Abstract
Genome-wide cancer mutation analyses are revealing an extensive landscape of functional mutations within the noncoding genome, with profound effects on the expression of long noncoding RNAs (lncRNAs). While the exquisite regulation of lncRNA transcription can provide signals of malignant transformation, we now understand that lncRNAs drive many important cancer phenotypes through their interactions with other cellular macromolecules including DNA, protein, and RNA. Recent advancements in surveying lncRNA molecular mechanisms are now providing the tools to functionally annotate these cancer associated transcripts, making these molecules attractive targets for therapeutic intervention in the fight against cancer.
Introduction
Cancer is fundamentally a genetic disease that alters cellular information flow to modify cellular homeostasis and promote growth. The discovery of a universal genetic code for protein coding genes produced countless breakthroughs in understanding how such mutations drive cancer, establishing the scientific principles on which the development of targeted therapies for malignancies are based, such as imatinib for BCR-ABL leukemia or vemurafenib for BRAFV600E melanoma. Furthermore, the clinical application of cancer exome sequencing has identified numerous protein coding mutations amenable to targeted therapy. However, the coding genome accounts for less than 2% of all sequences, and it has become apparent that aberrations within the noncoding genome drive important cancer phenotypes.
One of the most unexpected findings of the genomics era of biology is the extensive transcription of RNA from non-protein coding regions of the genome (Morris and Mattick, 2014; Rinn and Chang, 2012). Indeed the list of long noncoding RNAs (lncRNAs), which are functionally defined as transcripts >200 nucleotides in length with no protein coding potential, number in the tens of thousands, many of which are uniquely expressed in differentiated tissues or specific cancer types (Iyer et al., 2015). In fact, the number of lncRNA genes outnumbers protein-coding genes (Derrien et al., 2012) and more than 90% have no appreciable peptide products (Banfai et al., 2012; Guttman et al., 2013). It is now recognized that lncRNAs are exquisitely regulated and are restricted to specific cell types to a greater degree than mRNA (Cabili et al., 2011) and are frequently evolutionarily conserved function, secondary structure, and regions of microhomology, despite minimal overall sequence similarity (Hezroni et al., 2015; Quinn et al., 2016; Ulitsky et al., 2011). However, the function of the vast majority of these transcripts remains to be identified.
While the discovery of the universal genetic code has provided investigators the tools to know how genetic mutations result in functional protein defects, lncRNA biology has so far remained resistant to a rules-based, predicative framework for understanding how sequence impacts function. However, recurring molecular mechanisms for lncRNA are now being realized and emerging technologies are expanding investigators abilities to functionally annotate cancer-associated lncRNAs. In this perspective, we provide an overview of the current state of lncRNA biomarker identification in cancer phenotypes, catalog the molecular roles for lncRNAs in cellular processes, and review known roles for lncRNAs in cancer pathophysiology.
Identifying lncRNA signals in the cancer transcriptome
We now appreciate that mutations within the noncoding genome are major determinants of human diseases (Maurano et al., 2012). Indeed recurrent somatic noncoding mutations (Melton et al., 2015), epigenetic alterations (Roadmap Epigenomics et al., 2015), or somatic copy number alterations (Beroukhim et al., 2010) are implicated in multiple cancer types. Mutations in regulatory DNA can broadly affect transcription by altering enhancer and promoter activity or chromatin states, leading to differential lncRNA expression in cancer. Remarkably, several loci that are frequently mutated in cancer encode ultra-conserved noncoding sequences that are deregulated in cancer (Calin et al., 2007). Indeed, SNPs, copy number alterations, or mutations within the non-coding genome can dramatically alter lncRNA transcription (Table 1). The 8q24 locus is particularly richly populated with cancer-associated SNPs, several of which are implicated in the expression of cancer-associated lncRNAs, including CCAT2 in colorectal cancers and PCAT-1 in prostate cancer (Ling et al., 2015).
Table 1.
Cancer-Associated LncRNAs
| LncRNAs Associated with Common Cancer Genomic Alterations | |||
|---|---|---|---|
| LncRNA | Cancer Type | Genomic Alteration | References |
| PVT1 | Colorectal | 8q24 amplification | (Tseng et al., 2014) |
| PCAT-1 | Prostate | 8q24 SNPs | (Eeles et al., 2008; Prensner et al., 2011) |
| CCAT2 | Colorectal | 8q24 SNPs | (Ling et al., 2013b; Tomlinson et al., 2007) |
| PTCSC3 | Thyroid | rs944289 | (Jendrzejewski et al., 2012) |
| HULC | Hepatocellular | rs7763881 | (Liu et al., 2012) |
| ANRIL | Various | 9p21.3 SNPs | (Pasmant et al., 2011) |
| TERC | Oral cavity | 3q26 amplification | (Dorji et al., 2015) |
| GAS5 | Hepatocellular | rs145204276 | (Tao et al., 2015) |
| LncRNAs in Cancer Diagnosis and Monitoring | |||
| LncRNA | Cancer Type | Bioavailability of LncRNA | References |
| H19 | Gastric | Blood | (Zhou et al., 2015) |
| HULC | Hepatocellular | Blood | (Xie et al., 2013) |
| AA174084 | Gastric | Gastric secretions | (Shao et al., 2014) |
| PCA3 | Prostate | Urine | (Bussemakers et al., 1999) |
| SeCATs | Sezary | Tumor | (Lee et al., 2012) |
| SPRY4-IT1 | Melanoma | Tumor | (Khaitan et al., 2011) |
| Prognostic LncRNAs | |||
| LncRNA | Cancer Type | Prognostic Information | References |
| FAL1 | Ovarian | Poor prognosis | (Hu et al., 2014a) |
| HOTAIR | Breast | Increased risk of metastasis | (Gupta et al., 2010) |
| HOTTIP | Hepatocellular | Increased risk of progression | (Quagliata et al., 2014) |
| MEG3 | Meningioma | Associated with tumor grade and risk of progression | (Zhang et al., 2010) |
| NBAT-1 | Neuroblastoma | Good prognosis | (Pandey et al., 2014) |
| NKILA | Breast | Decreased risk of metastasis | (Liu et al., 2015) |
| SCHLAP1 | Prostate | Increased risk of metastasis | (Prensner et al., 2014b) |
| LncRNAs Predicting Therapeutic Responsiveness | |||
| LncRNA | Cancer Type | Therapeutic Agent | References |
| CCAT1 | Colorectal | BET inhibitors | (McCleland et al., 2016) |
| HOTAIR | Ovarian | Platinum chemotherapies | (Teschendorff et al., 2015) |
The catalog of noncoding genes has grown tremendously over the past few years in large part to the identification of extensive lncRNA transcription (Djebali et al., 2012; Harrow et al., 2012; Iyer et al., 2015), arising from active enhancers (Kim et al., 2010), promoters (Seila et al., 2008) and intergenic sequences. These discoveries have depended in large part upon new computation methods for transcriptome assembly and lncRNA annotation (Guttman et al., 2010; Mattick and Rinn, 2015).
It is now widely understood that lncRNAs, serving as signals of specific cellular states or read-outs of active cellular programs (Wang and Chang, 2011), could identify cellular pathologies such as cancer, provide prognostic value, or even inform therapeutic options for cancer patients (Table 1). Overexpression of the lncRNA HOTAIR in early stage, surgically resected breast cancer is highly predictive of progression to metastatic disease and overall survival (Gupta et al., 2010). Subsequent studies showed that HOTAIR deregulation is associated with cancer progression in 26 human tumor types (Bhan and Mandal, 2015) and can predict differential sensitivity of ovarian cancer patients to two of platinum chemotherapies, potentially guiding clinical decision making (Teschendorff et al., 2015). Differential display analysis of human prostate cancers identified Differential display 3, also known as PCA3, as a specific biomarker of prostate cancer (Bussemakers et al., 1999). Subsequently PCA3 was approved by the FDA for prostate cancer diagnosis, first instance of an FDA-approved test based on a lncRNA. As a biomarker of prostate cancer in the urine, it has become a useful, noninvasive test for prostate cancer with improved specificity, positive predictive value and negative predictive value compared to serum PSA testing (Wei et al., 2014). Similarly, analyses of gastric secretions from patients with gastric cancer identified lncRNA-AA174084 as a biomarker capable of differentiating between gastric cancer and benign disorders of the gastric epithelium (Shao et al., 2014). Recent genome wide approaches have identified thousands of lncRNAs that are differentially transcribed between normal tissues and tumors arising from the same organ (Brunner et al., 2012; Iyer et al., 2015; Yan et al., 2015), suggesting enormous potential for further development of lncRNAs biomarkers for specific cancer histologies.
LncRNA expression profiles can also identify clinically relevant cancer subtypes that predict tumor behavior and disease prognosis (Du et al., 2013). Overexpression of the lncRNA SChLAP1 in men treated with radical prostatectomy was associated with a 2.45 fold increased odds of developing metastatic progression, similar to the risk associated with high grade prostate cancer (Gleason 8–10 on histology) vs. low - intermediate grade disease (Gleason ≤7 on histology) (Prensner et al., 2014b). Additional studies have identified lncRNAs with specificity for prostate cancer primary tumor stage or associated with lymph node metastases (Bottcher et al., 2015). Furthermore, recent reports are indicating that lncRNAs may be predictive of responsiveness to specific types of cancer therapy. Expression of HOTAIR in ovarian cancer predicts poor survival following treatment with a carboplatin, yet this group had improved responses to cisplatin relative to HOTAIR-negative ovarian cancer patients (Teschendorff et al., 2015). Validation of these observations may provide a lncRNA biomarker for guiding the selection of platinum containing regimens in patients with ovarian cancer.
Differential regulation of lncRNAs relative to coding genes may underlie the high degree of tissue specific lncRNA expression. Intriguingly, genome wide characterization of lncRNA and protein coding promoters revealed that these classes of genes are under distinct regulatory regimes (Quinn and Chang, 2015). LncRNA promoters are enriched for A/T nucleotides and diminished in CpG patterns, contain distinct transcription factor binding sequences, and have a unique pattern of chromatin marks relative to protein coding genes (Alam et al., 2014). Furthermore, lncRNAs contain a higher density of methylation at transcriptional start sites (TSS) than at the TSS of protein coding genes, regardless of expression levels, highlighting the context-dependent manner of gene silencing by methylation (Sati et al., 2012). Finally, the Dicer-microRNA (miRNA) pathway and the Myc oncogene differentially regulate the expression of mRNA and lncRNAs (Zheng et al., 2014). Dicer production of miR-295 drives Myc expression, upon which Myc is recruited to lncRNA genes with a high density of Myc binding sites thereby activating lncRNA transcription initiation and elongation. Remarkably, lncRNA genes are substantially more sensitive to this Dicer-miRNA-Myc circuit than protein coding genes with Myc binding sites.
Molecular Mechanisms of Long Noncoding RNAs
In this section we highlight some of the recurring molecular mechanisms that govern how lncRNAs regulate cellular processes (Guttman and Rinn, 2012). Functionality in part begins with lncRNA cellular localization: nuclear lncRNAs are enriched for functionality involving chromatin interactions, transcriptional regulation, and RNA processing while cytoplasmic lncRNAs can modulate mRNA stability or translation and influence cellular signaling cascades (Batista and Chang, 2013). We also highlight recent innovations in molecular tools to survey lncRNA interactions with other cellular macromolecules and describe how these tools have dramatically accelerated the discovery of lncRNA functions (Chu et al., 2015a). These methods are now elucidating how lncRNAs elicit functional outcomes through interactions with DNA, chromatin, signaling and regulatory proteins, and a variety of cellular RNA species (Figure 1).
Figure 1. LncRNA mechanisms rely on interactions with cellular macromolecules.
(A) Chromatin bound lncRNAs can regulate gene expression by controlling local chromatin architecture (above) or directing the recruitment of regulatory molecules to specific loci (below).
(B) LncRNA interactions with multiple proteins can promote the assembly of protein complexes (above), or impair protein-protein interactions (below).
(C) mRNA interactions with lncRNA can recruit protein machinery involved in multiple aspects of mRNA metabolism to affect splicing, mRNA stability, or translation (above) or sequester miRNA away from target mRNA (below).
LncRNAs Localizing to Chromatin
LncRNA transcriptional regulation at the level of chromatin is widely observed mechanism that can involve activities affecting neighboring intrachromosomal genes in cis or targeting of gene in trans on different chromosomes (Huarte, 2015; Huarte et al., 2010; Sahu et al., 2015). LncRNAs are widely known to regulate genes in cis as enhancer associated RNAs (Orom et al., 2010), through transcriptional regulation (Dimitrova et al., 2014; Huarte et al., 2010; Trimarchi et al., 2014; Zhu et al., 2013), transcription factor trapping (Sigova et al., 2015), chromatin looping (Wang et al., 2011), and gene methylation (Schmitz et al., 2010) to name just a few mechanisms. LncRNAs also regulate distant genes through modulation of transcription factor recruitment (Hung et al., 2011; Yang et al., 2013b), chromatin modification (Wang and Chang, 2011), and serving as a scaffold for assembly of multiple regulatory molecules at single locus (Tsai et al., 2010).
One particularly well-characterized mechanism by which lncRNAs regulate gene expression both in cis and in trans involves interaction with chromatin to facilitate histone modification (Khalil et al., 2009). Xist, one of the first functionally annotated lncRNAs, regulates dosage compensation in female mammals by localizing to the X chromosome and recruiting multiple factors directly and indirectly to execute X chromosome inactivation (XCI) (Gendrel and Heard, 2011; Lee and Bartolomei, 2013). That human malignancies frequently have X aneuploidy suggested an important role for dosage compensation of X-linked genes in preventing malignant transformation (Pageau et al., 2007). Confirming this, female mice with Xist deletion in the hematopoietic compartment develop an aggressive myeloproliferative disorder with full penetrance (Yildirim et al., 2013).
While Xist coating of the inactive chromosome is required for X silencing, annotation of Xist interaction domains is a necessary prerequisite for clarifying the role of Xist. To define the genomic interactions of lncRNAs, Chu and colleagues developed chromatin isolation by RNA purification (ChIRP), in which short biotinylated oligonucleotides complementary to the lncRNA transcript purify chromatin bound by the target RNA (Chu et al., 2011). Similar methods including capture hybridization analysis of RNA targets (CHART) and RNA antisense purification (RAP) demonstrated that Xist initially binds gene rich islands on the X chromosome that are in close three dimensional proximity, then spreads to gene poor regions during de novo X chromosome inactivation (Engreitz et al., 2014; Simon et al., 2013; Simon et al., 2011).
Cancer transcriptional programs are also modulated by lncRNA recruitment to distant promoters and enhancers. ChIRP revealed that Paupar, a CNS restricted lncRNA located adjacent to PAX6, interacts with numerous promoters in order to regulate the cell cycle and maintain the dedifferentiated state of neuroblastoma (Vance et al., 2014). Nuclear enriched abundant transcript 1 (NEAT1), a component of nuclear paraspeckles (Clemson et al., 2009), is associated with an increased risk of biochemical progression and metastasis of prostate cancer and is a downstream transcriptional target of estrogen receptor alpha (Chakravarty et al., 2014). ChIRP revealed that NEAT1 localizes to promoters of genes involved in prostate cancer growth and increases chromatin marks of active transcription at these sites, driving androgen-independent prostate cancer growth. T helper 17 cells (Th17) are pro-inflammatory T cells, and the lncRNA RMRP was shown by ChIRP to co-associate with key Th17 transcription factor ROR-γt for inflammatory effector function (Huang et al., 2015). Nascent RNA transcripts in the vicinity of enhancers and promoters can also bind the transcription factor YY1, promoting local accumulation of YY1 in the vicinity of the regulatory sites, supporting the engagement of YY1 with these elements, and driving a positive feedback loop that maintains active transcription (Sigova et al., 2015).
LncRNA Interactions with Protein
LncRNAs interact with proteins to modulate protein function, regulate protein – protein interactions, or direct localization within cellular compartments. These interactions are central to determine lncRNA functional effects, yet characterizing protein – lncRNA has presented significant challenges. RNA chromatography has been successfully used to identify protein interactions for lncRNAs of interests as an initial handle for mechanistic characterization. In this approach proteins bound to genetically or biochemically modified bait RNA are retrieved by affinity purification and proteins identified by mass spectrometry or western blot. Using this strategy, Huarte and colleagues identified that the p53-inducible lincRNA-p21 bound to hnRNP-K to mediate gene repression in response to DNA damage (Huarte et al., 2010). Li and colleagues, using similar methods, identified that Apela RNA positively regulates DNA damage induced apoptosis in mouse embryonic stem cells by binding to hnRNPL, inhibiting its ability to sequester p53 away from the mitochondria (Li et al., 2015). While RNA chromatography has yielded important insights into lncRNA molecular mechanisms, artifacts resulting from non-physiologic interactions that take place with misfolded bait RNA or only in non-physiologic in vitro condition can complicate analyses. Recently, multiple groups developed methods to overcome these limitations. By coupling ChIRP and RAP with mass spectrometry, two different groups of investigators discovered dozens of novel protein components of the Xist complex that are involved in XCI (Chu et al., 2015b; McHugh et al., 2015). CHART-mass spectrometry has also been used to characterize the protein interaction landscape of NEAT1 and MALAT1 (West et al., 2014).
Complementary, protein-centric methods of characterizing ribonucleoprotein complexes have also identified unique lncRNA activities in cancer cells. Using RNA immunoprecipitation (RIP), two prostate specific lncRNAs, PCGEM1 and PRNCR1, were found to associate with androgen receptor in prostate cancer cells (Yang et al., 2013b) and promote transcriptional regulation by AR both in androgen sensitive and castrate resistant prostate cancer cell lines. However, another study of PCGEM1 and PRNCR1 suggests that the role of these lncRNAs in prostate cancer may be more limited (Prensner et al., 2014a). HOTAIR, initially found to define chromatin boundaries at the HOXD locus (Rinn et al., 2007), was reported to bind AR, which prevents binding to and subsequent MDM2 ubiquitinylation and protein degradation, thereby promoting AR transcriptional regulation and driving castrate-resistant prostate cancer (Zhang et al., 2015).
RNA Targets of LncRNA Actions
LncRNA modulation of RNA metabolism is an emerging theme with described roles in the control of mRNA stability, splicing, and translation. LncRNA Staufen-1 (STAU-1) mediates mRNA decay through interaction with a double stranded RNA in the 3′ UTR of select mRNAs. However, a subset of STAU-1 regulated mRNAs can only form the requisite double stranded RNA stem by duplexing with an Alu element of a cytoplasmic lncRNA (Gong and Maquat, 2011), thus a lncRNA can confer specificity on the STAU-1 pathway. STAU-1 – mRNA interactions are also modulated by terminal differentiation-induced noncoding RNA (TINCR), which drives epidermal differentiation and is down-regulated in poorly differentiated cutaneous squamous cell carcinomas (Kretz et al., 2013). Using ChIRP retrieval of TINCR and associated RNAs, TINCR was found to bind several mRNAs containing a 25 nucleotide TINCR Box motif. TINCR recruits STAU-1 to mRNA bearing the TINCR Box, and, somewhat unexpectedly, stabilizes these messages and facilitates the translation of several genes involved in keratinocyte differentiation. A similar technique, RNA antisense purification of RNA (RAP-RNA), identified a MALAT1 interaction with pre-mRNA that directs MALAT1 to localize at the proximal chromatin region of transcriptionally active genes (Engreitz et al., 2014), in line with prior observations that MALAT1 can regulate alternative splicing through interactions with serine/arginine splicing factors and pre-mRNA (Tripathi et al., 2010). LncRNA-p21 also interacts with JUNB and CTNNB1 mRNA to selectively impair translation of these messages (Yoon et al., 2012).
LncRNAs and pseudogene RNA have also been postulated to act as competing endogenous RNA (ceRNA) or “RNA sponges”, interacting with microRNAs in a manner that can sequester these molecules and reduce their regulatory effect on target mRNA (Tay et al., 2014). Transcription of the PTEN pseudogene, PTENP1 and other transcripts, while unable to serve as a template for PTEN translation, nonetheless can increase PTEN protein levels through an RNA-dependent mechanism that involves binding and sequestering PTEN targeting miRNA (Karreth et al., 2011; Poliseno et al., 2010; Tay et al., 2011). Similar ceRNA mechanisms have bee reported for lncRNAs H19 and HULC (Kallen et al., 2013; Keniry et al., 2012; Wang et al., 2010). However, quantitative analyses of miRNA and target mRNA abundance suggest that the ceRNA mechanism can only operate given appropriate stoichiometry between ceRNA and miRNAs (Denzler et al., 2014). Experimental analyses and knockout studies are thus needed beyond the presence of matching miRNA seed sequences to validate ceRNA mechanisms.
LncRNA Drivers of Cancer Phenotypes
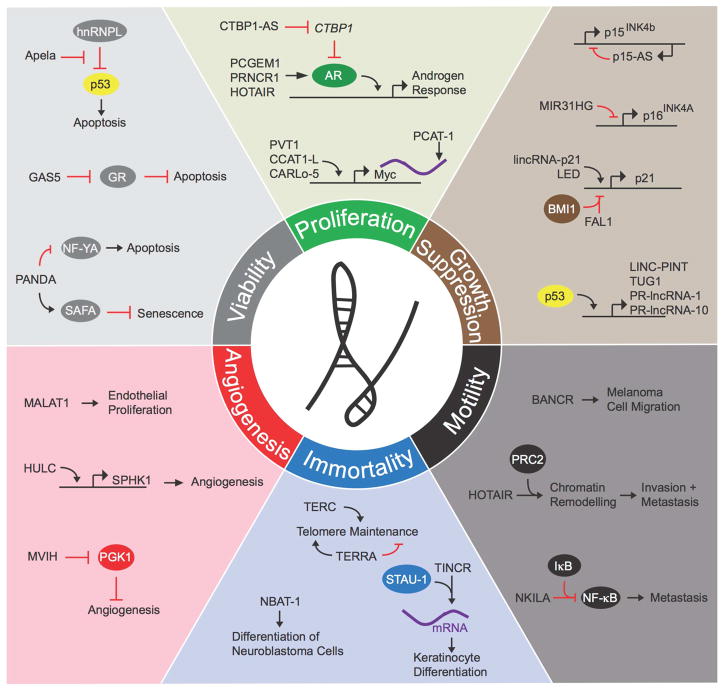
Cancer consists of a constellation of phenotypes resulting from (1) dysfunctional intrinsic cellular regulatory networks and (2) intercellular communication to generate the tumor microenvironment (Hanahan and Weinberg, 2000; Hanahan and Weinberg, 2011). Intracellular signaling networks are modulated in cancer to sustain proliferation, impair cytostatic and differentiation signals, enhance viability, and promote motility. As lncRNAs have primarily been studied in the context of intracellular networks, our review will focus on the role of lncRNAs in these specific cancer phenotypes. Indeed, each of the hallmarks of cancer, as described by Hanahan and Weinberg in 2000, is modulated by the activity of multiple lncRNAs (Figure 2).
Figure 2. LncRNAs in Cancer Phenotypes.
LncRNAs contribute to each of the six hallmarks of cancer (diagram adapted from Hanahan and Weinberg, 2000). Selected examples of lncRNAs and their molecular partners or genomic targets are shown for proliferation, growth suppression, motility, immortality, angiogenesis, and viability cancer phenotypes.
Proliferation Circuits
Multiple lncRNAs are downstream targets of chemokine and hormonal pathways (Xing et al., 2014). In T cell acute lymphoblastic leukemia, the Notch1 oncogene drives growth in part by inducing the lncRNA LUNAR1 in order to up-regulate insulin like growth factor 1 receptor expression and signaling (Trimarchi et al., 2014). Androgen signaling in prostate cancer also relies on a number of lncRNAs that are implicated in prostate cancer proliferation that act through direct interactions with the androgen receptor (Yang et al., 2013b; Zhang et al., 2015) or by inhibiting repressors of the androgen receptor (Takayama et al., 2013).
Amplification of the 8q24 locus is a well-characterized oncogenic event in many types of human malignancies resulting in MYC amplification, but multiple lines of evidence are now implicating lncRNAs in Myc-driven cancers. PVT1 is a lncRNA gene that is at the breakpoint of the t(2:8) translocation in Burkitt’s lymphoma that brings the human immunoglobulin enhancer to PVT1-MYC locus. In a mouse model of Myc oncogensis, single copy amplification of Myc alone was insufficient to enhance tumor formation whereas amplification of a multi-gene segment encompassing Myc and the lncRNA Pvt1 promoted efficient tumor development (Tseng et al., 2014). Co-amplification of PVT1 and MYC increased Myc protein levels while depletion of PVT1 in Myc-driven human colon cancer cells impaired proliferation. Furthermore, Myc transcription is activated in cis by the colon cancer associated lncRNA CCAT1, also known as CARLo-5, by facilitating long range interaction between Myc and an enhancer element (Kim et al., 2014; Xiang et al., 2014). The prostate specific lncRNA PCGEM1, also residing at the 8q24 locus, binds to Myc, and enhances Myc’s transcriptional activation of several genes involved in various metabolic processes required for growth of prostate cancer cells (Hung et al., 2014). Myc also targets numerous lncRNAs for transcriptional regulation (Zheng et al., 2014), which can in turn regulate cell cycle progression (Kim et al., 2015).
Tumor Suppressor Circuits
Recently several lncRNAs have been found to play an extensive role in modulating tumor suppressor and growth arrest pathways. The complement of lncRNA transcription is dynamically regulated under differing cell cycling conditions (Hung et al., 2011) and during senescence (Abdelmohsen et al., 2013). Several lncRNAs regulate the expression of key tumor suppressors from the CDKN2A/CDKN2B locus, which encodes p15INK4b, p16INK4a, and p14ARF. The antisense noncoding transcript p15-AS induces silencing of p15INK4b through heterochromatin formation (Yu et al., 2008) and elevated p15-AS expression is associated with low p15INK4b expression in leukemic cells. The lncRNA MIR31HG recruits Polycomb group proteins to the INK4A locus to repress its transcription during normal growth but is sequestered away from the INK4A locus during senescence (Montes et al., 2015). Expression of the tumor suppressor TCF21 is activated by its antisense RNA TARID, which recruits the GADD45a to the TCF21 promoter to facilitate demethylation (Arab et al., 2014).
Regulation of the p53 tumor suppressor pathway by lncRNAs has been a topic of especially intense interest. The maternally imprinted RNA MEG3 binds to p53 and activates p53-dependent transcription of a subset of p53-regulated genes (Zhou et al., 2007). Furthermore, the known complement of p53-regulated lncRNAs is growing rapidly pointing to widespread involvement of lncRNAs downstream of p53 activation (Huarte et al., 2010; Hung et al., 2011; Marin-Bejar et al., 2013; Sanchez et al., 2014; Younger et al., 2015; Zhang et al., 2014). Distant p53 bound enhancer regions generate enhancer RNAs that are required for p53’s regulation of multiple genes from a single enhancer site (Melo et al., 2013). Genome wide profiling of p53-regulated enhancer RNAs identified the p53 induced lncRNA LED, which interacts with and activates strong enhancers including a CKDN1A enhancer to supports cell cycle arrest by p53. LED is silenced in a subset of p53-wildtype leukemia cells indicating a possible tumor suppressor role for this lncRNA (Leveille et al., 2015). LncRNA-p21, which is induced by DNA damage in a p53-dependent manner, interacts with hnRNPK to regulate CDKN1A in cis and arrest the cell cycle in a p21-dependent manner (Dimitrova et al., 2014; Huarte et al., 2010). The lncRNA FAL1, located within a region of chromosome 1 with frequent amplification in cancer, recruits the chromatin repressor protein BMI-1 to multiple genes including CDKN1A in order to promote tumor cell proliferation (Hu et al., 2014a). Additional p53 pathway activities are also regulated by the p53 and DNA damage inducible lncRNA PANDA, which inhibits DNA damage induced apoptosis by binding to the transcription factor NF-YA and blocking its recruitment to pro-apoptotic genes (Hung et al., 2011). PANDA has also been shown to regulate senescence through and interaction with SAFA and PRC1 (Puvvula et al., 2014).
Viability Circuits
The selective advantage of tumor cells is driven by telomere maintenance, tolerance of nutrient stress, and in some cancers, preservation of an undifferentiated tumor cell population. LncRNA Growth arrest specific 5 (Gas5) is induced in cells arrested by nutrient deprivation or withdrawal of growth factors. Gas5 blocks glucocorticoid responsive gene expression by binding to the glucocorticoid receptor’s (GR’s) DNA binding domain and acting as a decoy (Hudson et al., 2014; Kino et al., 2010). This blockade of GR decreases expression of the cellular inhibitor of apoptosis 2 (Kino et al., 2010), thereby enhancing apoptosis under stressed conditions in normal cells. However, suppression of Gas5 in human breast cancer cells relative to adjacent normal breast tissue may support the enhanced viability of breast cancer cells in the nutrient poor tumor microenvironment (Mourtada-Maarabouni et al., 2009).
Tumor cell immortality requires telomere maintenance in order to avoid replicative senescence. Telomere RNA Component (TERC) is a critical component of the ribonucleoprotein telomerase complex and encodes the template for the hexanucleotide repeats that compose the telomere sequence (Feng et al., 1995). While the majority of human tumors maintain telomeres by overexpressing the reverse transcriptase TERT, single nucleotide polymorphisms at the TERC locus are associated with telomere lengthening and an increased risk of developing high-grade glioma (Walsh et al., 2014). Furthermore, TERC copy number gain strongly predicts the progression of premalignant oral cavity neoplasms to invasive cancer (Dorji et al., 2015). Meanwhile, Marek’s disease virus efficiently induces T cell lymphomas in chickens by expressing a viral telomerase RNA to promote telomere lengthening (Trapp et al., 2006). The telomeric repeat containing RNA (TERRA) transcribed from subtelomeric and telomeric DNA sequences exerts both telomerase-dependent and telomerase-independent effects on telomere maintenance (Rippe and Luke, 2015). One role for TERRA involves its dynamic regulation during the cell cycle, which regulates the exchange of single strand DNA binding protein RPA by POT-1 and thus telomere capping (Flynn et al., 2011). Cancer cells lacking the SWI/SNF component ATRX maintain persistent TERRA loci at telomeres as cells transition from S phase to G2. This results in persistent RPA occupancy on the single stranded telomeric DNA preventing telomerase-dependent telomere lengthening. These cells instead rely on the recombination-dependent alternative pathway of telomere lengthening which requires ATR, rendering ATRX-deficient cancer cells highly sensitive to ATR inhibitors (Flynn et al., 2015).
A recent report has also identified a role for a lncRNAs in the maintenance of genome stability. In the century since Boveri first described aneuploidy, chromosome aberrations have become one of the defining features of cancer. The lncRNA NORAD has been shown exert a profound effect on chromosomal stability (Lee et al., 2016). NORAD, which is an abundant lncRNA sequesters PUMILIO proteins away from target mRNA, including those for genes involved in mitosis, DNA repair and replication. PUMILIO proteins negatively regulate these mRNAs by decreasing mRNA stability and translation. Thus, NORAD−/− cells have hyperactive PUMILIO and subsequently develop genomic instability and aneuploidy.
LncRNAs are extensively involved in stem cell maintenance and differentiation circuits (Flynn and Chang, 2014). That cancer cells can co-opt these circuits by modulating lncRNAs is now appreciated. LncTCF7 recruits the SWI/SNF complex to the TCF7 promoter to activate Wnt signaling in order to maintain liver cancer stem cell self-renewal (Wang et al., 2015). Cutaneous squamous cell carcinoma cells repress the lncRNA TINCR, which is required for keratinocyte differentiation by stabilizing differentiation-associated mRNAs through recruitment of STAU-1 (Kretz et al., 2013). The lncRNA NBAT-1 has also been observed to promote neuronal differentiation in neuroblastoma cells through regulation of the neuron specific transcription factor NRSF/REST, and repression of this lncRNA is associated with high-risk neuroblastoma (Pandey et al., 2014).
Motility Circuits
Overexpression of MALAT1, an evolutionarily conserved, abundant nuclear lncRNA, was found to predict a high risk of metastatic progression in early stage non-small cell lung cancer patients (Ji et al., 2003). While MALAT1 loss of function in mouse revealed that it is a non-essential gene in development or for adult normal tissue homeostasis (Nakagawa et al., 2012; Zhang et al., 2012), depletion of MALAT1 in lung carcinoma cells impairs cellular motility in vitro and metastasis in mice (Gutschner et al., 2013), suggesting that MALAT1 overexpression in cancer may drive gain of function phenotypes not observed during normal tissue development or homeostasis.
Multiple cancer-associated lncRNAs have been implicated in regulating cancer invasion and metastases (Flockhart et al., 2012; Hu et al., 2014b; Huarte, 2015). TGF-β was found to induce the expression of lncRNA-ATB in hepatocellular carcinoma (HCC) cells which facilitated epithelial to mesenchymal transition (EMT), cellular invasion, and organ colonization by HCC cells by two distinct RNA - RNA interactions (Yuan et al., 2014). LncRNA-ATB competitively binds miR-200 to activate the expression of ZEB1 and ZEB2 during EMT, while interactions with IL-11 mRNA enhances Stat3 signaling to promote metastasis. The breast cancer associated lncRNA BCAR4 connects extracellular CCL22 to non-canonical Gli2 signaling by bindings the transcription factors SNIP1 and PNUTS, stimulating cell migration and metastasis (Xing et al., 2014). LncRNAs also mediate metastasis programs through chromatin deregulation. Overexpression of the HOX associated lncRNA HOTAIR in breast cancer reprograms the chromatin landscape genome-wide via recruitment of PRC2, enforcing a mesenchymal cellular phenotype which promotes breast cancer metastasis (Gupta et al., 2010), and is associated with poor prognosis in other malignancies as well (Kogo et al., 2011). The prostate cancer lncRNA SChLAP1, associated with poor prognosis and metastatic progression (Prensner et al., 2014b), promotes prostate cancer invasion and metastasis by disrupting the metastasis suppressing activity of the SWI/SNF complex (Prensner et al., 2013).
Recent identification of metastasis suppressing lncRNAs has opened a new perspective into a link between the tumor microenvironment and lncRNA modulation of the metastasis phenotype. The lncRNA NKILA, which is induced by NF-κB in response to inflammatory signaling, mediates a negative feedback loop suppressing NF-κB signaling by binding the cytoplasmic NF-κB/IκB complex and preventing IκB phosphorylation, NF-κB release, and nuclear localization (Liu et al., 2015). NKILA suppression in human breast cancer is linked to metastatic dissemination and poor prognosis. The lncRNA LET connects the hypoxia response to metastasis. Hypoxia-induced histone deacetylase 3 suppresses the LET promoter, impairing its expression and facilitating nuclear factor 90 accumulation and hypoxia-induced cellular invasion (Yang et al., 2013a).
Therapeutic Promise in Targeting LncRNAs
LncRNAs, as highly tissue-specific drivers of cancer phenotypes, are now set to become prime targets for cancer therapy. The development of RNA targeting therapeutics provides tremendous opportunities to modulate lncRNAs for anti-cancer purposes. While several strategies have been successfully employed to deplete lncRNAs, a prior knowledge of lncRNA cellular localization is critical for selecting the appropriate strategy to achieve robust lncRNA modulation (Lennox and Behlke, 2016). siRNAs, upon loading in the RISC complex in the cytoplasm, can efficiently deplete cytoplasmic lncRNAs, however these molecules have had variable success in targeting predominantly nuclear lncRNAs. Antisense oligonucleotides (ASOs), on the other hand, are chimeric RNA:DNA oligonucleotides that direct RNase H to cleave complimentary target mRNA or lncRNA (Ideue et al., 2009) and can robustly deplete the transcripts regardless of their cellular localization.
ASOs have already demonstrated success in modulating coding genes involved in a variety of benign disorders, lymphoma, and solid tumors (Buller et al., 2015; Gaudet et al., 2014; Hong et al., 2015; Meng et al., 2015), and are poised for use in lncRNA targeted therapeutics (Ling et al., 2013a). Preclinical studies are demonstrating the therapeutic efficacy of ASOs targeting cancer-associated lncRNAs. For example, Malat1 may be a viable therapeutic target in some cancers, such as in the mouse MMTV-PyMT breast cancer model where Malat1 drives tumor growth and metastasis. In this model, ASOs targeting Malat1 have demonstrated therapeutic efficacy in vivo by promoting cystic differentiation, increased cell adhesion, and decreased migration (Arun et al., 2016).
Inhibiting lncRNA function by RNA depletion need not be the only mechanism of targeting lncRNAs for cancer therapy. New classes of RNA-based therapeutics (Kole et al., 2012) hold promise for modulating the activity of lncRNAs in diverse ways. Splice switching oligonucleotides could be utilized to excise lncRNA exons that encode necessary functional domains. Alternatively, steric blocking oligonucleotides may interfere with the interaction of lncRNAs with their binding partners. Future technical innovations in modulating lncRNAs in vivo along with increased insights into lncRNA pathways in cancer biology will offer further opportunities for lncRNA targeting therapeutics.
Emerging Paradigms and Future Directions
The vast majority of studies in cancer-associated lncRNAs have focused on the cellular effects of altered transcript abundance of lncRNAs. However, structural approaches to evaluating lncRNA functions have revealed that even single nucleotide polymorphisms can alter local RNA structure at functionally relevant sites involved in miRNA or protein binding (Wan et al., 2014). Application of whole transcriptome RNA structural analyses in cancer may reveal functional consequences of SNPs or somatic mutations within cancer associated lncRNAs (Chu et al., 2015a; Spitale et al., 2015). Furthermore, the emerging evidence that some putative lncRNAs may encode short, translated open reading frames (Anderson et al., 2015; Ingolia et al., 2014), and that coding RNAs can exert translation-independent functional roles (Li et al., 2015), suggests that distinction between mRNA and lncRNA may be less absolute than once thought.
Cataloging the physiologic role of cancer associated lncRNAs will require a transition in investigative approaches from lncRNA annotation and molecular or cellular characterization to animal genetic models of cancer (Sauvageau et al., 2013), as recently suggested (Li and Chang, 2014). Organismal models will be especially critical for elucidating the emerging physiologic roles for noncoding RNA in intercellular signaling, inflammation, angiogenesis, and immune modulation, which are central factors in the cooperation of cancer cells and stromal cells required to support tumor growth and development (Bernard et al., 2012; Lu et al., 2015; Michalik et al., 2014; Satpathy and Chang, 2015; Yuan et al., 2012).
Acknowledgments
We apologize to colleagues whose work was not able to be included in this review due to space limitations. This work was supported by NIH (R01-CA118750 and R01-ES023168 to H.Y.C.). HYC is a founder of Epinomics and was a member of the scientific advisory board of RaNa Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelmohsen K, Panda A, Kang MJ, Xu J, Selimyan R, Yoon JH, Martindale JL, De S, Wood WH, 3rd, Becker KG, Gorospe M. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging cell. 2013;12:890–900. doi: 10.1111/acel.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam T, Medvedeva YA, Jia H, Brown JB, Lipovich L, Bajic VB. Promoter analysis reveals globally differential regulation of human long non-coding RNA and protein-coding genes. PloS one. 2014;9:e109443. doi: 10.1371/journal.pone.0109443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Molecular cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes & development. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome research. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard JJ, Cowing-Zitron C, Nakatsuji T, Muehleisen B, Muto J, Borkowski AW, Martinez L, Greidinger EL, Yu BD, Gallo RL. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nature medicine. 2012;18:1286–1290. doi: 10.1038/nm.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochimica et biophysica acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher R, Hoogland AM, Dits N, Verhoef EI, Kweldam C, Waranecki P, Bangma CH, van Leenders GJ, Jenster G. Novel long non-coding RNAs are specific diagnostic and prognostic markers for prostate cancer. Oncotarget. 2015;6:4036–4050. doi: 10.18632/oncotarget.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome biology. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller HR, Bethune C, Bhanot S, Gailani D, Monia BP, Raskob GE, Segers A, Verhamme P, Weitz JI, Investigators FAT. Factor XI antisense oligonucleotide for prevention of venous thrombosis. The New England journal of medicine. 2015;372:232–240. doi: 10.1056/NEJMoa1405760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer research. 1999;59:5975–5979. [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nature communications. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long non-coding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nature structural & molecular biology. 2015a;22:29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015b;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Molecular cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Molecular cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, Jacks T. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Molecular cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorji T, Monti V, Fellegara G, Gabba S, Grazioli V, Repetti E, Marcialis C, Peluso S, Di Ruzza D, Neri F, Foschini MP. Gain of hTERC: a genetic marker of malignancy in oral potentially malignant lesions. Human pathology. 2015;46:1275–1281. doi: 10.1016/j.humpath.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nature structural & molecular biology. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nature genetics. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome research. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell stem cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suva ML, Benes CH, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet D, Brisson D, Tremblay K, Alexander VJ, Singleton W, Hughes SG, Geary RS, Baker BF, Graham MJ, Crooke RM, Witztum JL. Targeting APOC3 in the familial chylomicronemia syndrome. The New England journal of medicine. 2014;371:2200–2206. doi: 10.1056/NEJMoa1400284. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Heard E. Fifty years of X-inactivation research. Development. 2011;138:5049–5055. doi: 10.1242/dev.068320. [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer research. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature biotechnology. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large noncoding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell reports. 2015;11:1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, Fowler N, Zhou T, Schmidt J, Jo M, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Science translational medicine. 2015;7:314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer cell. 2014a;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer research. 2014b;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo FW, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517–522. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Huarte M. The emerging role of lncRNAs in cancer. Nature medicine. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Pickard MR, de Vera IM, Kuiper EG, Mourtada-Maarabouni M, Conn GL, Kojetin DJ, Williams GT, Ortlund EA. Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nature communications. 2014;5:5395. doi: 10.1038/ncomms6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, Kung HJ. A long noncoding RNA connects c-Myc to tumor metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature genetics. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideue T, Hino K, Kitao S, Yokoi T, Hirose T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. Rna. 2009;15:1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell reports. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature genetics. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R, de la Chapelle A. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8646–8651. doi: 10.1073/pnas.1205654109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Molecular cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature cell biology. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer research. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Cui R, Jeon YJ, Fadda P, Alder H, Croce CM. MYC-repressed long noncoding RNAs antagonize MYC-induced cell proliferation and cell cycle progression. Oncotarget. 2015;6:18780–18789. doi: 10.18632/oncotarget.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Cui R, Jeon YJ, Lee JH, Lee JH, Sim H, Park JK, Fadda P, Tili E, Nakanishi H, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4173–4178. doi: 10.1073/pnas.1400350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Science signaling. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer research. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nature reviews Drug discovery. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Ungewickell A, Bhaduri A, Qu K, Webster DE, Armstrong R, Weng WK, Aros CJ, Mah A, Chen RO, et al. Transcriptome sequencing in Sezary syndrome identifies Sezary cell and mycosis fungoides-associated lncRNAs and novel transcripts. Blood. 2012;120:3288–3297. doi: 10.1182/blood-2012-04-423061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic acids research. 2016;44:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveille N, Melo CA, Rooijers K, Diaz-Lagares A, Melo SA, Korkmaz G, Lopes R, Akbari Moqadam F, Maia AR, Wijchers PJ, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nature communications. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends in cell biology. 2014;24:594–602. doi: 10.1016/j.tcb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gou H, Tripathi BK, Huang J, Jiang S, Dubois W, Waybright T, Lei M, Shi J, Zhou M, Huang J. An Apela RNA-Containing Negative Feedback Loop Regulates p53-Mediated Apoptosis in Embryonic Stem Cells. Cell stem cell. 2015;16:669–683. doi: 10.1016/j.stem.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nature reviews Drug discovery. 2013a;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome research. 2013b;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, Calin GA. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–5011. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J, Zhang Y, Chen J, Shen H, Hu Z. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PloS one. 2012;7:e35145. doi: 10.1371/journal.pone.0035145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z, Li J, Ye L, Zhang X. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1) Oncotarget. 2015 doi: 10.18632/oncotarget.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, Huang L, Moreno I, Navarro A, Monzo M, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome biology. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nature structural & molecular biology. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, Mesh K, Lorenzana E, Chopra VS, Segal E, Watanabe C, Haley B, Mayba O, Yaylaoglu M, Gnad F, Firestein R. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. The Journal of clinical investigation. 2016;126:639–652. doi: 10.1172/JCI83265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Molecular cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nature genetics. 2015;47:710–716. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circulation research. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- Montes M, Nielsen MM, Maglieri G, Jacobsen A, Hojfeldt J, Agrawal-Singh S, Hansen K, Helin K, van de Werken HJ, Pedersen JS, Lund AH. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nature communications. 2015;6:6967. doi: 10.1038/ncomms7967. [DOI] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nature reviews Genetics. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. Rna. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pageau GJ, Hall LL, Ganesan S, Livingston DM, Lawrence JB. The disappearing Barr body in breast and ovarian cancers. Nature reviews Cancer. 2007;7:628–633. doi: 10.1038/nrc2172. [DOI] [PubMed] [Google Scholar]
- Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature biotechnology. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature genetics. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Sahu A, Iyer MK, Malik R, Chandler B, Asangani IA, Poliakov A, Vergara IA, Alshalalfa M, Jenkins RB, et al. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014a;5:1434–1438. doi: 10.18632/oncotarget.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, Magi-Galluzzi C, Mehra R, Sahu A, Siddiqui J, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. The Lancet Oncology. 2014b;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvvula PK, Desetty RD, Pineau P, Marchio A, Moon A, Dejean A, Bischof O. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nature communications. 2014;5:5323. doi: 10.1038/ncomms6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nature reviews Genetics. 2015;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Zhang QC, Georgiev P, Ilik IA, Akhtar A, Chang HY. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes & development. 2016;30:191–207. doi: 10.1101/gad.272187.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K, Luke B. TERRA and the state of the telomere. Nature structural & molecular biology. 2015;22:853–858. doi: 10.1038/nsmb.3078. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics C. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends in cancer. 2015;1:93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Segura V, Marin-Bejar O, Athie A, Marchese FP, Gonzalez J, Bujanda L, Guo S, Matheu A, Huarte M. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nature communications. 2014;5:5812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sati S, Ghosh S, Jain V, Scaria V, Sengupta S. Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic acids research. 2012;40:10018–10031. doi: 10.1093/nar/gks776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes & development. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, Urano T, Fujimura T, Takagi K, Takahashi S, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. The EMBO journal. 2013;32:1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Hu S, Wang S, Zhou X, Zhang Q, Wang C, Zhao X, Zhou W, Zhang S, Li C, et al. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis. 2015;36:1136–1143. doi: 10.1093/carcin/bgv099. [DOI] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Lee SH, Jones A, Fiegl H, Kalwa M, Wagner W, Chindera K, Evans I, Dubeau L, Orjalo A, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome medicine. 2015;7:108. doi: 10.1186/s13073-015-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nature genetics. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- Trapp S, Parcells MS, Kamil JP, Schumacher D, Tischer BK, Kumar PM, Nair VK, Osterrieder N. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. The Journal of experimental medicine. 2006;203:1307–1317. doi: 10.1084/jem.20052240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP. The long non-coding RNA Paupar regulates the expression of both local and distal genes. The EMBO journal. 2014;33:296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, Kollmeyer T, Kosel ML, Molinaro AM, McCoy LS, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nature genetics. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, Chang HY. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic acids research. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell stem cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Wei JT, Feng Z, Partin AW, Brown E, Thompson I, Sokoll L, Chan DW, Lotan Y, Kibel AS, Busby JE, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2014;32:4066–4072. doi: 10.1200/JCO.2013.52.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Molecular cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell research. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed research international. 2013;2013:136106. doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Molecular cell. 2013a;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013b;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Molecular cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger ST, Kenzelmann-Broz D, Jung H, Attardi LD, Rinn JL. Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic acids research. 2015;43:4447–4462. doi: 10.1093/nar/gkv284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D, Mo YY, Yu J. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell reports. 2015;13:209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell reports. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell death & disease. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN, Klibanski A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer research. 2010;70:2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Do BT, Webster DE, Khavari PA, Chang HY. Dicer-microRNA-Myc circuit promotes transcription of hundreds of long non-coding RNAs. Nature structural & molecular biology. 2014;21:585–590. doi: 10.1038/nsmb.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long noncoding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Scientific reports. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. The Journal of biological chemistry. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Rowley MJ, Bohmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Molecular cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]