Abstract
Background
The aim of this meta-analysis was to determine whether genetic polymorphisms in the osteoprotegerin (OPG) gene contribute to increased risk of cardiovascular disease (CVD).
Material/Methods
Electronic databases were searched carefully without any language restriction. Analyses of data were conducted using STATA software. Odds ratios (OR) and 95% confidence intervals (95%CI) were also calculated.
Results
Seven clinical case-control studies that enrolled 1170 CVD patients and 1194 healthy subjects were included. The results indicated that OPG gene polymorphism might be closely associated with susceptibility to CVD, especially for rs2073617 T>C and rs2073618 G>C polymorphisms. Ethnicity-stratified analysis indicated that genetic polymorphism in the OPG were closely related with the pathogenesis of CVD among Asians (all P<0.001), but no obvious relationship was found among Caucasians (all P>0.05).
Conclusions
Our meta-analysis provided quantitative evidence that OPG gene polymorphism may be closely related to an increased risk of CVD, especially for rs2073617 T>C and rs2073618 G>C polymorphisms.
MeSH Keywords: Cardiovascular Diseases; Meta-Analysis; Polymorphism, Genetic; RANK Ligand
Background
Cardiovascular disease (CVD), a leading cause of death worldwide, refers to any disease that affects the human cardiovascular system, primarily including cardiac diseases, vascular diseases of the brain and kidney, and peripheral arterial disease [1]. It is reported that CVD accounts for approximately 30% of all deaths worldwide, and the mortality rates of CVD vary between developed countries and low- and middle-income countries [2]. Furthermore, it has been estimated that approximately 82,600,000 American adults have 1 or more types of CVD, and about half of those people are over 60 years old [3]. Typically, CVD is a multifactorial disease caused by a combination of genetic and non-genetic factors [4,5]. Though massive environmental factors have been established in CVD, including smoking, unhealthy diet, reduced physical activity, obesity, stress, diabetes, hypertension, behavior, and lifestyle changes, genetic factors also have been demonstrated to be vital in the pathogenesis of CVD [4,6–8]. Growing evidence has indicated that genetic polymorphisms of some special genes play vital roles in the pathogenesis of CVD [9,10]. During the past decades, epidemiological studies have documented that osteoprotegerin (OPG), which is normally produced in vascular smooth muscle and endothelial cells, might be strongly related to the development of CVD [5,11].
OPG is a secreted basic glycoprotein that is a part of tumor necrosis factor (TNF) receptor super-family [12]. OPG was observed to be synthesized by osteoblasts and is found in a variety of organs and tissues, such as kidney, heart, lung, bone, and vessel wall [13]. By binding to the receptor activator of nuclear factor-γB ligand (RANKL), OPG can modulate bone resorption, functioning as a decoy receptor. It is also important for the survival, differentiation, and activation of osteoclasts and can regulate osteoblast-osteoclast cross-talks and bone homeostasis [14]. Recently, serum OPG level was demonstrated to be correlated with the severity of atherosclerosis, particularly with the progression of CVD [5,15]. This was partly because arterial wall calcification may be related to reduced bone mineral density and increased incidence of fractures [16,17]. Bone metabolism involves alternate cycles of bone resorption and formation, and the RANKL-OPG system may be beneficial in the maturation of osteoclasts [18]. The potential relationship between OPG and calcification in arterial walls was investigated in several experimental studies, which illustrated that OPG gene knockout mice also have aortic calcification and osteoporosis at the same time [19]. In this regard, OPG may play a crucial role in arterial wall calcification, and thus contribute to the development of CVD. More importantly, genetic polymorphisms related to alteration in expression of OPG may also be predisposing factors for the risk of CVD [11,14]. The human OPG gene is located on chromosome 8q23–24; it spans approximately 29 kb and consists of 5 exons [20]. Many reports have focused on the relation between genetic polymorphisms in OPG gene and susceptibility to CVD, especially for rs2073617 T>C and rs2073618 G>C polymorphisms [13,21,22]. However, the results from relevant studies were inconclusive and contradictory [23,24]. Therefore, we performed this meta-analysis to examine the connection between OPG genetic polymorphisms and pathogenesis of CVD.
Material and Methods
Literature search and selection criteria
The Cochrane Library Database, MEDLINE, EMBASE, the Chinese Biomedical Database (CBM), and Web of Science databases were searched carefully without any language restriction. The following keywords and MeSH terms were used: (“genetic polymorphism” or “SNP” or “variation” or “single nucleotide polymorphism” or “polymorphism” or “mutation” or “variant”) and (“osteoprotegerin” or “TNFRSF11B” or “OPG” or “osteoclastogenesis inhibitory factor” or “follicular dendritic cell-derived receptor-1” or “FDCR-1 protein” or “tumor necrosis factor receptor 11b”) and (“coronary artery disease” or “acute coronary syndrome” or “myocardial infarction” or “coronary arteriosclerosis” or “MI” or “CAD” or “CHD” or “ischemic heart disease” or “ACS” or “cardiovascular disease” or “CVD”). Moreover, a manual search was carried out to acquire other potential studies.
Inclusion criteria were: (1) studies focus on the relations between OPG gene polymorphism and susceptibility to CVD; (2) all patients should be diagnosed with any type of cardiovascular diseases, such as coronary artery disease (CAD), acute coronary syndrome (ACS), and ischemic heart disease (IHD); (3) the genotype frequencies of control subjects should follow Hardy-Weinberg equilibrium (HWE); (4) sufficient data about frequency of OPG polymorphisms must be provided. Articles that did not fit these inclusion criteria were eliminated.
Data extraction
The following information from included studies was extracted by 2 authors: first author’s surname, publication year, language, geographical location, subject source, study design, total number of cases, sample size, the frequency of single-nucleotide polymorphisms (SNPs), detection method of genotypes, and type of disease.
Statistical analysis
Statistical data were analyzed with STATA statistical software (Version 12.0, College Station, TX, USA). Odds ratios (OR) and corresponding 95% confidence intervals (95%CI) were estimated. The significance of pooled data and ORs were evaluated by Z test. Heterogeneity between reports was analyzed using the Cochran’s Q-statistic and I2 tests [25]. A P value <0.05 or I2 >50% means studies were heterogeneous, in which case the random-effects model was used; otherwise, the fixed-effects model was used. Meta-regression and subgroup analyses were also applied to explore sources of heterogeneity. Sensitivity analysis was also performed. Potential publication bias was examined using Funnel plots and Egger’s test [26].
Results
Study selection and characteristics of included studies
Initially, our search strategy obtained 184 articles. We screened the titles and abstracts and then removed 90 articles. After reviewing the full texts, we excluded 81 articles. In addition, 2 studies were eliminated for lack of data integrity (Figure 1). Finally, 7 clinical case-control studies that enrolled 1170 CVD patients and 1194 healthy subjects were enrolled [13,14,22–24,27,28]. The publication years of enrolled studies ranged from 2004 to 2012. Overall, 5 studies were performed among Asians, and the other 2 studies were among Caucasians. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), Minisequencing, TaqMan assay, and PCR-ligase detection reaction (PCR-LDR) method were implemented for genotyping. Two genetic polymorphisms (rs2073617 T>C and rs2073618 G>C) in the OPG gene were analyzed in our meta-analysis. Genotype frequencies of controls were all in HWE (all P>0.05). The characteristics and methodological quality of enrolled studies are demonstrated in Table 1.
Figure 1.
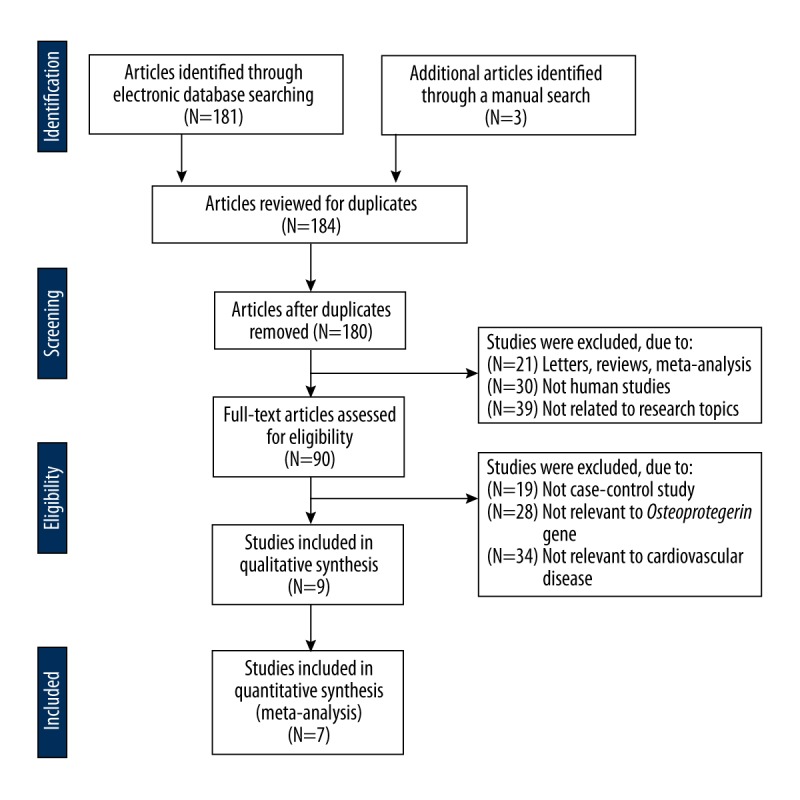
Flow chart shows study selection procedure. Seven case-control studies were included in this meta-analysis.
Table 1.
Main characteristics and methodological quality of all eligible studies.
| First author | Year | Language | Disease | Number | Gender (M/F) | Age (years) | Genotype methods | SNP | NOS score | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Case | Control | |||||||
| Luo ZR [28] | 2012 | Asians | ACS | 360 | 360 | 216/144 | 210/150 | 66.0 ±5.0 | 60.0 ±10.0 | PCR–RFLP | rs2073617 T>C | 8 |
| rs2073618 G>C | ||||||||||||
| Hong WL [27] | 2012 | Asians | ACS | 69 | 65 | – | – | 70.1 ±10.9 | 63.7 ±10.8 | PCR–LDR | rs2073618 G>C | 5 |
| Celczynska-Bajew L [22] | 2011 | Caucasians | CAD | 31 | 30 | 0/31 | 0/30 | 66 (39~82) | 71 (56~84) | Minisequencing | rs2073618 G>C | 6 |
| Fang K [23] | 2010 | Asians | CHD | 150 | 150 | 106/44 | 95/55 | 64.0 ±10.0 | 58.0 ±10.0 | PCR–RFLP | rs2073617 T>C | 7 |
| rs2073618 G>C | ||||||||||||
| Xu L [24] | 2009 | Asians | CHD | 48 | 102 | – | – | – | – | PCR–RFLP | rs2073617 T>C | 5 |
| Ohmori R [13] | 2006 | Asians | CAD | 405 | 126 | 405/0 | 126/0 | 63.0 ±9.0 | 59.0 ±10.0 | TaqMan assay | rs2073617 T>C | 8 |
| rs2073617 T>C | ||||||||||||
| Soufi M [14] | 2004 | Caucasians | CAD | 107 | 361 | 107/0 | 361/0 | 58.5 ±8.9 | – | TaqMan assay | rs2073617 T>C | 7 |
| rs2073618 G>C | ||||||||||||
M – male; F – female; SNP – single-nucleotide polymorphisms; NOS – Newcastle-Ottawa Scale; CAD – coronary artery disease; ACS – acute coronary syndrome; CHD – coronary heart disease; PCR-RFLP – polymerase chain reaction-restriction fragment length polymorphism; PCR-LDR – PCR-ligase detection reaction.
Quantitative data synthesis
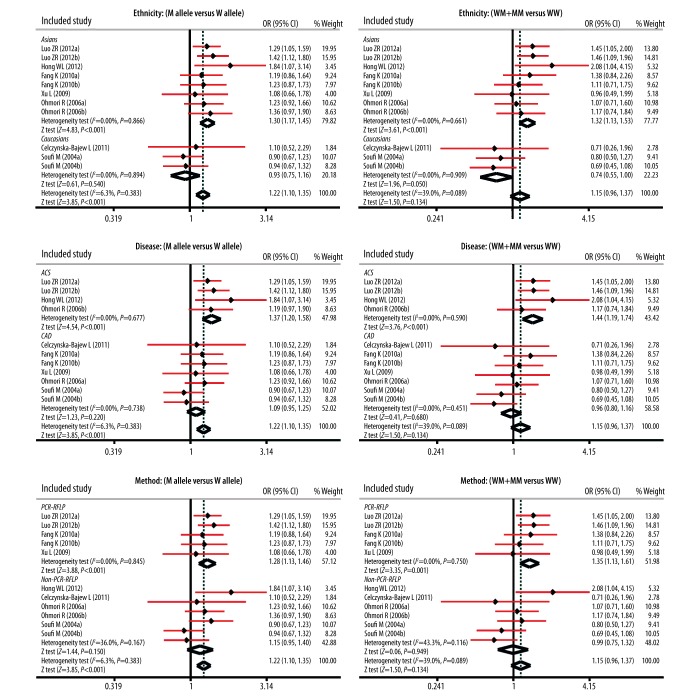
As presented in Figure 2, our study showed that OPG rs2073617 T>C and rs2073618 G>C polymorphisms had statistical significance in the allele model (rs2073617 T>C: OR=1.19, 95%CI: 1.06~1.35, P=0.005; rs2073618 G>C: OR=1.26, 95%CI: 1.03~1.55, P=0.026), but not in dominant models (both P>0.05). In Table 2, we summarized the results for the correlations between OPG gene polymorphism and susceptibility of CVD. Our findings demonstrated that OPG genetic polymorphisms were associated with an increased risk of CVD (M allele vs. W allele: OR=1.22, 95%CI: 1.10~1.35, P<0.001; MM vs. WW+WM: OR=1.49, 95%CI: 1.24~1.79, P<0.001; MM vs. WW: OR=1.59, 95%CI: 1.30~1.94, P<0.001; MM vs. WM: OR=1.52, 95%CI: 1.20~1.92, P<0.001; respectively.
Figure 2.
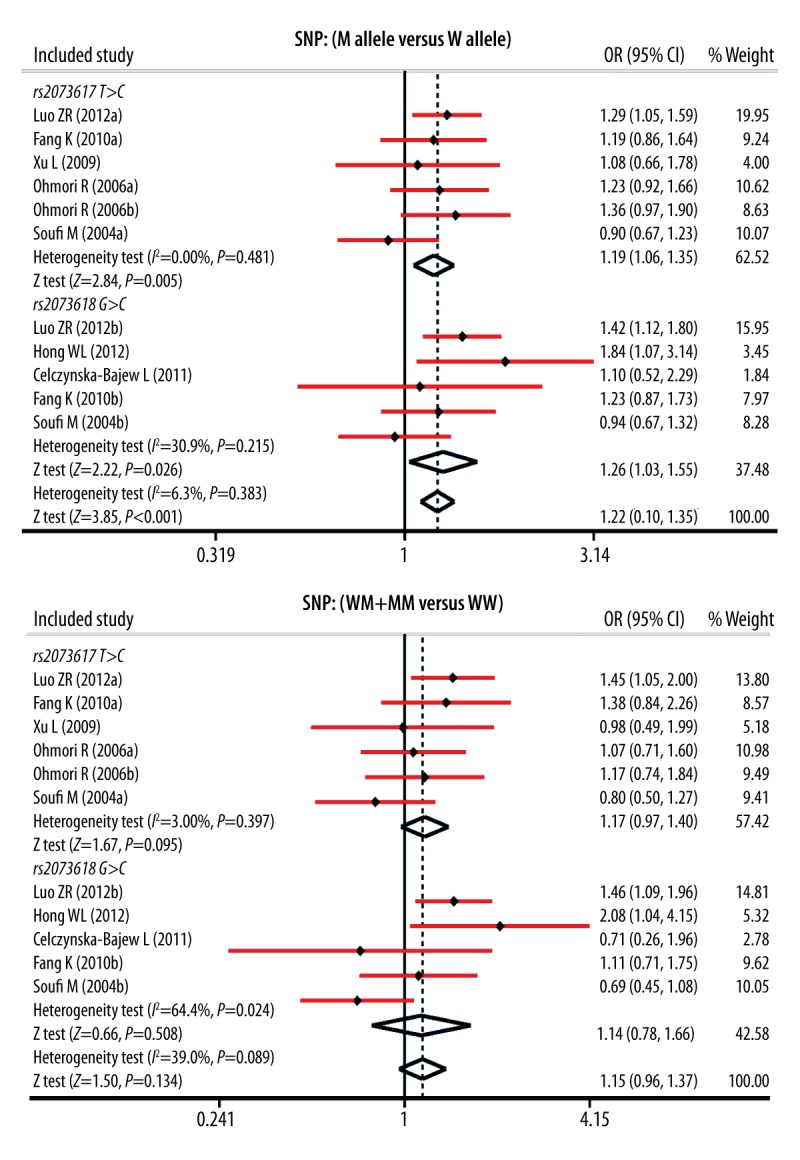
Forest plots for the influences of OPG genetic polymorphisms and cardiovascular disease under the allele and dominant models.
Table 2.
Meta-analysis of the relationships of opg gene with the cardiovascular disease.
| M allele vs. W allele (allele model) | WM + MM vs. WW (dominant model) | MM vs. WW + WM (recessive model) | MM vs. WW (homozygous model) | MM vs. WM (heterozygous model) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| Overall | 1.22 | 1.10–1.35 | <0.001 | 1.15 | 0.96–1.37 | 0.134 | 1.49 | 1.24–1.79 | <0.001 | 1.59 | 1.30–1.94 | <0.001 | 1.52 | 1.20–1.92 | <0.001 |
| SNP type | |||||||||||||||
| rs2073617 T>C | 1.19 | 1.06–1.35 | 0.005 | 1.17 | 0.97–1.40 | 0.095 | 1.37 | 1.07–1.75 | 0.013 | 1.46 | 1.12–1.91 | 0.006 | 1.34 | 1.02–1.78 | 0.039 |
| rs2073618 G>C | 1.26 | 1.03–1.55 | 0.026 | 1.14 | 0.78–1.66 | 0.508 | 1.92 | 1.36–2.71 | <0.001 | 1.88 | 1.32–2.69 | 0.001 | 1.96 | 1.35–2.84 | <0.001 |
| Ethnicity | |||||||||||||||
| Asians | 1.30 | 1.17–1.45 | <0.001 | 1.32 | 1.13–1.53 | <0.001 | 1.54 | 1.25–1.89 | <0.001 | 1.74 | 1.38–2.19 | <0.001 | 1.41 | 1.13–1.77 | 0.002 |
| Caucasians | 0.93 | 0.75–1.16 | 0.540 | 0.74 | 0.55–1.00 | 0.050 | 1.48 | 0.82–2.67 | 0.192 | 1.16 | 0.73–1.86 | 0.526 | 2.01 | 0.88–4.56 | 0.096 |
| Disease | |||||||||||||||
| ACS | 1.37 | 1.20–1.58 | <0.001 | 1.44 | 1.19–1.74 | <0.001 | 1.59 | 1.21–2.08 | 0.001 | 1.88 | 1.39–2.53 | <0.001 | 1.43 | 1.04–1.97 | 0.028 |
| CAD | 1.09 | 0.95–1.25 | 0.220 | 0.96 | 0.80–1.16 | 0.680 | 1.42 | 1.10–1.82 | 0.007 | 1.37 | 1.04–1.81 | 0.024 | 1.60 | 1.13–2.26 | 0.008 |
| Genotype method | |||||||||||||||
| PCR-RFLP | 1.28 | 1.13–1.46 | <0.001 | 1.35 | 1.13–1.61 | 0.001 | 1.39 | 1.10–1.76 | 0.007 | 1.62 | 1.24–2.11 | <0.001 | 1.25 | 0.97–1.61 | 0.084 |
| Non-PCR-RFLP | 1.15 | 0.95–1.40 | 0.150 | 0.99 | 0.75–1.32 | 0.949 | 1.72 | 1.23–2.40 | 0.002 | 1.59 | 1.10–2.29 | 0.014 | 1.93 | 1.32–2.82 | 0.001 |
OPG – osteoprotegerin; W – wild allele; M – mutant allele; WW – wild homozygote; WM – heterozygote; MM – mutant homozygote; OR – odds ratio; 95%CI – 95% confidence interval; SNP – single-nucleotide polymorphisms; CAD – coronary artery disease; ACS – acute coronary syndrome; PCR-RFLP – polymerase chain reaction-restriction fragment length polymorphism.
To comprehensively evaluate the influence of OPG genetic polymorphisms on the pathogenesis of CVD, we also carried out subgroup analysis based on ethnicity, disease, and genotyping method. Ethnicity-stratified subgroup analysis showed that genetic polymorphisms in the OPG gene were closely related with the development of CVD among Asians (all P<0.001; respectively) but not among Caucasians (all P>0.05) (Figure 3). Furthermore, the results of subgroup analysis base on type of disease and genotyping method illustrated that there was a positive correlation in most subgroups.
Figure 3.
Subgroup analyses for the influences of OPG genetic polymorphisms and cardiovascular disease under the allele and dominant models.
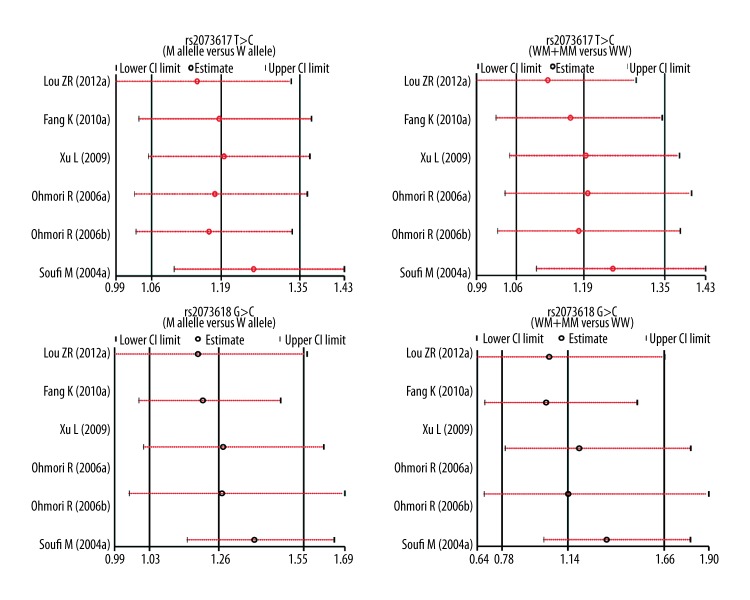
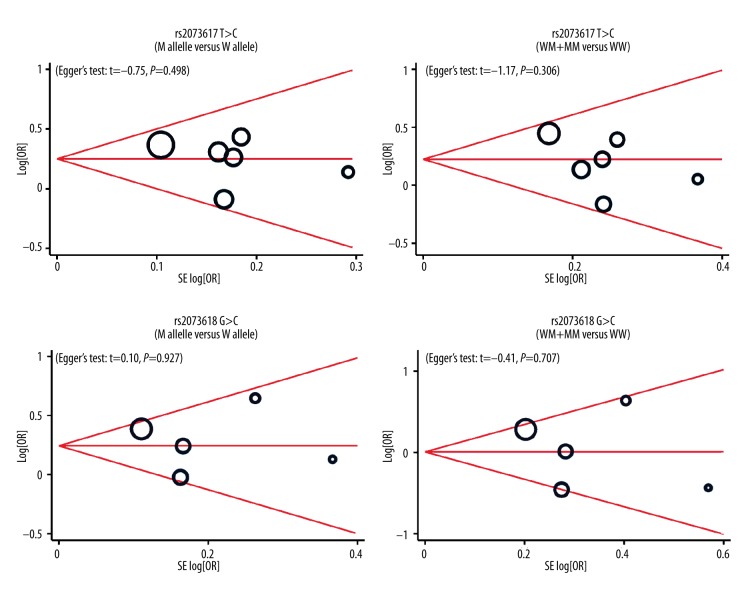
Univariate and multivariate meta-regression analyses indicated that ethnicity may be a potential source of heterogeneity (Table 3). Sensitivity analysis revealed the overall pooled estimates were not be influenced by any single study (Figure 4). Funnel plots revealed no presence of obvious asymmetry (Figure 5). No publication bias was shown in Egger’s test (all P>0.05).
Table 3.
Univariate and multivariate meta-regression analyses of potential source of heterogeneity.
| Heterogeneity factors | Coefficient | SE | z | P | 95%CI | |
|---|---|---|---|---|---|---|
| LL | UL | |||||
| Publication year | ||||||
| Univariate | 0.036 | 0.015 | 2.34 | 0.019 | 0.006 | 0.067 |
| Multivariate | 0.029 | 0.041 | 0.71 | 0.480 | −0.051 | 0.109 |
| SNP type | ||||||
| Univariate | 0.064 | 0.110 | 0.58 | 0.559 | −0.151 | 0.280 |
| Multivariate | 0.074 | 0.113 | 0.65 | 0.513 | −0.148 | 0.296 |
| Ethnicity | ||||||
| Univariate | −0.334 | 0.124 | −2.69 | 0.007 | −0.576 | −0.091 |
| Multivariate | −0.293 | 0.182 | −1.61 | 0.107 | −0.650 | 0.063 |
| Disease | ||||||
| Univariate | −0.233 | 0.098 | −2.37 | 0.018 | −0.426 | 0.040 |
| Multivariate | −0.076 | 0.139 | −0.55 | 0.586 | −0.347 | 0.196 |
| Genotyping method | ||||||
| Univariate | −0.121 | 0.100 | −1.21 | 0.226 | −0.317 | 0.075 |
| Multivariate | 0.210 | 0.203 | 1.04 | 0.300 | −0.187 | 0.607 |
SE – standard error; 95%CI – 95% confidence interval; UL – upper limit; LL – lower limit; SNP – single-nucleotide polymorphisms.
Figure 4.
Sensitivity analysis for the influences of OPG genetic polymorphisms and cardiovascular disease under the allele and dominant models.
Figure 5.
Funnel plot of publication biases on the relationships between genetic polymorphisms and cardiovascular disease under the allele and dominant models.
Discussion
Our findings showed that both rs2073617 and rs2073618 polymorphisms in the OPG gene were related with an increased risk of CVD, suggesting that OPG genetic polymorphism may be involved in the development of CVD. However, the exact mechanism of OPG genetic polymorphism in the progression of CVD is still unclear. Clinically, OPG is a vital factor in bone remodeling, and in addition to its role in the skeletal system, it also is crucial in the development of inflammatory diseases and vascular disease [29,30]. Furthermore, it is well established that the RANK/RANKL/OPG system plays an important role in many metabolic pathways and it is involved in the regulation of bone and endothelial metabolism [22]. Since OPG knockout mice concurrently exhibit arterial calcification of the great arteries, OPG has become a critical candidate gene for the development of CVD [31]. Moreover, elevated OPG concentrations have been documented to be related with the severity of unstable angina, peripheral artery disease and heart failure, vulnerable carotid plaques, symptomatic carotid stenosis, and acute myocardial infarction [32–34]. Consequently, we hypothesized that OPG genetic polymorphism, which may lead to alteration of OPG expression, might be related to the pathogenesis of CVD. The polymorphisms of T950C (rs2073617 T>C) and G1181C (rs2073618 G>C) in the OPG gene have been investigated in our meta-analysis [13,35]. The G1181C polymorphism, located in the upstream region of exon 1, encodes the signal peptide of the OPG and causes a lysine substituted to asparagine, and the T950C polymorphism is on the promoter of OPG [36]. Sequence variations in the OPG promoter in cooperation with polymorphisms in the exon encoding for the signal peptide of this secretory protein may function synergistically in regulation of intracellular trafficking, transcription, or secretion of OPG protein [14]. Our results are consistent with a previous study that revealed that subjects with C allele in the promoter region (TC and CC) may have higher circulating levels of OPG; moreover, genetic variations in the OPG may confer an increased risk of CVD [35]. Ohmori et al. also supported that the 950TC/1181GC and 950CC/1181CC haplotypes were more prevalent in men with CAD than those without CAD [13]. Ethnicity-stratified analysis suggested that OPG polymorphism was obviously related to the development of CVD among Asians, revealing that ethnicity difference may be a causative factor influencing heterogeneity between studies. The above results demonstrate unambiguously that genetic polymorphisms in the OPG might be affected by ethnic differences closely related to the pathogenesis of CVD.
There were also some limitations which should be acknowledged. At first, owing to the small number of studies, we did not obtain all desired information from all documents. Consequently, our general findings may lack broad applicability and should be considered preliminary. Secondly, we could not obtain original data from the enrolled reports. However, our study had distinct inclusion criteria in the literature search. Future study results based on more rigorous statistical analyses may result in more credible conclusions.
Conclusions
In brief, our meta-analysis provided quantitative evidence that OPG genetic polymorphisms might be closely related to an increased risk of CVD, especially for rs2073617 T>C and rs2073618 G>C polymorphisms. Thus, OPG genetic polymorphisms may be considered as a potential candidate in early prediction of CVD. Nevertheless, due to several study limitations, large sample-size reports with more integral data are needed to acquire a more representative statistical analysis.
Footnotes
Competing interests
None.
Source of support: Departmental sources
References
- 1.Fuster V, Kelly BB. Promoting cardiovascular health in the developing world: A critical challenge to achieve global health. Washington (DC): 2010. [PubMed] [Google Scholar]
- 2.Gaziano TA, Bitton A, Anand S, et al. Growing epidemic of coronary heart disease in low- and middle-income countries. Curr Probl Cardiol. 2010;35:72–115. doi: 10.1016/j.cpcardiol.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roger VL, Go AS, Lloyd-Jones DM, et al. Heart disease and stroke statistics – 2011 update: a report from the American Heart Association. Circulation. 2011;123:e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mozaffarian D, Wilson PW, Kannel WB. Beyond established and novel risk factors: lifestyle risk factors for cardiovascular disease. Circulation. 2008;117:3031–38. doi: 10.1161/CIRCULATIONAHA.107.738732. [DOI] [PubMed] [Google Scholar]
- 5.Van Campenhout AGolledge J. Osteoprotegerin, vascular calcification and atherosclerosis. Atherosclerosis. 2009;204:321–29. doi: 10.1016/j.atherosclerosis.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahlof B. Cardiovascular disease risk factors: epidemiology and risk assessment. Am J Cardiol. 2010;105:3A–9A. doi: 10.1016/j.amjcard.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Libby P, Okamoto Y, Rocha VZ, et al. Inflammation in atherosclerosis: transition from theory to practice. Circ J. 2010;74:213–20. doi: 10.1253/circj.cj-09-0706. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Yang H, Zou X, et al. Analysis of the CYP2C19 genetic polymorphism in Han and Uyghur patients with cardiovascular and cerebrovascular diseases in the Kashi area of Xinjiang. Med Sci Monit. 2014;20:2213–18. doi: 10.12659/MSM.892475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pac-Kozuchowska E, Krawiec P. Cholesterol ester transfer protein (CETP) gene polymorphism and selected parameters of lipid metabolism in children from families with history of cardiovascular system diseases. Med Sci Monit. 2013;19:818–25. doi: 10.12659/MSM.889550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nybo M, Rasmussen LM. The capability of plasma osteoprotegerin as a predictor of cardiovascular disease: a systematic literature review. Eur J Endocrinol. 2008;159:603–8. doi: 10.1530/EJE-08-0554. [DOI] [PubMed] [Google Scholar]
- 12.Chollet ME, Brouland JP, Bal dit Sollier C, et al. Evidence of a colocalisation of osteoprotegerin (OPG) with von Willebrand factor (VWF) in platelets and megakaryocytes alpha granules. Studies from normal and grey platelets. Br J Haematol. 2010;148:805–7. doi: 10.1111/j.1365-2141.2009.07989.x. [DOI] [PubMed] [Google Scholar]
- 13.Ohmori R, Momiyama Y, Taniguchi H, et al. Association between osteoprotegerin gene polymorphism and coronary artery disease in Japanese men. Atherosclerosis. 2006;187:215–17. doi: 10.1016/j.atherosclerosis.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 14.Soufi M, Schoppet M, Sattler AM, et al. Osteoprotegerin gene polymorphisms in men with coronary artery disease. J Clin Endocrinol Metab. 2004;89:3764–68. doi: 10.1210/jc.2003-032054. [DOI] [PubMed] [Google Scholar]
- 15.Tousoulis D, Siasos G, Maniatis K, et al. Serum osteoprotegerin and osteopontin levels are associated with arterial stiffness and the presence and severity of coronary artery disease. Int J Cardiol. 2013;167:1924–28. doi: 10.1016/j.ijcard.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Sennerby U, Melhus H, Gedeborg R, et al. Cardiovascular diseases and risk of hip fracture. JAMA. 2009;302:1666–73. doi: 10.1001/jama.2009.1463. [DOI] [PubMed] [Google Scholar]
- 17.Pennisi P, Signorelli SS, Riccobene S, et al. Low bone density and abnormal bone turnover in patients with atherosclerosis of peripheral vessels. Osteoporos Int. 2004;15:389–95. doi: 10.1007/s00198-003-1550-9. [DOI] [PubMed] [Google Scholar]
- 18.Silva IBranco JC. Rank/Rankl/opg: literature review. Acta Reumatol Port. 2011;36:209–18. [PubMed] [Google Scholar]
- 19.Collin-Osdoby P. Regulation of vascular calcification by osteoclast regulatory factors RANKL and osteoprotegerin. Circ Res. 2004;95:1046–57. doi: 10.1161/01.RES.0000149165.99974.12. [DOI] [PubMed] [Google Scholar]
- 20.Morinaga T, Nakagawa N, Yasuda H, et al. Cloning and characterization of the gene encoding human osteoprotegerin/osteoclastogenesis-inhibitory factor. Eur J Biochem. 1998;254:685–91. doi: 10.1046/j.1432-1327.1998.2540685.x. [DOI] [PubMed] [Google Scholar]
- 21.Nehring P, Mrozikiewicz-Rakowska B, Sobczyk-Kopciol A, et al. Osteoprotegerin gene rs2073617 and rs3134069 polymorphisms in type 2 diabetes patients and sexspecific rs2073618 polymorphism as a risk factor for diabetic foot. Pol Arch Med Wewn. 2013;123:176–82. doi: 10.20452/pamw.1684. [DOI] [PubMed] [Google Scholar]
- 22.Celczynska Bajew L, Horst Sikorska W, Bychowiec B, et al. The effects of osteoprotegerin (OPG) gene polymorphism in patients with ischaemic heart disease on the morphology of coronary arteries and bone mineral density. Kardiol Pol. 2011;69:573–78. [PubMed] [Google Scholar]
- 23.Fang K, Zeng Z. Relavance of osteoprotegerin gene with coronary heart disease. Med Information. 2010;23:5–5. 7. [Google Scholar]
- 24.Xu L, Xiang GD, Tang Y, et al. Relationship between polymorphism of osteoprotegerin gene promoter 950T/C and vascular complications in type 2 diabetic patients. Chin J Diabetes. 2009;17:806–7+10. [Google Scholar]
- 25.Zintzaras EIoannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–73. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- 26.Peters JL, Sutton AJ, Jones DR, et al. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–80. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 27.Hong WL, Guo XG, Jiao QP, et al. Correlation of osteoprotegerin gene polymorphism G1181C and osteoprotegerin serum levels with the presence of acute coronary syndrome. Anhui Med Phamaceutical J. 2012;16:775–77. [Google Scholar]
- 28.Luo ZR. Correlation between osteoprotegerin gene rs2073617T/C, rs2073618G/C polymorphism and acute coronary syndromes in Fujian area. Chin J Clinicians (Electronic Edition) 2012;6:4203–6. [Google Scholar]
- 29.Turk N, Cukovic-Cavka S, Korsic M, et al. Proinflammatory cytokines and receptor activator of nuclear factor kappaB-ligand/osteoprotegerin associated with bone deterioration in patients with Crohn’s disease. Eur J Gastroenterol Hepatol. 2009;21:159–66. doi: 10.1097/MEG.0b013e3283200032. [DOI] [PubMed] [Google Scholar]
- 30.Trouvin AP, Goeb V. Receptor activator of nuclear factor-kappaB ligand and osteoprotegerin: maintaining the balance to prevent bone loss. Clin Interv Aging. 2010;5:345–54. doi: 10.2147/CIA.S10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo C, Hu F, Zhang S, et al. Association between osteoprotegerin gene polymorphisms and cardiovascular disease in type 2 diabetic patients. Genet Mol Biol. 2013;36:177–82. doi: 10.1590/S1415-47572013005000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadoglou NP, Gerasimidis T, Golemati S, et al. The relationship between serum levels of vascular calcification inhibitors and carotid plaque vulnerability. J Vasc Surg. 2008;47:55–62. doi: 10.1016/j.jvs.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 33.Song TJ, Kim J, Yang SH, et al. Association of plasma osteoprotegerin levels with stroke severity and functional outcome in acute ischaemic stroke patients. Biomarkers. 2012;17:738–44. doi: 10.3109/1354750X.2012.727027. [DOI] [PubMed] [Google Scholar]
- 34.Sandberg WJ, Yndestad A, Oie E, et al. Enhanced T-cell expression of RANK ligand in acute coronary syndrome: possible role in plaque destabilization. Arterioscler Thromb Vasc Biol. 2006;26:857–63. doi: 10.1161/01.ATV.0000204334.48195.6a. [DOI] [PubMed] [Google Scholar]
- 35.Straface G, Biscetti F, Pitocco D, et al. Assessment of the genetic effects of polymorphisms in the osteoprotegerin gene, TNFRSF11B, on serum osteoprotegerin levels and carotid plaque vulnerability. Stroke. 2011;42:3022–28. doi: 10.1161/STROKEAHA.111.619288. [DOI] [PubMed] [Google Scholar]
- 36.Pitocco D, Zelano G, Gioffre G, et al. Association between osteoprotegerin G1181C and T245G polymorphisms and diabetic charcot neuroarthropathy: a case-control study. Diabetes Care. 2009;32:1694–97. doi: 10.2337/dc09-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]