ABSTRACT
Free fetal DNA (ffDNA) in maternal plasma has now become a valuable source for noninvasive prenatal diagnosis. Being able to accurately identify the size of ffDNA in maternal plasma is essential for a noninvasive prenatal diagnosis. Furthermore, it is important to investigate the molecular characteristics related to apoptosis which gives rise to ffDNA. We investigated the fragment size of ffDNA in each sample more precisely, using both Y-STR and SRY primers, in 20 maternal plasma samples from the 17th to 39th weeks of gestation. PCR was conducted with Y-STR and SRY primers which can be used to amplify 100–524 bp fragments. In samples from 10 pregnant women carrying male fetuses, the maximum fragment size detected by Y-STR and SRY primers ranged from 219 to 313 bp. As a result, the mean average maximum fragment size of free fetal DNA detected by Y-STR and SRY primers was 286±28 bp. The Y-STR alleles detected in each maternal plasma DNA sample were all in agreement with the results of their cord blood samples. We concluded that the fragment size of ffDNA comprises 2 nucleosomal complexes or less, but not exceeding 3.
Key Words: Free fetal DNA, Prenatal diagnosis, Electrophoresis, Size fractionation
INTRODUCTION
A significant proportion of cell-free DNA is bound to protein molecules as nucleosomes. Circulating DNA fragments are mainly sized in multiples of the nucleosomal DNA.1,2) The presence of fragmented DNA is reliable evidence of apoptosis, and this sign is widely used as a qualitative marker of cell death (apoptosis).3,4)
The size characteristics of free fetal DNA (ffDNA) fragments from apoptosis have been analyzed. According to the work reported by Chan et al.,5) the median percentages of fetal-derived DNA in maternal plasma with sizes >193 and > 313 bp were 20 and 0%, respectively, while Li Ying et al.6) also reported that fragment sizes of less than –0.3 kb were enriched by fetal DNA.
Free fetal DNA concentration in maternal plasma increases as pregnancy advances, and placental apoptosis has been reported to increase with gestational age.7-9)
However, the precise maximum fragment size of ffDNA in maternal plasma throughout the pregnancy period has not yet been precisely reported.
We investigated the size distribution of ffDNA and related this to apoptosis-associated fragmentation using both Y-chromosomal short-tandem repeat (Y-STR) and SRY primers. Evaluating the fragment size of ffDNA is essential for choosing the target genes for noninvasive prenatal diagnosis and investigating the molecular characteristics related to the apoptosis of ffDNA.
We used a commercial AmpFlSTR® Y-filerTM kit (Applied Biosystems, CA, USA), which can detect the size distributions of Y-STR fragments ranging from 100 to 325 bp, including 17 Y-STR loci. We also examined whether a > 313-bp fragment size was detectable in samples in which the Y-STR loci were determined by performing PCR using SRY primers (193, 313, 392 and 524 bp), as reported by Chan et al.5) By combining these Y-STR and SRY primers, we were able to investigate the fragment size of ffDNA more precisely.
MATERIALS AND METHODS
Twenty pregnant women in their 17th to 39th weeks of gestation without pregnancy-associated complications were enrolled. Ethical approval for this project was obtained from the Saitama Medical University Ethics Committee, and all samples were obtained with informed consent.
Plasma preparation
Seven milliliters of peripheral blood was collected into tubes containing EDTA. The samples were first centrifuged at 3,000 g for 5 minutes at room temperature, and the plasma was then collected and subjected to further centrifugation at 10,000 g for 5 minutes. We also collected cord blood samples (2 mL) from each pregnant woman after delivery.
DNA extraction
Total plasma DNA was extracted with a QIA amp Mini Blood kit (Qiagen) according to the manufacturer’s instructions. The plasma DNA isolated from 1 mL of maternal plasma was eluted in a final volume of 50 uL of AE buffer. Furthermore, we extracted DNA from 1 mL of cord blood (whole blood) to confirm our results after delivery.
PCR reaction
17 Y-STR loci multiplex amplification
Using maternal plasma samples, the 17 Y-STR loci were co-amplified using the AmpFlSTR® Y-filerTM kit (Applied Biosystems, CA, USA, PCR Reagents Lot No.: 0608005, Ampli Taq Gold DNA Polymerase Lot No.: 0606079, Allelic Ladder Lot No.: 0604002) according to the manufacturer’s instructions.
Extracted cord blood DNA was diluted to 0.1 ng/uL in distilled water, and the PCR conditions were the same as those above, except for the number of PCR cycles, which was 30 after the initial denaturation step.
SRY gene amplification
SRY gene amplification was performed using samples in which Y-STR loci were found in maternal plasma. SRY primers (193, 313, 392 and 524 bp) were applied. Primers were used as previously reported by Chan et al. (F: 5’-AAA GGC AAC GTC CAG GAT AGA G-3’; R1: 5’-TGT AAT TTC TGT GCC TCC TGG A-3, amplicon size 193 bp; R2: 5’-ACT TCG CTG CAG AGT GTA CCG AA-3’, amplicon size 313 bp; R3: 5’-TAA GTG GCC TAG CTG GTG CTC-3’, amplicon size 392 bp R4: 5’-ATG TTA CCC GAT TGT CCT ACA GC-3, amplicon size 524 bp). The forward primers were labeled at the 5’ end with the fluorescent dye 5-FAM (Applied Biosystems). PCR was carried out with a reaction volume of 20 uL containing 2 uL of 10x PCR Gold Buffer (Applied Biosystems), 1.0 uM of each primer, 1.75 mM of MgCl2 (Perkin Elmer), 0.2 mM of dNTPs (Applied Biosystems), 2.0 U of Ampli Taq GoldTM (Applied Biosystems), and 10 uL of DNA. Following preheating at 95°C for 11 minutes, 40 cycles of amplification at 94°C for 1 minute, 55°C for 1 minute, 72°C for 1 minute, and a final extension of 60°C for 45 minutes were carried out in a GeneAmp PCR System 9700 (Applied Biosystems).
Detection of PCR products with capillary electrophoresis
Following the manufacturer’s instructions, each sample mixture was prepared by adding 1.5 uL of PCR products to 24 uL of formamide, and 1 uL of internal-size standard ROX-500 (Applied Biosystems). The sample mixture was denatured at 95°C for 3 minutes, and then kept at 4°C in ice water. The results were analyzed using the GeneMapperTM ID Software Version 3.2 package (Applied Biosystems). A peak detection threshold of 50 RFUs (relative fluorescence units) was used for the SRY-specific peak, and 150 RFUs was the level for Y-STR allele designation.
RESULTS
Among plasma samples from 20 pregnant women, Y-STR loci were identified in 10. These ten mothers bore male infants, while the women carrying female fetuses showed no such peaks. Comparing the Y-STR genotyping results of maternal plasma samples carrying male fetuses with the results of their cord blood samples after delivery, the Y-STR alleles detected in each maternal plasma DNA sample were all concordant with the results of their cord blood samples (Table 1). All the ffDNAs of Y-STR observed in maternal plasma were detected in cord blood.
Based on the above 10 samples, the maximum fragment size detected by Y-STR genotyping of each sample ranged from 219 to 302 bp, with a mean of 275±26 bp (Table 1). Combining Y-STR and SRY genotyping, the detected maximum fragment sizes of those 10 samples ranged from 219 to 313 bp, with a mean of 286±28 bp (Figure 1 and Table 2).
Table 1.
| No. | Gestation (week). | Y-STR genotyping | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DYS 393 |
DYS 456 |
Y GATAH4 |
DYS 458 |
DYS 389I |
DYS 391 |
DYS 19 |
DYS 437 |
DYS 390 |
DYS 439 |
DYS 438 |
DYS 635 |
DYS 385a |
DYS 385b |
DYS 389II |
DYS 448 |
DYS 392 |
|||
| * | 100- 130 |
103- 120 |
120- 140 |
130- 155 |
140- 165 |
150- 175 |
175- 210 |
180- 200 |
190- 225 |
195- 225 |
225- 250 |
245- 270 |
240- 315 |
250- 275 |
280- 325 |
290- 325 |
|||
| 1 | 17 | A | 13 (118) |
15 (111) |
11 (133) |
15 (134) |
13 (154) |
10 (162) |
(-) |
14 (185) |
24 (214) |
12 (212) |
10 (233) |
(-) |
13 (265) |
(-) |
30 (276) |
18 (286) |
11 (302) |
| B | 13 | 15 | 11 | 15 | 13 | 10 | 17 | 14 | 24 | 12 | 10 | 21 | 13 | 17 | 30 | 18 | 11 | ||
| 2 | 19 | A | - | - | 11 (133) |
- | - | - | - | 15 (189) |
- | - | - | 19 (241) |
- | - | - | - | - |
| B | 13 | 15 | 11 | 18 | 12 | 10 | 15 | 15 | 23 | 11 | 10 | 19 | 12 | 18 | 29 | 19 | 12 | ||
| 3 | 20 | A | 13 (119) |
15 (111) |
- | 15 (134) |
- | 10 (162) |
- | 14 (185) |
- | - | - | - | - | - | - | 17 (280) |
- |
| B | 13 | 15 | 12 | 15 | 13 | 10 | 15 | 14 | 24 | 11 | 10 | 21 | 13 | 16 | 31 | 17 | 12 | ||
| 4 | 22 | A | 13 (119) |
15 (111) |
12 (137) |
20 (155) |
14 (159) |
10 (163) |
15 (194) |
14 (185) |
22 (207) |
12 (212) |
13 (248) |
- | - | - | - | 18 (286) |
- |
| B | 13 | 15 | 12 | 20 | 14 | 10 | 15 | 14 | 22 | 12 | 13 | 20 | 10 | 19 | 19 | 18 | 13 | ||
| 5 | 23 | A | 12 (115) |
16 (115) |
12 (137) |
21 (159) |
- | - | - | - | 25 (219) |
13 (216) |
- | - | - | - | - | - | - |
| B | 12 | 16 | 12 | 21 | 12 | 10 | 17 | 14 | 25 | 13 | 10 | 22 | 12 | 20 | 29 | 19 | 13 | ||
| 6 | 20 | A | 14 (123) |
15 (111) |
11 (133) |
- | 12 (150) |
10 (162) |
- | 14 (185) |
24 (214) |
- | - | - | - | - | 29 (272) |
18 (286) |
- |
| B | 14 | 15 | 11 | 16 | 12 | 10 | 15 | 14 | 24 | 12 | 10 | 21 | 12 | 14 | 29 | 18 | 14 | ||
| 7 | 23 | A | 13 (119) |
15 (111) |
12 (137) |
14 (130) |
12 (150) |
11 (166) |
16 (198) |
14 (185) |
- | 12 (212) |
10 (233) |
- | - | - | - | 18 (286) |
- |
| B | 13 | 15 | 12 | 14 | 12 | 11 | 16 | 14 | 23 | 12 | 10 | 21 | 13 | 13 | 28 | 18 | 14 | ||
| 8 | 24 | A | 12 (115) |
15 (111) |
12 (137) |
18 (146) |
12 (150) |
10 (162 |
- | - | 24 (214) |
- | 11 (238) |
23 (257) |
- | - | 28 (268) |
- | - |
| B | 12 | 15 | 12 | 18 | 12 | 10 | 16 | 14 | 24 | 12 | 11 | 23 | 14 | 16 | 28 | 20 | 13 | ||
| 9 | 24 | A | 13 (119) |
15 (111) |
12 (137) |
19 (151) |
14 (159) |
10 (162) |
- | 14 (185) |
- | 12 (212) |
13 (248) |
- | - | - | - | 18 (286) |
- |
| B | 13 | 15 | 12 | 19 | 14 | 10 | 15 | 14 | 22 | 12 | 13 | 20 | 10 | 21 | 29 | 18 | 13 | ||
| 10 | 39 | A | 12 (115) |
15 (111) |
- | 17 (142) |
13 (154) |
10 (163) |
- | 14 (185) |
- | 12 (212) |
11 (238) |
21 (249) |
- | - | - | 18 (286) |
12 (305) |
| B | 12 | 15 | 11 | 17 | 13 | 10 | 15 | 14 | 23 | 12 | 11 | 21 | 13 | 18 | 31 | 18 | 12 | ||
Y-STR and SRY genotyping of 10 pregnant women carrying male fetuses, and results of their corresponding cord blood samples. These Y-STR loci represent a paternally inherited haploid transmission pattern and reside on male-specific DNA; female DNA is not reactive.
A), results for detected alleles and allele sizes (bp) using maternal plasma DNA. The maximum fragment size detected by Y-STR genotyping of each sample is emphasized in bold font.
B), results of detected alleles of cord blood samples.
*Allelic size ranges (bp)
- Not detected
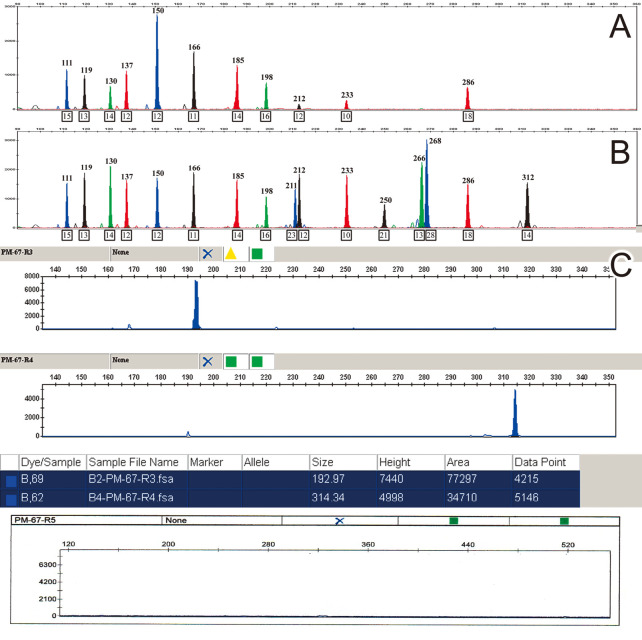
Fig. 1.
Electropherograms of PCR amplicons generated with primer pairs of Y-STR and SRY gene (sample 7 in Table. 1).
A) illustrates Y-STR genotyping results using maternal plasma DNA from pregnant women carrying a male fetus.
B) indicates Y-STR genotyping result from the cord blood sample. The detected Y-STR alleles were all in agreement with results of the maternal plasma sample.
C) shows CE peak heights detected by each primer of the SRY gene. In this sample, PCR amplicon of the R5 primer (amplicon size 392 bp) was not detected. The maximum fragment detected in this sample was 313 bp.
Table 2.
| No. | Gestation (weeks) |
SRY genotyping | Maximum fragment size detected by Y-STR and SRY primers |
|||
|---|---|---|---|---|---|---|
| 193 bp | 313 bp | 392 bp | 524 bp | |||
| 1 | 17 | + | + | – | – | 313 |
| 2 | 19 | + | + | – | – | 313 |
| 3 | 20 | + | – | – | – | 280 |
| 4 | 22 | + | – | – | – | 286 |
| 5 | 23 | + | – | – | – | 219 |
| 6 | 20 | + | – | – | – | 286 |
| 7 | 23 | + | + | – | – | 313 |
| 8 | 24 | + | – | – | – | 268 |
| 9 | 24 | + | – | – | – | 286 |
| 10 | 39 | + | – | – | – | 305 |
SRY genotyping of 10 pregnant women (same as in Table 1).
Combining Y-STR and SRY genotyping, the detected maximum fragment size of these 10 samples ranged from 219 to 313 bp, with a mean of 286±28 bp.
DISCUSSION
The origin of circulating nucleic acids is presently unknown. Although previous studies have demonstrated that serum or plasma DNA levels are often increased in patients with various malignancies and in those with several benign diseases such as infections, sepsis, trauma, stroke, and autoimmune disease,10-13) the plasma DNA concentrations of those patients have been found to be closely correlated with the levels of circulating nucleosomes, which are products of apoptosis.14) These findings suggest that increased rates of cell death events such as apoptosis or necrosis are considered the main sources of circulating fragmented DNA.
Free fetal DNA concentrations in maternal plasma increase as pregnancy advances and have also been found to correlate with the maternal human chorionic gonadotropin concentrations produced by syncytiotrophoblasts.7,8,15) Furthermore, ffDNA concentrations are significantly elevated in fetal aneuploidy and preeclampsia.16-19) However, the precise clearance and release mechanisms of ffDNA also remain unknown.
Nucleosomal complexes consist of histones with ~146 bp of DNA on the outside.20) Plasma DNA from cancer patients contains fragments of various lengths, ranging from small (180-bp or shorter mononucleosomes, and those twice, three times, and four times longer than the mononucleosomes) due to apoptosis as well as very large fragments (>10,000 bp) due to necrosis. In various pathological conditions including cancer, plasma DNA has been shown to circulate mainly in the form of mononucleosomes.21-23) However, ffDNA cannot be visualized directly by electrophoresis because of the small amount of ffDNA in maternal plasma (0.39–11.4%)8) as well as the presence of maternal DNA in the background.
Chan et al.5) conducted PCR for ffDNA using six SRY primers varying in size from 107 to 524 bp, and demonstrating that product sizes of 313 bp or longer accounted for less than 1%, with most products being shorter than 193 bp. Jorgez et al.24) conducted real-time PCR for a quantitative comparison of SRY genes between male fetuses and adult males, and revealed a significant difference at 100–300 bp, but not above that size, throughout the pregnancy period. Thus, it has been reported that 300-bp or shorter ffDNA fragments are present in abundance. Here, we used a Y-STR kit that can evaluate sizes in 4-bp increments between 100 and 325 bp to examine the presence of 200–300-bp-sized fragments in individual cases and the ffDNA status during apoptosis.
Although our results showed the mean maximum fragment size of ffDNA, 286 bp, to be caused by the apoptosis of 2 nucleosomal complexes, we found no fragment sizes indicating apoptosis of more than 3 nucleosomal complexes. We therefore concluded that the fragment size of ffDNA comprises 2 nucleosomal complexes or less throughout the pregnancy period.
This was a new finding, concerning the molecular characteristics of ffDNA, a potentially important clue to improving future prenatal diagnosis.
REFERENCES
- 1).Kerr JF, Winterford CM, Harmon B. Apoptosis: its significance in cancer and cancer therapy. Cancer, 1994; 73: 2013–2026. [DOI] [PubMed]
- 2).Lichtenstein AV, Melkonyan HS, Tomei LD, Umansky SR. Circulating nucleic acids and apoptosis. Ann N Y Acad Sci, 2001; 945: 239–249. [DOI] [PubMed]
- 3).Hacker G. The morphology of apoptosis. Cell Tissue Res, 2000; 301: 5–17. [DOI] [PubMed]
- 4).Nath R, Scott M, Nadimpalli R, Gupta R, Wang KK. Activation of apoptosis-linked caspase(s) in NMDA-injured brains in neonatal rats. Neurochem Int, 2000; 36: 119–126. [DOI] [PubMed]
- 5).Chan KCA, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, Lo KW, Huang DW, Lo YM. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem, 2004; 50: 88–92. [DOI] [PubMed]
- 6).Li Y, Zimmermann B, Rusterholz C, Kang A, Holzgreve W, Hahn S. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin Chem, 2004; 50: 1002–1011. [DOI] [PubMed]
- 7).Ariga H, Ohto H, Busch MP, Imamura S, Watson R, Reed W, Lee TH. Kinetics of fetal cellular and cell-free DNA in the maternal circulation during and after pregnancy: implications for noninvasive prenatal diagnosis. Transfusion, 2001; 41: 1524–1530. [DOI] [PubMed]
- 8).Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet, 1998; 62: 768–775. [DOI] [PMC free article] [PubMed]
- 9).Smith SC, Baker PN, Symonds EM. Placental apoptosis in normal human pregnancy. Am J Obstet Gynecol, 1997; 177: 57–65. [DOI] [PubMed]
- 10).Rumore PM, Steinman CR. Endogenous circulating DNA in systemic lupus erythematosus: occurrence as multimeric complexes bound to histone. J Clin Invest, 1990; 86: 69–74. [DOI] [PMC free article] [PubMed]
- 11).Zeerleder S, Zwart B, Wuillemin WA, Aarden LA, Groeneveld AB, Caliezi C, van Nieuwenhuijze AE, van Mierlo GJ, Eerenberg AJ, Lämmle B, Hack CE. Elevated nucleosome levels in systemic inflammation and sepsis. Crit Care Med, 2003; 31: 1947–1951. [DOI] [PubMed]
- 12).Lo YM, Rainer TH, Chan LY, Hjelm NM, Cocks RA. Plasma DNA as a prognostic marker in trauma patients. Clin Chem, 2000; 46: 319–323. [PubMed]
- 13).Rainer TH, Wong LK, Lam W, Yuen E, Lam NY, Metreweli C, Lo YM. Prognostic use of circulating plasma nucleic acid concentrations in patients with acute stroke. Clin Chem, 2003; 49: 562–569. [DOI] [PubMed]
- 14).Holdenrieder S, Stieber P, Chan LY, Geiger S, Kremer A, Nagel D, Lo YM. Cell-free DNA in serum and plasma: comparison of ELISA and quantitative PCR. Clin Chem, 2005; 51: 1544–46. [DOI] [PubMed]
- 15).Ohashi Y, Miharu N, Honda H, Samura O, Ohama K. Correlation of fetal DNA and human chorionic gonadotropin concentrations in second-trimester maternal serum. Clin Chem, 2002; 48: 386–368. [PubMed]
- 16).Lo YM, Lau TK, Zhang J, Leung TN, Chang AM, Hjelm NM, Elmes RS, Bianchi DW. Increased fetal DNA concentrations in the plasma of pregnant women carrying fetuses with trisomy 21. Clin Chem. 1999; 45: 1747–1751. [PubMed]
- 17).Lo YM, Leung TN, Tein MS, Sargent IL, Zhang J, Lau TK, Haines CJ, Redman CW. Quantitative abnormalities of fetal DNA in maternal serum in preeclampsia. Clin Chem, 1999; 45: 184–188. [PubMed]
- 18).Zhong XY, Bürk MR, Troeger C, Jackson LR, Holzgreve W, Hahn S. Fetal DNA in maternal plasma is elevated in pregnancies with aneuploid fetuses. Prenat Diagn, 2000; 20: 795–798. [DOI] [PubMed]
- 19).Zhong XY, Laivuori H, Livingston JC, Ylikorkala O, Sibai BM, Holzgreve W, Hahn S. Elevation of both maternal and fetal extracellular circulating deoxyribonucleic acid concentrations in the plasma of pregnant women with preeclampsia. Am J Obstet Gynecol, 2001; 184: 414–419. [DOI] [PubMed]
- 20).Luger K. Structure and dynamic behavior of nucleosomes. Curr Opin Genet Dev, 2003; 13: 127–135. [DOI] [PubMed]
- 21).Fournié GJ. Circulating DNA and lupus nephritis. Kidney Int, 1988; 3: 487–497. [DOI] [PubMed]
- 22).Le Lann AD, Fournié GJ, Boissier L, Toutain PL, Benoist H. In vitro inhibition of natural killer mediated lysis by chromatin fragments. Cancer Immunol Immun, 1994; 39: 185–192. [DOI] [PMC free article] [PubMed]
- 23).Duvall E, Wyllie AH. Death and the cell. Immunol Today, 1986; 7: 115–119. [DOI] [PubMed]
- 24).Jorgez CJ, Bischoff FZ. Improving enrichment of circulating fetal DNA for genetic testing: size fractionation followed by whole gene amplification. Fetal Diagn Ther, 2009; 25: 314–319. [DOI] [PMC free article] [PubMed]