOS camera. For each treatment condition and actinomycin time point, ≥ 150 cells were analyzed for mean FITC intensity. Cellular FITC values were adjusted for background fluorescence by subtracting the mean extra-cellular pixel value. Image analysis was performed using Nikon Elements
Luminescent apoptosis assays
Caspase-3/7 activity was assessed in MYC-ER HMECs and breast cancer cell lines by incubating Caspase-Glo 3/7 Reagent with cells in triplicate wells of a 96-well plate and measuring luminescence with a plate reader (Molecular Devices). Luminescence was normalized using cell numbers determined by Hoeschst3321 staining of a duplicate plate, followed by nuclei counting using the Celígo Imaging Cell Cytometer (Brooks).
Tumorigenicity and Metastasis Assays
SUM159 breast cancer cells were transduced with pINDUCER11-shBUD31-3’UTR virus and analyzed by flow cytometry to confirm >98% transduction. 8 × 106 transduced cells were injected with matrigel (BD Biosciences) subcutaneously into the flank of four-week-old female athymic nude Foxn1-nu mice (Harlan Labs). Mice were maintained on sucrose water (−dox) or sucrose water with dox (+dox). Tumor volume was measured using calipers, and once tumors achieved 150 mm3, mice were randomized onto sucrose water (−dox) or sucrose water with dox (+dox).
For mixed population experiments, MDA-MB-231-LM2 breast cancer cells were individually transduced with pINDUCER11-shRNAs targeting the indicated genes at an M.O.I. appropriate to transduce all cells (1.3–1.5). The individual populations were mixed in vitro and expanded prior to injection. For mixing experiments shRNAs, populations were mixed at equal ratios. 3 × 106 or 2 × 105 mixed population cells were injected subcutaneously into the right flank or into the lateral tail vein of four-week-old female athymic nude Foxn1-nu mice (Harlan Labs), respectively. Subcutaneous tumor volume was measured with calipers over time. Mice were randomized onto sucrose water (−dox) or sucrose water with dox (+dox) after tumors exceeded 150 mm3. Lung metastatic progression was monitored and quantified using noninvasive bioluminescence as described previously19. When tumors reached 1000 mm3 or the total luminescence flux reached 1 × 109, genomic DNA from dissected tumors or lungs were harvested using the QIAamp DNA mini kit (Qiagen). qPCR was performed with SYBR Green PCR Master Mix (Life Technologies) using manufacturer’s recommendations and the following primers. Experimental target CT values were normalized to the TRE CT values, and NCOR2 was used as a negative control.
| BUD31 | F: 5’-TGGAAGACATCTGCGTGGTATT-3’ |
| R: 5’-CGCGCAAACCTAAAGGCATA-3’ | |
| SF3B1 | F: 5’-GCCGTATCATTAGTACGCCATA-3’ |
| R: 5’-TCGATCCTAGGACGGGGTAT-3’ | |
| MYC | F: 5’-GCCGGCCATATTTTCACTTC-3’ |
| R: 5’-CACACCTACCGAAAAACAAAC-3’ | |
| NCOR2 | F: 5’-AACTTCCGGTGCTGTCGTTT |
| R: 5’-CGCGTCCTAGGTAATACGACTCA-3’ | |
| TRE: | F: 5’-TGTACGGTGGGAGGCCTATATAA |
| R: 5’-GCGTCTCCAGGCGATCTG-3’ |
For SD6 drug infusion studies, 3 × 106 or 2 × 105 MDA-MB-231-LM2 breast cancer cells were injected into the flank or lateral tail vein of four-week-old female athymic nude Foxn1-nu mice (Harlan Labs), respectively. For mice with subcutaneous tumors, jugular vein catheters (SAI Infusion Technologies) were surgically implanted into each mouse 13–16 days after injection, and were randomized to receive vehicle (n=11) or (n=10) infusion. Tail vein-injected mice were randomized to receive vehicle (n=7) or SD6 (n=6) infusion 1 day after tail vein injections. Animals received daily infusions of vehicle (10% [(2-hydroxypropyl)-β-cyclodextrin] dissolved in 50 mM Na 2 HPO 4 /NaH 2 PO 4, pH 7.4) or 50 mg/kg of SD6 for 20 consecutive days (subcutaneous cohort) or 10 consecutive weekdays (tail vein cohort). Mice were infused via jugular catheter at a rate of 3.5 µL/minute with a Fusion 200 Touch Syringe Pump (SAI Infusion Technologies). The total volume infused did not exceed 500 µL per day. Subcutaneous tumor volumes were monitored with calipers, and lung metastatic progression was monitored with noninvasive bioluminescence. Mice were sacrificed once tumors reached 2000 mm3 or the total luminescence flux reached 1 × 109. In progression-free survival analyses, progression is defined as 5-fold increase in pulmonary bioluminescence relative to initial values or 4-fold increase in subcutaneous tumor volume relative to its volume at time of randomization.
Investigators responsible for monitoring and measuring the xenografts of individual tumors were not blinded. Simple randomization was used to allocate animals to experimental groups. All animal studies were performed in accordance with institutional and national animal regulations. Animal protocols were approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine.
Power analysis was used to determine appropriate sample size to detect significant changes in animal survival, which were based on previous survival analyses in our laboratory. All animals were included in analyses.
Pooled shRNA screens in breast cancer cell lines
Pooled shRNA screens were performed on 68 breast cancer lines and four non-malignant immortalized mammary epithelial lines, essentially as described38. Briefly, cells are infected with a lentiviral shRNA library at a MOI of 0.3, and passaged under standard conditions. At 4 and 8 doublings, respectively, DNA is isolated and hybridized to a customized chip to assess shRNA dropout. A detailed description of the results of these screens will be published separately (R. M., A. S., and B.G.N., manuscript in preparation).
Correlation between MYC dependency and spliceosome dependency
First, to calculate MYC dependency scores using likely on-target hairpins, we used assay observations associated with 3 hairpins incorporated into the first ATARIS39 solution for MYC. MYC dependency scores were generated using a hierarchical linear model, with pooled shRNA screen observations as the independent variable and two regression covariates: initial signal intensity (with coefficient β0) and linear time-course dropout trend (with coefficient β1). The dropout trend is calculated for each cell line separately, resulting in a per-cell-line MYC dropout score (the value of coefficient β1).
Second, the MYC dependency score was used in a hierarchical linear model to search for associations with the essentiality of other genes (such as spliceosome encoding genes). This model uses pooled shRNA screen observations as the independent variable and three regression covariates: initial assay signal intensity (with coefficient β0), linear time-course dropout trend (with coefficient β1), and an interaction term between dropout trend and MYC dependency score (with coefficient βM). The p-value associated with the interaction term βM is used to determine whether a significant association exists. A detailed description of this approach will be published elsewhere (A. S., R. M., and B.G.N., manuscript in preparation).
A summary statistic using results from the single-gene analyses was used to test the significance of the association between MYC dependency and the essentiality of a gene set. For a gene set containing genes g, we calculate the gene set summary statistic as
where sign() and pvalue() indicate the values associated with the regression coefficient βM. The resulting metric, termed a siMEM (mixed-effect model) score, indicates the significance and correlation between sensitivity to MYC-shRNAs and sensitivity to a group of shRNAs targeting a gene set (such as those targeting the spliceosome). A gene set (ex. spliceosome genes) for which a substantial number of genes are significantly associated with MYC dependency, and all with the same direction (sign) of association, will have a large positive score.
When calculated for the gene set consisting of the core spliceosome, this value summarizes the direction and strength of the significance observed across genes in the spliceosome. To determine whether this observation is significant, the same statistic is calculated for 100,000 randomly drawn gene sets of the same size as the core spliceosome, yielding the null distributions of gene set summary statistics in Figures 4b–c.
Statistical analysis
All experiments were performed on biological replicates unless otherwise specified. Sample size for each experimental group/condition is reported in the appropriate figure legends and methods. For cell culture experiments, sample size was not predetermined, and all samples were included in analyses. For significance testing, analyses were chosen if data met the assumptions of the tests. Data was checked for comparable variance prior to statistical analysis. Statistically significant differences between control and experimental groups were determined using two-tailed unpaired Student’s t-test, one-way ANOVA with Tukey-Kramer minimum significant difference test, Mann-Whitney test, Kolmogorov-Smirnov test, Wilcoxon test, permutation-based test of significance, and log-rank test as indicated in the appropriate figure legend and methods text.
Extended Data
Extended Data Figure 1. Validation of BUD31 as a MYC-synthetic lethal gene in HMECs.
a, qRT-PCR analysis of BUD31 mRNA level (mean±s.d., n=3 biological replicates). b, Clonogenicity of MYC-ER HMECs with or without MYC hyperactivation or BUD31 depletion (mean±s.e.m., n=4 biological replicates, **P<0.01, two-tailed Student’s t-test). c, Caspase-3/7 activation by caspase luminescence assay (mean±s.e.m., n=3, ***P<0.001, one-way ANOVA). d, FLAG-tagged protein levels in MYC-ER HMECs in which vinculin was used as a loading control.
Extended Data Figure 2. BUD31 interacts with core spliceosomal factors and is required for spliceosomal assembly and pre-mRNA splicing.
a, 134 core spliceosomal proteins are listed. Proteins in red are shown to interact with BUD31, as discovered by Flag-BUD31 IP-MS and BUD31 bimolecular fluorescence complementation (BiFC). b, Heat map of BUD31-interacting spliceosomal proteins, organized by spliceosome sub-complexes. A black-green color scale depicts normalized BiFC interaction values between spliceosomal proteins and negative control protein (technical replicates in 2 left lanes) and BUD31 (technical replicates in 2 right lanes). c, Spliceosomal snRNPs (colored circles) interact in a stepwise manner to excise intronic sequences from pre-mRNA. snRNPs with proteins identified from the BUD31 IP-MS are noted (blue outline) to be BUD31-associated. d, Co-immunoprecipitation of Flag-BUD31 for non-spliceosomal proteins. Input and IP blots probed by EIF2S1 and EIF3I were taken at different exposures to minimize background signal. e, Interaction between N-YFP-tagged BUD31 and C-YFP-tagged spliceosomal (DDX46) or cytoplasmic proteins (TRIM9, SOCS2, EPHA8) was assessed by cellular fluorescence (mean±s.e.m., n=3 technical replicates). f, Nuclear extracts with or without BUD31 knockdown were incubated with pre-mRNA substrate, and RT-PCR of unspliced RNA (top) and spliced RNA (bottom) was performed, using primers at the indicated arrows (left). BUD31 protein levels in the nuclear extracts were normalized to vinculin expression (middle) and quantified (right). g, Radioactively labeled pre-mRNA (MINX) was incubated with nuclear extracts with or without BUD31 depletion. RNA purified from the splicing reaction was run on a denaturing gel and imaged by autoradiography. The identities of prominent bands are based on size. * denotes putative intron-lariat band. h, After in vitro splicing was performed as described previously, products were electrophoresed on native gel, and spliceosome complexes were visualized by autoradiography. Complex A and non-specific H complexes are labeled. i, Phosphorimager quantification of the ratio of RNA in complex A compared to that in complex H. j, Interaction between N-YFP-tagged wild-type (WT) or mutant BUD31 and C-YFP-tagged splicing factors was assessed by cellular fluorescence (mean±s.e.m., n=2 technical replicates, ***P<0.001, two-tailed Student’s t-test).
Extended Data Figure 3. HMECs with oncogenic activation of HER2 and EGFR do not require BUD31.
a, Cell number changes in HMECs with inducible shBUD31 and constitutive HER2 or EGFR expression (mean±s.e.m.; n=4 technical replicates; *P<0.05; N.S., not statistically significant; two-tailed Student’s t-test). HER2 and EGFR protein is normalized to vinculin (right). b, MYC protein levels in HMECs with constitutive HER2 or EGFR expression. c, MYC induction by tamoxifen in MYC-ER HMECs does not increase cell proliferation over time (mean±s.e.m., n=8 technical replicates).
Extended Data Figure 4. Partial knockdown of core splicing factors is MYC-synthetic lethal in HMECs.
a–d, mRNA levels for core splicing factors (a) SF3B1, (b) U2AF1, (c) EFTUD2, and (d) SNRPF were evaluated by qRT-PCR (mean ±s.d., n=3 technical replicates). e–i, Caspase-3/7 luminescence in MYC-ER HMECs with (e–h) partial suppression of core spliceosomal proteins or (i) spliceosome inhibitor SD6 (mean±s.e.m., n=3 technical replicates, ***P<0.001, one-way ANOVA).
Extended Data Figure 5. BUD31 loss in MYC-hyperactivated cells destabilizes mRNA.
a–b, MYC-ER HMECs with inducible shBUD31 treated with actinomycin D for 5 hours were labeled with oligo(dT)25 LNA probes via fluorescence in situ hybridization. Cellular FITC intensity was assessed within (a) cellular and (b) nuclear regions (DAPI+). Data are represented as the difference in cellular FITC intensity between 0 hour and 5 hours of actinomycin D treatment in each cell state (mean±s.e.m., n=150, ***P<0.001, two-tailed Student’s t-test).
Extended Data Figure 6. BUD31 depletion in MYC-hyperactivated cells enhances intron retention and decreases expression of cell-essential genes.
In MYC-hyperactive cells, 17 representative genes display increased intron retention and decreased steady-state RNA levels after BUD31 knockdown. Depletion of these genes by shRNA decreased cell viability (mean barcode abundance±s.e.m.). 2-fold decrease in barcode abundance is noted by the dashed red line. All values are reflective of 3 biological replicates, and genes are color-coded based on their GO-term annotation.
Extended Data Figure 7. MYC-dependent breast cancer cells require BUD31 for in vitro and in vivo growth.
a, Relative cell number of SUM159 cells with doxycycline-inducible shBUD31 in vitro (mean±s.e.m., n=8 technical replicates, ***P<0.001, two-tailed Student’s t-test). b, Caspase-3/7 luminescence in BUD31-depleted SUM159 cells (mean±s.e.m., n=3 technical replicates, ***P<0.001, two-tailed Student’s t-test). c–d, SUM159 cells engineered with dox-inducible shBUD31 were subcutaneously transplanted into mice and randomized onto dox treatment (−dox n=10, +dox n=9). Loss of BUD31 SUM159 xenografts (c) inhibits tumor growth (mean±s.e.m., *** P<0.001 at day 21, two-tailed Student’s t-test) and (d) prolongs progression-free survival in nude mice (P-value, log-rank test).
Extended Data Figure 8. BUD31 depletion does not affect levels of MYC protein.
a, MYC protein levels in MYC-ER HMECs with inducible shBUD31 expression normalized to vinculin expression. To confirm specificity of MYC antibody, human mammary epithelial cells without the MYC-ER construct (HMEC) were engineered to express inducible shMYC. b, MYC protein levels in SUM159 and LM2 cells with inducible shBUD31 normalized to vinculin expression. To confirm specificity of MYC antibody, SUM159 cells were engineered to express inducible shMYC.
Extended Data Figure 9. Core splicing factors EFTUD2 and SNRPF are required for MYC-dependent LM2 breast cancer tumor growth.
Schematic for in vivo barcode-based competition assay. LM2 cells transduced with inducible shRNAs targeting negative control genes or candidate genes were mixed at an equal ratio. This mixed population was transplanted into mice, and tumors were allowed to form in the presence or absence of dox. At the experimental endpoint, genomic DNA was isolated for comparisons of relative barcode (shRNA) abundance in tumor genomic DNA.
Extended Data Figure 10. Spliceosome inhibitor SD6 inhibits MYC-dependent cancer cells in vitro and in vivo.
a, MYC-dependent breast cancer cells (SUM159, LM2) and MYC-normal immortalized epithelial cells (F7, HME1) were cultured with SD6 at low density and analyzed for clonogenic growth. b, MYC-repressible human B-cell line P493-6 was treated with or without 100 nM SD6 in the absence or presence of MYC-hyperactivation for four days, and cells were counted for relative cell number changes (mean±s.e.m., n=3 biological replicates, ***P<0.001, one-way ANOVA). c, Kaplan-Meier survival analysis of nude mice with pulmonary seeding of LM2 cells treated with or without SD6 for 10 days (vehicle n=7, SD6 n=6, P-value by log-rank test).
Acknowledgments
We would like to thank J. Rosen, S. Butler, K. Neugebauer, and members of T.F.W, C.A.S., and T.A.C. laboratories for helpful comments, and P. Yu for bioinformatics support. The authors also acknowledge the joint participation by Adrienne Helis Melvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine for cancer research. The Dan L. Duncan Cancer Center Shared Resources was supported by the NCI P30CA125123 Center Grant and provided technical assistance including Cell-Based Assay Screening Service (Dan Liu), Genomic and RNA Profiling Resource (Lisa White), Biostatistics & Informatics Shared Resource (Susan Hilsenbeck) and Cytometry and Cell Sorting (Joel Sederstrom; P30 AI036211 and S10 RR024574). T.Y.-T.H. was supported by NIH pre-doctoral fellowship (NCI 1F30CA180447) and CPRIT training grant (RP101499). M.O. and R.J.B. were supported by The Gillson Longenbaugh Foundation. R.J.B. was supported by Alex's Lemonade Stand Foundation. Work in the Golding lab was supported by NIH grant R01 GM082837, NSF grants PHY 1430124 (PFC: Center for the Physics of Living Cells) and PHY 1147498 (CAREER), and Welch Foundation grant Q-1759. T.F.W. was supported by CPRIT (RP120583), the Susan G. Komen for the Cure (KG090355), the NIH (1R01CA178039-01 and U54-CA149196) and the DOD Breast Cancer Research Program (BC120604).
Footnotes
AUTHOR CONTRIBUTIONS
T.Y.-T.H., N.N., R.M., C.S.B., G.V.E., T.S., S.J.K., S.T., K.L.K., J.D.H., K.S., R.J.B., S.O.S., A.J., C.L., M.O. performed the experiments. L.M.S., A.S., R.D.V., A.R., C.A.S. performed statistical analyses. I.G., S.Y.J., J.R.N., X.Z., T.A.C., T.R.W., B.G.N., C.A.S., T.F.W. devised or supervised experiments. T.Y.-T.H. and T.F.W. wrote the manuscript.
RNAseq data sets have been deposited in NCBI Gene Expression Omnibus under accession number GSE66182.
The authors declare no competing financial interests.
REFERENCES
- 1.Eilers M, Eisenman RN. Myc's broad reach. Genes & development. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabo A, Amati B. Genome recognition by MYC. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a014191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harbor perspectives in medicine. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruggero D. The role of Myc-induced protein synthesis in cancer. Cancer research. 2009;69:8839–8843. doi: 10.1158/0008-5472.CAN-09-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessler JD, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masciadri B, et al. Characterization of the BUD31 gene of Saccharomyces cerevisiae. Biochemical and biophysical research communications. 2004;320:1342–1350. doi: 10.1016/j.bbrc.2004.05.228. [DOI] [PubMed] [Google Scholar]
- 11.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 13.Lagisetti C, et al. Optimization of antitumor modulators of pre-mRNA splicing. Journal of medicinal chemistry. 2013;56:10033–10044. doi: 10.1021/jm401370h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene. 2003;22:5707–5711. doi: 10.1038/sj.onc.1206800. [DOI] [PubMed] [Google Scholar]
- 15.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh CM, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015 doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- 17.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nature reviews. Molecular cell biology. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 18.Mezquita P, Parghi SS, Brandvold KA, Ruddell A. Myc regulates VEGF production in B cells by stimulating initiation of VEGF mRNA translation. Oncogene. 2005;24:889–901. doi: 10.1038/sj.onc.1208251. [DOI] [PubMed] [Google Scholar]
- 19.Minn AJ, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. The Journal of clinical investigation. 2005;115:44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Cosimo S, Baselga J. Management of breast cancer with targeted agents: importance of heterogeneity. Nature reviews. Clinical oncology. 2010;7:139–147. doi: 10.1038/nrclinonc.2009.234. [corrected] [DOI] [PubMed] [Google Scholar]
- 21.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabo A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walz S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin CJ, et al. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell reports. 2012;1:325–333. doi: 10.1016/j.celrep.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nature genetics. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. The New England journal of medicine. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubert CG, et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes & development. 2013;27:1032–1045. doi: 10.1101/gad.212548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adler AS, et al. An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth. Genes & development. 2014;28:1068–1084. doi: 10.1101/gad.237206.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157:1088–1103. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu YC, et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PloS one. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 31.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuhmacher M, et al. Control of cell growth by c-Myc in the absence of cell division. Current biology : CB. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 33.Malovannaya A, et al. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zapp ML, Berget SM. Evidence for nuclear factors involved in recognition of 5' splice sites. Nucleic acids research. 1989;17:2655–2674. doi: 10.1093/nar/17.7.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic acids research. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Echeverria GV, Cooper TA. Muscleblind-like 1 activates insulin receptor exon 11 inclusion by enhancing U2AF65 binding and splicing of the upstream intron. Nucleic acids research. 2014;42:1893–1903. doi: 10.1093/nar/gkt1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolken L, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. Rna. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer discovery. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao DD, et al. ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome research. 2013;23:665–678. doi: 10.1101/gr.143586.112. [DOI] [PMC free article] [PubMed] [Google Scholar]