Abstract
Collapsin response mediator protein 4 (CRMP4), a member of the CRMP family, is involved in the pathogenesis of neurodevelopmental disorders such as schizophrenia and autism. Here, we first compared layer thickness of the olfactory bulb between wild‐type (WT) and CRMP4‐knockout (KO) mice. The mitral cell layer (MCL) was significantly thinner, whereas the external plexiform layer (EPL) was significantly thicker in CRMP4‐KO mice at postnatal day 0 (PD0) compared with WTs. However, differences in layer thickness disappeared by PD14. No apoptotic cells were found in the MCL, and the number of mitral cells (MCs) identified with a specific marker (i.e. Tbx21 antibody) did not change in CRMP4‐KO neonates. However, DiI‐tracing showed that the length of mitral cell apical dendrites was greater in CRMP4‐KO neonates than in WTs. In addition, expression of CRMP4 mRNA in WT mice was most abundant in the MCL at PD0 and decreased afterward. These results suggest that CRMP4 contributes to dendritic elongation. Our in vitro studies showed that deletion or knockdown of CRMP4 resulted in enhanced growth of MAP2‐positive neurites, whereas overexpression of CRMP4 reduced their growth, suggesting a new role for CRMP4 as a suppressor of dendritic elongation. Overall, our data suggest that disruption of CRMP4 produces a temporary alteration in EPL thickness, which is constituted mainly of mitral cell apical dendrites, through the enhanced growth of these dendrites.
Keywords: brain development, CRMP4‐knockout mice, dendrite development, neurodevelopmental disorders
Introduction
Members of the collapsin response mediator protein (CRMP) family (CRMP1‐5) were originally identified as intracellular signaling mediators of Sema3A‐induced growth cone collapse (Goshima et al. 1995), revealing their involvement in axonal guidance (CRMP2, Arimura et al. 2005; CRMP4, Khazaei et al. 2014). In addition, CRMPs are also involved in neural migration (CRMP1, Yamashita et al. 2006; CRMP2, Ip et al. 2011), generation and survival of newborn neurons in the adult mouse brain (CRMP1, 2, and 5, Veyrac et al. 2005; CRMP5, Veyrac et al. 2011), decision on number and bifurcation of dendrites (CRMP3, Quach et al. 2008; CRMP4, Niisato et al. 2012), and dendritic spine development and plasticity (CRMP1, Yamashita et al. 2007; CRMP5, Yamashita et al. 2011). All these data indicate that CRMPs play important roles in the formation of neuronal circuits not only during development but also in adult neurogenesis.
In agreement with the role of CRMPs in brain development, previous studies have suggested the involvement of CRMPs in the pathogenesis of neurodevelopmental disorders such as schizophrenia and autism spectrum disorders (ASD) (Edgar et al. 2000; Johnston‐Wilson et al. 2000; Charrier et al. 2003; Hong et al. 2005; Beasley et al. 2006; Bader et al. 2012; Braunschweig et al. 2013; Yamashita et al. 2013). CRMP2 gene polymorphism and altered expression levels of CRMP1 or CRMP2 have been reported in schizophrenia (Edgar et al. 2000; Johnston‐Wilson et al. 2000; Hong et al. 2005; Beasley et al. 2006; Bader et al. 2012). In patients diagnosed with ASD, CRMP1 and CRMP2 have been identified as two marker antigens in maternal autoantibody‐related autism (Braunschweig et al. 2013). In addition to CRMP1 and CRMP2, CRMP4 has been suggested to be involved in psychiatric disorders because CRMP4 forms heteromers with CRMP1 and CRMP2 (Rembutsu et al. 2008). Despite the important roles of CRMPs in normal development and pathogenesis of developmental disorders, the functions of CRMPs, particularly CRMP4, have not been elucidated.
Numerous studies have shown morphological alterations in brain structures in patients with developmental disorders such as ASD or schizophrenia either by analyzing postmortem brain samples or using optical brain imaging techniques in patients (Kuperberg et al. 2003; Mukaetova‐Ladinska et al. 2004; Hadjikhani et al. 2005; Hardan et al. 2006; Rimol et al. 2010; Kubota et al. 2011; Dennis & Thompson, 2013; Ecker et al. 2013; Williams et al. 2013). Interestingly, our previous study revealed that CRMP4 mRNA is broadly distributed in the mouse brain during the early postnatal period, with a strong expression in the olfactory bulb (OB), hippocampus, and several cortical regions (Tsutiya & Ohtani‐Kaneko, 2012). In addition, we recently reported that CRMP4‐KO neonates showed impaired olfactory ability, abnormal OB neuronal activities, and higher levels of glutamate receptor 1 (GluR1) and GluR2 expression in the OB than WT pups did (Tsutiya et al. 2015). Olfactory dysfunction and elevated GluRs expression are relevant to neurodevelopmental disorders including schizophrenia and ASD (Pilpel et al. 2009; Hrdlicka et al. 2011; May et al. 2011; Wilson et al. 2011; Clepce et al. 2013; Moberg et al. 2013; Zhang & Abdullah, 2013). In this study, we investigated whether deletion of CRMP4 induced morphological alterations to the developing OB. In addition, we investigated the underlying mechanisms of such alterations using in vivo and in vitro approaches.
Materials and methods
Animals
CRMP4‐KO mice (Accession no. CDB0637K: http://www.cdb.riken.jp/arg/mutant%20mice%20list.html) were established as previously described (Niisato et al. 2012). Age‐matched wild‐type (WT) mice were used as controls. Animals were maintained on a 12‐h light/dark cycle with food and water available ad libitum. Experiments were performed under a protocol that was reviewed and approved by the Institutional Animal Care and Use Committee of the University of Tokyo and Toyo University.
Morphological analysis of the olfactory bulb using Nissl stain, apoptotic cell detection, and immunohistochemistry
For morphological analysis of the OB, we first examined the difference in OB size between WT and CRMP4‐KO mice (n = 3 each) at postnatal day (PD) 0. Animals were deeply anesthetized by hypothermia, the dorsal part of the skull was removed (Fig. 1A), and then the length and width of the OB was rapidly measured using a vernier caliper (Mitutoyo Co., Kanagawa, Japan). The brain was subsequently removed from the skull and fixed with 4% paraformaldehyde in 0.1 m phosphate buffer saline (PB, pH 7.3), and then images of the OBs were captured using a stereoscopic microscope (SZX7, Olympus, Tokyo, Japan) after fixation for 2 h (Fig. 1B). According to a previous report (Schmouth et al. 2012), we quantitated the surface area of the OB using the Adobe photoshop CS4 (version 11.0, Adobe System Inc., San Jose, CA, USA).
Figure 1.
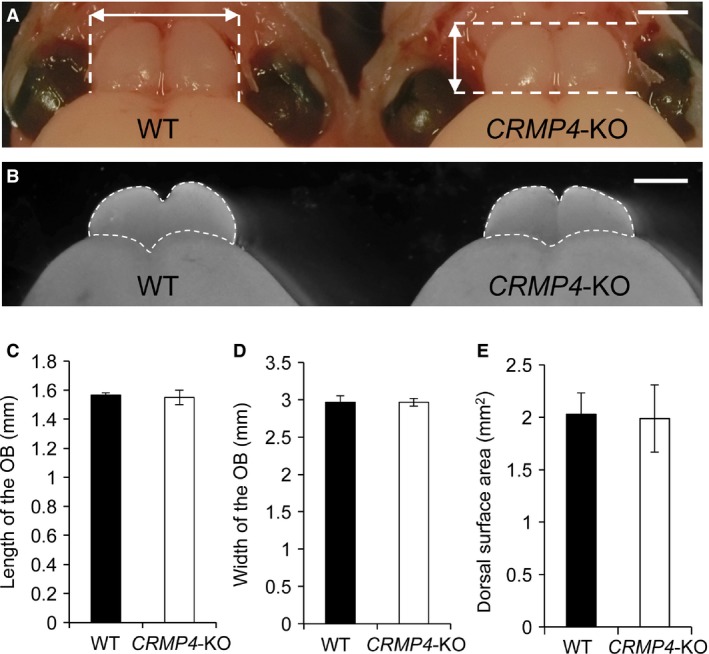
Comparison of olfactory bulbs (OBs) of wild‐type (WT) and collapsin response mediator protein 4 (CRMP4)‐knockout (KO) neonates. (A,B) Representative images of OBs from WT and CRMP4‐KO mice at PD0 before and after removal from the skull (A and B, respectively). Length and width were measured using a vernier caliper as indicated by arrows in (A). After removal from the skull and fixation, images of the OB were acquired, and its dorsal surface area was quantified as indicated by dotted lines in B. (C‐E) Mean length, width, and surface area of OBs were not significantly different between WT and CRMP4‐KO neonates. Student's t‐test was performed to compare the means. Bars indicate mean ± SE. Scale bars: 1 mm (A,B).
For Nissl staining, OBs from male WT and CRMP4‐KO mice of different postnatal stages were used: PD0 (WT, n = 7; CRMP4‐KO, n = 5), PD7 (WT, n = 4; CRMP4‐KO, n = 3), PD14 (WT, n = 3; CRMP4‐KO, n = 3), and PD21 (WT, n = 4; CRMP4‐KO, n = 4). Animals were deeply anesthetized by hypothermia (PD0) or pentobarbital (PD7, 14, and 21). The brains of neonates were rapidly removed and stored in 4% paraformaldehyde in 0.1 m PB overnight at 4 °C. Mice at other stages of development were transcardially perfused with saline, followed by treatment with 4% paraformaldehyde in PB. Brains were removed, post‐fixed in 4% paraformaldehyde in PB overnight at 4 °C, rinsed in phosphate‐buffered saline (PBS), cryoprotected at 4 °C in 10, 20, and 30% sucrose solutions in PB for 8–12 h each, and embedded in optimal cutting temperature (OCT) compound (Sakura Finetek Japan Co., Ltd., Tokyo, Japan). In all cases, serial frontal sections (20‐μm) through the OB were cut on a cryostat (CM‐3050‐S, Leica Microsystems, Wetzlar, Germany) in five parallel series, and thaw‐mounted on MAS‐coated glass slides (Matsunami Glass Ind., Ltd., Osaka, Japan). One of the series of OB sections from each animal was stained with Cresyl violet, and images of sections located 100 μm rostral to the front of the accessory OB were obtained with a microscope (Observer.D1, Carl Zeiss AG). We measured the thickness of the external plexiform layer (EPL) and mitral cell layer (MCL) at 80 different points in each section using axiovision microscopy Software (Carl Zeiss).
Another series of OB sections from PD0 WT and CRMP4‐KO mice was stained to detect apoptotic cells using an apoptosis in situ Detection Kit (Wako Pure Chemical Industries, Ltd.) following the instructions of the manufacturer. An additional series was treated with the DNase provided in the apoptosis kit and then stained as a positive control for DNA fragmentation. After the staining procedures, apoptotic cells were examined under a microscope (Observer.D1, Carl Zeiss AG).
Another series of OB sections from PD0 WT and CRMP4‐KO animals was used for the immunohistochemical identification of mitral cells (MCs). Sections were blocked with blocking solution for 1 h at room temperature. Then, sections were incubated with the primary antibody overnight at 4 °C. The primary antibody used was a rabbit polyclonal antibody against Tbx21 (1 : 10 000 dilution; provided by Y. Yoshihara, Riken), which is a molecular marker for MCs in the OB (Faedo et al. 2002). After rinsing with PBS, sections were incubated for 2 h at room temperature with Alexa Fluor 488‐conjugated goat anti‐rabbit IgG (Life Technologies Corp.), diluted 1 : 500 in blocking solution, and subjected to nuclear staining with Höechst 33 258 (Sigma‐Aldrich Japan Co., Ltd.). Finally, after several rinses with PBS, the sections were coverslipped using ProLong Gold antifade reagent (Life Technologies Corp.). Sections located 100 μm rostral to the front of the accessory OB were used to measure the thickness of the MCL, count the number of Tbx21‐positive MCs, and calculate MC density. To measure thickness and count MCs, images of 16 different areas within the MCL were obtained with a confocal microscope (LSM 5, Carl Zeiss AG). A grid (minimum scale = 20 μm) was superimposed on the respective images, and the average thickness of the MCL was calculated from the measurement at 10 grid lines that crossed the layer. The mean number of MCs was calculated from the number of Tbx21‐positive cells per 100 μm lengths of MCL line and the density of Tbx21‐positive MCs was calculated from the number of MCs and area of the respective regions.
Retrograde labeling of mitral cells
For retrograde labeling of MCs, WT and CRMP4‐KO (n = 3 each), PD0 male mice were used. After decapitation under deep anesthesia by hypothermia, brains were quickly removed from the cranium and stored in 4% paraformaldehyde in 0.1 m PB (pH 7.3) at 4 °C until use. Using a fine needle, crystals of 1,1′‐dioctadecyl‐3,3,3′,3′‐tetramethylindocarbocyanine perchlorate (DiI, Life Technologies Corp.) were implanted in the lateral olfactory tract, at the level of the anterior olfactory nucleus. Brains were then stored in the same fixative overnight at 37 °C, and embedded in 5% agarose gel. A vibratome (Microslicer, D.S.K./Dosaka EM Co., Ltd., Kyoto, Japan) was used to cut 100‐μm‐thick coronal sections of the brains. To reduce potential bias from regional variations in the orientation of MC dendrites (Buonviso et al. 1991), only sections from the middle portion of the OB where all bulbar layers were visible as complete concentric structures were used for qualitative and quantitative analyses. Sections were mounted on MAS‐coated glass slides, coverslipped using Prolong Gold antifade reagent, and observed using a confocal microscope. Serial optical images of MCs were taken at 1‐μm intervals in the Z‐axis (Z‐stacks) with a high magnification lens (64×), and the morphometric analysis of MCs was performed using LSM5 pascal software (Carl Zeiss). For this analysis, DiI‐labeled MCs with cell bodies located in the MCL and apical dendrites that could be traced into the glomerular layer (GL), were chosen. Area of the soma and length of primary apical dendrites of DiI‐labeled MCs (WT and CRMP4‐KO, n = 38 and 32, respectively) were measured and compared between WT and CRMP4‐KO mice. Tufted cells, which were distinguished from MCs because of the location of their cell bodies, were not included in this analysis.
In situ hybridization
To analyze the precise expression of CRMP4 mRNA in the OB of WT mice during prenatal and postnatal development, we performed in situ hybridization at embryonic day 15 (E15), PD0, PD7, PD14, and adult (8‐week‐old) mice.
Two or three male mice at each developmental stage were used. Animals were deeply anesthetized by hypothermia (E15 and PD0) or with pentobarbital (PD7, 14, and adults). Mice at E15 and PD0 were euthanized by cervical dislocation, and their brains were rapidly removed and stored in 4% paraformaldehyde in 0.1 m PB (pH 7.3) overnight at 4 °C. PD7, 14, and adult mice were transcardially perfused, and their brains were removed and post‐fixed as described above. Five parallel serial frontal cryosections (E15, 15‐μm thick; PD0, 7, 14, and adults, 20‐μm thick) through the OB were cut and thaw‐mounted on MAS‐coated glass slides. One of the five parallel series was used for in situ hybridization as previously described (Tsutiya & Ohtani‐Kaneko, 2012). We used a digoxigenin (DIG)‐labeled CRMP4 RNA probe generated from a cDNA containing 1792 bp of Rattus norvegicus CRMP4 (GenBank accession no. AF389425.1). After treatment with an alkaline phosphatase‐conjugated anti‐DIG antibody (1 : 1000; Roche Diagnostics, Tokyo, Japan), sections were developed with a chromogen solution until a visible signal was detected.
Primary cultures of OB cells from WT and CRMP4‐KO mice and CRMP4‐KO cells transfected with CRMP4 expression vector
Primary cultures were performed according to our previously reported procedure with slight modifications (Wu et al. 2011). Briefly, OBs were removed from neonatal WT and CRMP4‐KO mice (PD0), the tissue was dissociated with papain (Worthington Biochemical Corp., NJ, USA), and then gently triturated through a pipette. Dissociated cells were suspended in a culture medium consisting of Dulbecco's modified Eagles' medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U mL–1 penicillin G, and 25 μg mL–1 streptomycin (both from Meiji Seika Pharma Co. Ltd, Tokyo, Japan). The resulting cell suspension was plated at a density of 1.0 × 105 cells/well in 8‐well Falcon culture slides (Becton‐Dickinson) with bottoms coated with 0.1% polyethylenimine (Sigma‐Aldrich Japan Co., Ltd.). Cultures were maintained in 5% CO2 at 37 °C. The OB cells were fixed at 3 days in vitro (3 DIV) with 4% paraformaldehyde for 20 min at room temperature, and then immunocytochemically stained with antibodies against microtubule‐associated protein 2 (MAP2).
Next, we examined whether the expression of CRMP4 in cultured OB neurons from CRMP4‐KO mice rescues the morphological alteration. To this end, we constructed CRMP4 expression vector (pEGFP‐CRMP4 vector). In brief, the cDNA fragment containing the full‐length coding region of mouse CRMP4 gene was inserted into the EcoRI and XhoI sites of the pEGFP‐N vectors (Clontech Laboratories, Inc., Mountain View, CA, USA). The pEGFP‐CRMP4 vectors or pEGFP‐N vector (provided by H. Okado, Tokyo Metropolitan Institute of Medical Science) were transfected into the cultured OB cells removed from CRMP4‐KO mice using Lipofectamine 3000 (Invitrogen) on 1 DIV following the manufacturer's protocol. The OB cells were fixed on 3 DIV with 4% paraformaldehyde for 20 min at room temperature and then immunocytochemically stained with antibodies against MAP2. Transfected cells with these vectors were confirmed by GFP expression.
For immunostaining to detect MAP2, the fixed cells were incubated with blocking solution for 1 h at room temperature and then with the primary antibody overnight at 4 °C. The primary antibody used was rabbit anti‐MAP2 polyclonal antibody (1 : 1000 dilution, ab32454, Abcam). After rinsing with PBS, the cells were incubated for 2 h at room temperature with Alexa Fluor 555‐conjugated goat anti‐rabbit IgG (Life Technologies Corp.), diluted 1 : 500 in blocking solution, and subjected to nuclear staining with Höechst 33 258 (Sigma‐Aldrich Japan Co., Ltd.). Finally, after several rinses in PBS, they were coverslipped using ProLong Gold antifade reagent (Life Technologies Corp.).
The longest length of MAP2‐positive dendrites was measured on each putative MC, according to our previous study (Wu et al. 2011). In brief, we measured the length of the longest MAP2‐positive dendrite of each putative MC whose soma was larger than 100 μm2 and with at least one thick dendrite, to discriminate MCs from other cells such as granule cells. Image acquisition and measurements were performed using an LSM5 PASCAL laser microscope and its imaging software.
In vitro knockdown and overexpression of CRMP4 in HT22 cells
We also performed in vitro CRMP4 loss‐of‐function and gain‐of‐function experiments on the length of dendrites using the HT22 murine hippocampal cell line. First, to silence the CRMP4 expression, HT22 cells were transfected with a small interference RNA (siRNA) against CRMP4 using RNAiMAX (Invitrogen). The nucleotide sequences used for CRMP4 siRNAs were as follows: forward 5′‐CGCAUUAAAGCAAGGAGGATT‐3′, reverse 5′‐UCCUCCUUGCUUUAAUGCGTT‐3′. Nonspecific siRNA (Block‐iT™ Alexa Fluor® Red Fluorescent Control, Invitrogen) was used as a control to determine transfection efficiency.
HT22 cells suspended in DMEM supplemented with 10% FBS were plated at a density of 2.0 × 106 cells mL–1 or 1.0 × 104 cells mL–1 on culture flasks or glass bottom dishes (35 mm2; MatTek Corp., Ashland, MA, USA), respectively, with RNAiMax containing siRNA. Cell cultures were maintained in an incubator in 5% CO2 humidified atmosphere at 37 °C. After transfection for 24 h, the incubation medium was replaced. HT22 cells cultured in flasks were used to analyze CRMP4 expression using real‐time RT‐PCR as previously described (Tsutiya & Ohtani‐Kaneko, 2012). HT22 cells cultured in 35‐mm dishes were used for morphological analysis. Images of HT22 cells transfected with siRNAs were obtained with a microscope (Observer.D1, Carl Zeiss AG). Unpolarized HT22 cells were excluded from this analysis. Neurite length was measured using axiovision microscopy Software (Carl Zeiss). Average neurite length of each group was calculated measuring the length of all neurites.
Next, we performed a CRMP4 gain‐of‐function experiment. HT22 cells were suspended in serum culture medium containing DMEM supplemented with 10% FBS, 50 U mL–1 penicillin G, and 25 μg mL–1 streptomycin, and plated at a density of 1.0 × 104 cells mL–1 on 35‐mm glass‐bottom dishes. Cultures were maintained in 5% CO2 at 37 °C. After cells had been cultured for 24 h, the incubation medium was replaced (DMEM supplemented with 10% FBS), and cells were transfected with pEGFP‐CRMP4 vector using Lipofectamine 2000 (Invitrogen). After 2 h of incubation in 5% CO2 at 37 °C, the incubation medium was replaced, and HT22 cells were incubated for 24 h. After that, images of HT22 cells were obtained with a microscope (Observer.D1, Carl Zeiss). Polarized GFP‐positive HT22 cells were used for analysis, and the average neurite length of cells was calculated. HT22 cells transfected with pEGFP‐N were used as controls. Each experiment was repeated eight times.
For MAP2 immunocytochemistry, HT22 cells suspended in DMEM supplemented with 10% FBS were plated at a density of 1.0 × 104 cells mL–1 on glass bottom dishes. Cells were cultured for 24 h at 37 °C, and then fixed and stained immunocytochemically with a rabbit polyclonal anti‐MAP2 antibody (1 : 1000 dilution; ab32454, Abcam) overnight at 4 °C. An Alexa Fluor 488‐conjugated goat anti‐rabbit IgG (1 : 500 dilution) was used as secondary antibody for 2 h at room temperature. After rinsing, cells were observed under a confocal microscope (LSM 5, Carl Zeiss AG).
Statistical analysis
Student's t‐test and one‐way analysis of variance (anova) followed by Tukey's honest significant difference (HSD) post hoc test for multiple comparisons were used to determine the statistical differences. A P‐value < 0.05 was considered statistically significant.
Results
Deletion of CRMP4 alters thickness of olfactory bulb layers
First, we compared the size of OBs of WT and CRMP4‐KO neonates (Fig. 1). As shown in Fig. 1A, we measured the length and width of the OB using a vernier caliper immediately after the dorsal part of the skull was removed to avoid artifacts in measurements caused by its removal. In addition, we measured the dorsal surface area of the OB after fixation (Fig. 1B) according to a previous report (Schmouth et al. 2012). As shown in Fig. 1C–E, the mean length, width, and dorsal surface areas of CRMP4‐KO neonate OBs were not significantly different from those of WTs (length, 1.57 ± 0.02 vs. 1.55 ± 0.05 mm, P = 0.35; width, 2.97 ± 0.09 vs. 2.97 ± 0.05 mm, P = 0.30; dorsal surface area, 2.03 ± 0.20 vs. 1.99 ± 0.32 mm2, P = 0.91, WT vs. CRMP4‐KO, respectively, Student's t‐test, n = 3 each).
Then, we examined whether disrupting the CRMP4 gene would alter OB layering during development (Fig. 2A–H,Aʹ–Hʹ). Nissl staining revealed that the OB of WT and CRMP4‐KO mice had the same fundamental layout of layers: olfactory nerve layer, GL, EPL, MCL, internal plexiform layer (IPL), and granule cell layer (GCL). However, the thickness of the MCL and EPL was different between WT and CRMP4‐KO mice at PD0 (Fig. 2A–B,Aʹ–Bʹ). We then compared the thickness of these two layers between WT and CRMP4‐KO animals during postnatal development. The thickness of the MCL in CRMP4‐KO mice was significantly decreased compared with WT mice at PD0 and PD7 (Fig. 2I,J, PD0, P < 0.001; PD7, P < 0.05, Student's t‐test). We did not observe differences in MCL thickness between WT and CRMP4‐KO after PD14 (Fig. 2K,L). In contrast, the EPL was thicker in CRMP4‐KO than in WT mice at PD0 (Fig. 2M, P < 0.01, Student's t‐test). After PD7, EPL thickness gradually increased with age in both WT and CRMP4‐KO mice (Fig. 2C–H,Cʹ–Hʹ). There were no significant differences in EPL thickness between WT and CRMP4‐KO mice after PD7 (Fig. 2N–P).
Figure 2.
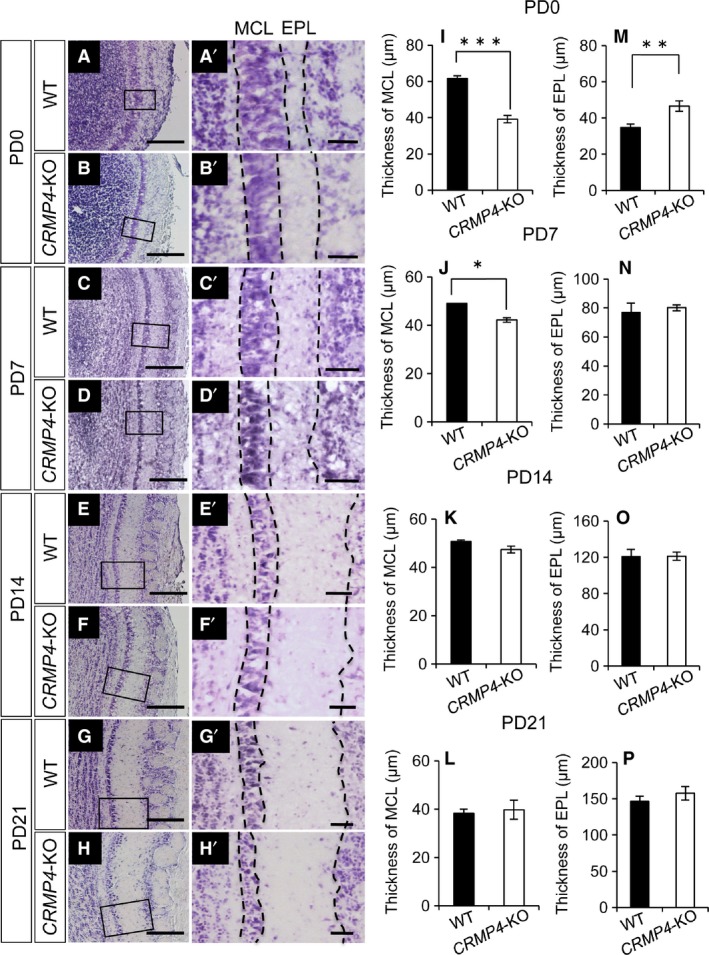
Morphometric analysis of layer thickness in the OB of WT and CRMP4‐KO mice at postnatal day (PD) 0 to PD21. (A‐H) Representative images of Nissl‐stained OB sections of WT (A,C,E,G) and CRMP4‐KO mice (B,D,F,H). (A′‐H′) Higher magnification of boxed areas in A–H. (I‐L) Graphs showing the thickness of the mitral cell layer (MCL) at PD0 (I), PD7 (J), PD14 (K), and PD21 (L). (M‐P) Graphs showing the thickness of the external plexiform layer (EPL) at PD0 (M), PD7 (N), PD14 (O), and PD21 (P). All results are expressed as the mean ± SE. Student's t‐test was performed to compare the means, *P < 0.05, **P < 0.01 and ***P < 0.001. Scale bars: 100 μm (A‐H), 20 μm (A′‐H′).
Reduced thickness of the mitral cell layer in CRMP4‐KO neonates is produced by an increase in density of mitral cells
Previous studies have described the presence of granule cells (GCs) in the MCL (Imamura et al. 2006; Batista‐Brito et al. 2008). To confirm that the altered MCL thickness observed in CRMP4‐KO mice was not caused by the GCs in the MCL, but by MCs, the latter were immunohistochemically stained with an antibody against Tbx21, a marker for MC nucleus. As shown in Fig. 3A and B, the Tbx21‐positive layer was significantly thinner in CRMP4‐KO mice than in WT mice at PD0.
Figure 3.
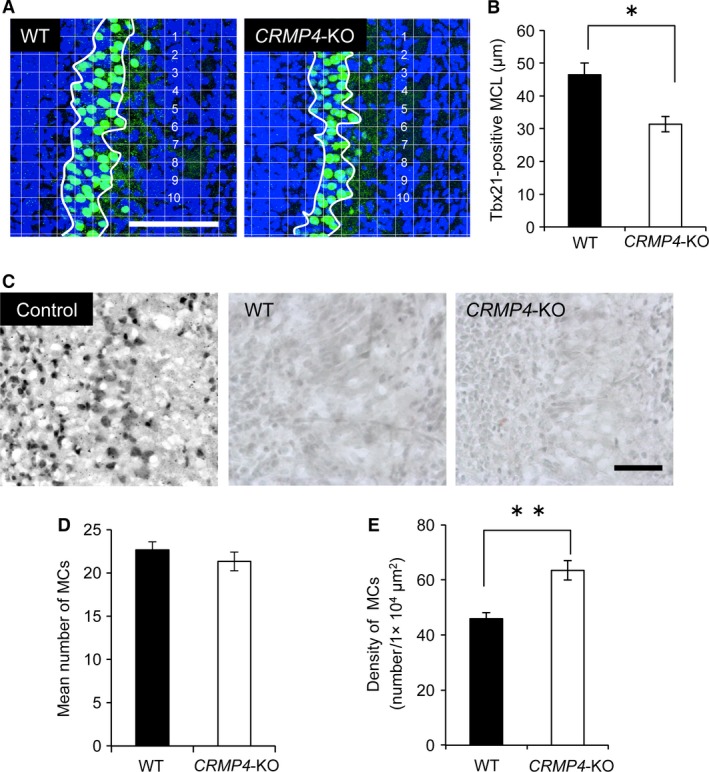
Morphometric analysis of mitral cell layer (MCL) thickness and mitral cell (MC) number in WT and CRMP4‐KO mice at PD0. (A) Representative micrographs showing Tbx21‐positive nuclei in the olfactory bulb (OB; green). Nuclei were stained with Höechst 33 258 (blue). Appropriate grids (20 × 20 μm) were applied to the images, and average thickness of the MCL was calculated from the measurement at 10 grid lines that crossed the layer. (B) Bar graph shows that at PD0 the thickness of the MCL in CRMP4‐KO mice was significantly decreased compared with WT mice. (C) Detection of apoptotic cells in the OB. TUNEL‐positive cells are observed in sections treated with DNase (Control) but sections through the OB of WT and CRMP4‐KO at PD0 did not show apoptotic cells. (D) Bar graph showing that the mean number of MCs in CRMP4‐KO mice was not significantly different from that in WT mice. (E) Bar graph showing that density of MCs in CRMP4‐KO neonates was significantly higher than in WT mice. Bars indicate mean ± SE. Student's t‐test was performed to compare the means, *P < 0.05 and **P < 0.01. Scale bars: 100 μm (A), 50 μm (C).
Previous studies have reported the involvement of CRMP4 in the regulation of apoptosis (Franken et al. 2003; Liu et al. 2009; Duplan et al. 2010). To examine whether a deficiency in CRMP4 led to increased apoptosis in the MCL, which would result in a thinner MCL in CRMP4‐KO neonates, we stained apoptotic cells in the OB of both animals at PD0. No apoptotic cells were found in the MCL of either WT or CRMP4‐KO neonates (Fig. 3C).
We further examined the number of MCs identified using the Tbx21 antibody in the MCL. Our results show that the mean number of Tbx21‐positive MCs did not differ between WT and CRMP4‐KO neonates (Fig. 3D). As expected from the difference in thickness of the MCL and the unchanged number of MCs between WT and CRMP4‐KO neonates, the mean density of Tbx21‐positive MCs was significantly higher in the MCL of CRMP4‐KO than WT mice (Fig. 3A,E, P < 0.05, Student's t‐test). These results indicate that the decreased thickness of the MCL in CRMP4‐KO mice is not a result of a decrease in the number of MCs but of an increase in the density of these cells.
Increased length of apical dendrites may contribute to thicker external plexiform layer in CRMP4‐KO neonates
Apical dendrites of MCs are the major constituent of the EPL and we therefore elucidated why the EPL is thicker in CRMP4‐KO neonates than in WT by comparing the length of apical dendrites of MCs in CRMP4‐KO and WT mice at PD0. To label the apical dendrites of MCs, we placed a DiI crystal in the lateral olfactory tubercle, which resulted in the retrograde labeling of these cells (Fig. 4A, inset). Numerous DiI‐labeled MCs were found in the ventral portion of the OB as reported previously (Imamura et al. 2011). Then, we measured the soma area and length of apical dendrites of MCs located in this region. Our results showed that there were no differences in soma area of DiI‐labeled MCs between the WT and CRMP4‐KO mice (Fig. 4B). However, the average length of primary apical dendrites of DiI‐labeled MCs was significantly greater in CRMP4‐KO neonates than in WT (Fig. 4C, P < 0.001, Student's t‐test), suggesting that the thickness of the EPL in the former animals was a result of an increased elongation of MC apical dendrites around the neonatal period. Regarding basal dendrites, MCs in WT and CRMP4‐KO mice were still immature and had a very few basal dendrites at PD0 (Fig. 4A), as reported previously (Matsutani & Yamamoto, 2000); therefore, the length of basal dendrites was not measurable in either animal at PD0.
Figure 4.
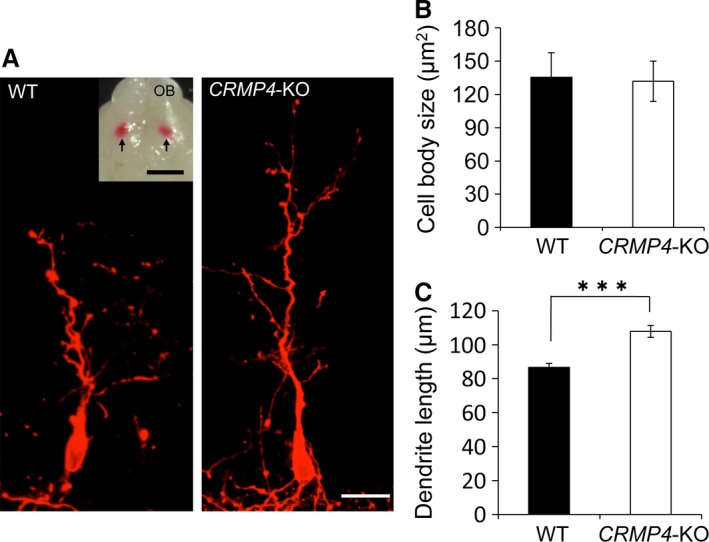
Tracing of mitral cells (MCs) with DiI in WT and CRMP4‐KO mice at PD0. (A) Representative micrographs showing retrogradely labeled MCs in WT and CRMP4‐KO mice. (Inset) Ventral view of a brain injected with DiI in the olfactory tract. Arrows indicate the points where DiI crystals were implanted. (B) Bar graph showing that there were no differences in average soma area of MCs between the two genotypes at PD0. (C) MCs in CRMP4‐KO mice had longer apical dendrites than WT at PD0. Bars indicate mean ± SE. Student's t‐test was performed to compare the means, ***P < 0.001. Scale bars: 20 μm (A), 2 mm (A inset).
Mitral cells strongly express CRMP4 mRNA at PD0
CRMP4 mRNA expression from E15 to adult was examined using in situ hybridization, and the results show dramatic changes during the pre‐ and post‐natal development (Fig. 5). At E15, when OB layering is not yet distinguishable, as reported in previous works (Mair et al. 1982; Malun & Brunjes, 1996), a diffuse and weak CRMP4 mRNA signal was observed in the OB, except in its central part (Fig. 5A). At PD0, the layers of the OB were differentiated (Fig. 5B) and the CRMP4 mRNA signal was stronger in the MCL than in any other layer (arrowheads in Fig. 5B); while scattered signal was also detected in the GL (arrows in Fig. 5B,Bʹ). The cells that showed strong CRMP4 mRNA signal were MCs with Tbx‐21‐positive nuclei (Fig. 5C,Cʹ). At PD7, CRMP4 mRNA signal in the MCL became weaker than in the previous stage (arrowheads in Fig. 5D), whereas it was strong in the GL (arrows in Fig. 5D). In addition, a weak signal was also observed in the rostral migratory stream (RMS) at PD7 (white arrow in Fig. 5D). At PD14 and later stages, there was weak CRMP4 mRNA signal in the MCL (arrowheads in Fig. 5E,F) as well as in the RMS (white arrows in Fig. 5E and F).
Figure 5.
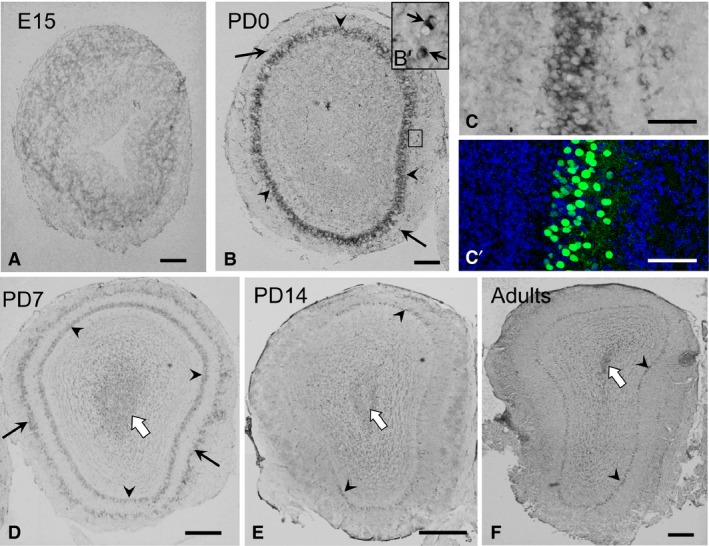
Expression and localization patterns of CRMP4 mRNA in the olfactory bulb (OB) during development. (A,B,D–F) Representative images of CRMP4 mRNA hybridization signal in the OB at embryonic day (E) 15 (A), PD0 (B), PD7 (D), PD14 (E), and adult (8‐week‐old) mice (F). (Bʹ) Higher magnification of the boxed area in B. (C,C′) Images of hybridization signal at PD0, and an adjacent section immunohistochemically stained with Tbx21 antibody (green), respectively. Tbx‐21 is a specific marker of mitral cell nuclei. Arrowheads and arrows indicate the mitral cell and glomerular layers, respectively. White arrows in D,E, and F indicate hybridization signal in the rostral migratory stream. Scale bars: 200 μm (A,B,D–F), 50 μm (B′,C).
These results show that CRMP4 mRNA expression changes dramatically during development, with the strongest signal appearing in the MCs at PD0. These results, together with the observation that CRMP4 deletion enhanced elongation of apical dendrites of MCs, suggest that CRMP4 gene expression in MCs affects elongation of apical dendrites of these cells.
Deletion of CRMP4 affects dendritic elongation of putative MCs in vitro
We examined whether CRMP4 expression affects dendritic elongation using primary cultures of OB cells from WT and CRMP4‐KO mice, and those from CRMP4‐KO mice transfected with pEGFP or pEGFP‐CRMP4 vectors (Fig. 6A). Our attempts to identify the cultured MCs using specific antibodies against MCs (Tbx21 and Pcdh21) that can stain MCs in vivo were unsuccessful. Therefore, we used putative MCs in cultured OB cells according to the morphological criteria used in our previous study (Wu et al. 2011).
Figure 6.
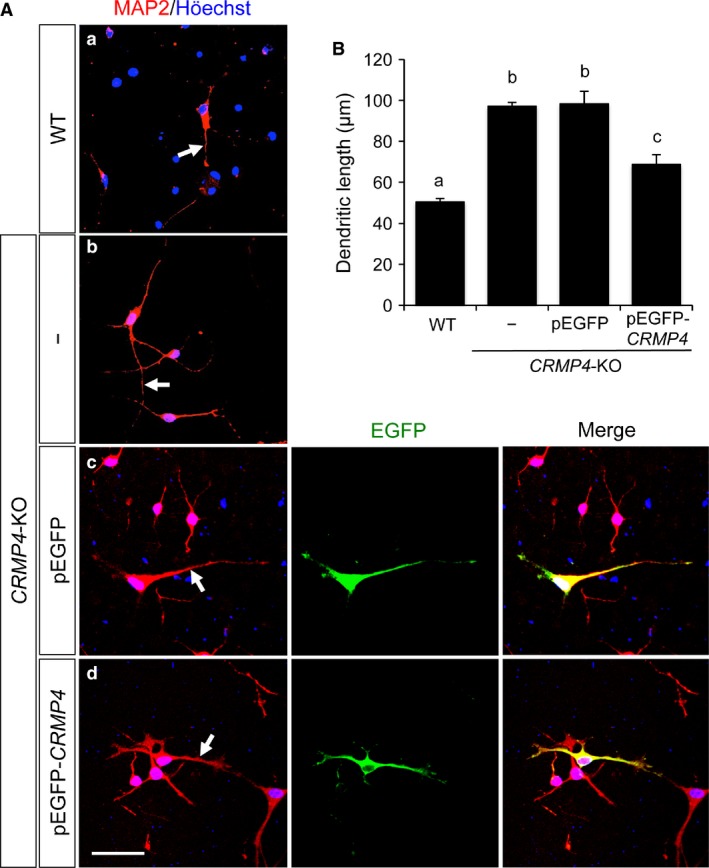
Effect of CRMP4 expression on dendritic elongation of cultured mitral cells (MCs). (A) Representative images of cultured OB cells from WT (a) and CRMP4‐KO mice (b‐d). Dendrites were stained with anti‐MAP2 antibody (red) and nuclei were stained with Höechst 33 258 (blue). Length of the longest MAP2‐positive dendrite of each putative MC was measured (arrows in a‐d). In pEGFP or pEGFP‐CRMP4 vector transfected groups (c and d), the longest MAP2‐immunostained dendrite (red) of each EGFP‐labeled putative MC (green) was used to measure dendritic length. Putative MCs were morphologically identified as those with large somata (>100 μm) and at least one thick dendrite. Scale bar: 50 μm. (B) Bar graph showing the average length of the longest MAP2‐positive dendrite of each putative MC. In WT and untransfected CRMP4‐KO OB cells (WT and −, respectively), MAP2‐positive putative MCs were analyzed. In pEGFP or pEGFP‐CRMP4 vector transfected groups (pEGFP and pEGFP‐CRMP4, respectively), MAP2‐positive putative MCs showing EGFP expression were analyzed. Bars indicate mean ± SE; n = 41–163 neurons. Different letters above the graph indicate statistical significance (P < 0.05) using one‐way anovas followed by Tukey's HSD post hoc test for multiple comparisons.
Dendrites were immunostained with an anti‐MAP2 antibody, and the length of the longest MAP2‐positive dendrite (dendritic length) was measured on each putative MC (arrows in Fig. 6A‐a–d). In the pEGFP or pEGFP‐CRMP4 vector transfected groups, MAP2‐immunostained dendrites of EGFP‐labeled putative MCs were used for the analysis (Fig. 6‐c, center, and ‐d, center). The dendritic length of the putative MCs from CRMP4‐KO mice (Fig. 6A‐b) was significantly longer than that from the WT mice (Fig. 6A‐a and B, WT and CRMP4‐KO, 50.51 ± 1.51 and 97.24 ± 1.73 μm, respectively). The dendritic length of the CRMP4‐KO cells transfected with pEGFP vector (Fig. 6A‐c) was not significantly different from that of the untransfected CRMP4‐KO cells (Fig. 6A‐b and B, CRMP4‐KO and CRMP4‐KO + pEGFP, 97.24 ± 1.73 and 98.37 ± 6.04 μm, respectively). In contrast, the CRMP4‐KO cells transfected with pEGFP‐CRMP4 vector (Fig. 6A‐d) had significantly shorter dendrites than those transfected with pEGFP vector (Fig. 6A‐c,B, CRMP4‐KO + pEGFP and CRMP4‐KO + pEGFP‐CRMP4, 98.37 ± 6.04 and 68.74 ± 4.83 μm, respectively). However, the dendritic length of the putative CRMP4‐KO MCs transfected with pEGFP‐CRMP4 vector remained significantly longer than that from the WT MCs (Fig. 6B).
Knockdown and overexpression of CRMP4 affects MAP2‐positive neurite elongation in HT22 cells
We observed an increase in the dendritic length of cultured putative MCs from CRMP4‐KO mice. In addition, their transfection with the pEGFP‐CRMP4 vector decreased their dendritic length. These data support the notion that CRMP4 gene expression in MCs affects the elongation of the apical dendrites of these cells. We also performed in vitro culture studies using hippocampal mouse neuronal cells (HT22 cells) for further assessment of the effect of CRMP4 genes on dendritic outgrowth because HT22 cells express CRMP4 mRNA and their neurites have MAP2‐immunoreactivity (Fig. 7A,B).
Figure 7.
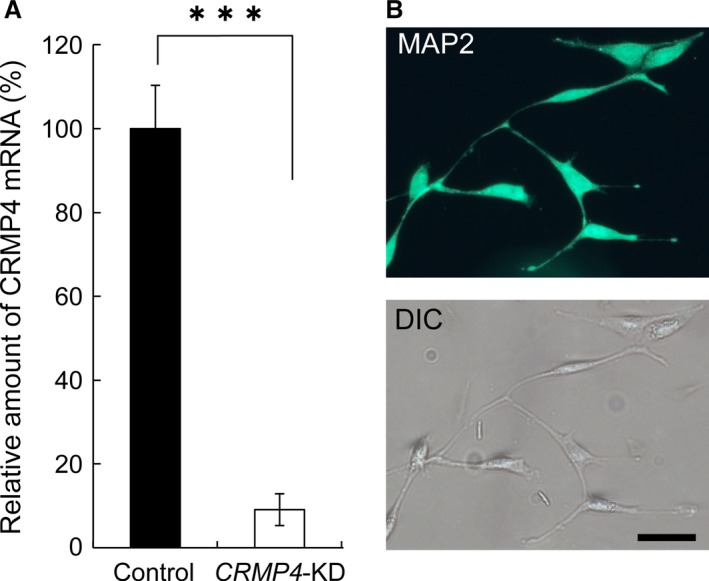
Expression of CRMP4 mRNA and MAP2 in HT22 cells. (A) HT22 cells express CRMP4 mRNA, whereas transfection with CRMP4 siRNA (CRMP4‐KD) significantly decreased its expression. (B) (Upper) Representative micrograph of HT22 cells immunohistochemically stained with an MAP2 antibody (green). (Bottom) Differential interference contrast image of the same cells. Neurites of HT22 cells show immunoreactivity to MAP2. Bars in (A) indicate mean ± SE. Student's t‐tests were performed to compare the means, ***P < 0.001. Scale bar: 50 μm.
HT22 cells were transfected with either a CRMP4 siRNA (knockdown, CRMP4‐KD) or a pEGFP‐CRMP4 (overexpression) vector, and the length of neurites was measured. CRMP4‐KD significantly increased the mean length of neurites, compared with cells transfected with control siRNA (control and CRMP4‐KD, 62.1 ± 3.6 and 79.9 ± 5.5 μm, P = 0.017, respectively, Student's t‐test, n = 8 each, Fig. 8A,B). Alternatively, when HT22 cells were transfected with pEGFP‐CRMP4 or pEGFP vectors, neurite growth of pEGFP‐CRMP4‐transfected cells was suppressed compared with pEGFP‐transfected cells used as controls (Fig. 8C). The average neurite length of pEGFP‐CRMP4 cells was significantly shorter than that of the controls (control and CRMP4, 57.1 ± 4.6 and 43.0 ± 1.3 μm, P = 0.018, respectively, Student's t‐test, n = 8 each, Fig. 8D). These results also show a suppressive regulatory role for CRMP4 in dendritic elongation.
Figure 8.
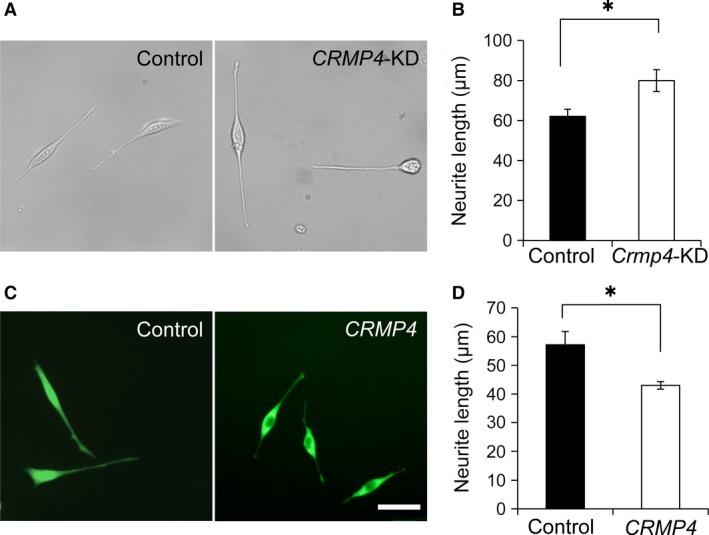
Effect of CRMP4‐knockdown and ‐overexpression on neurite elongation in HT22 cells. (A) Representative images of HT22 cells transfected with control siRNA (Control) or CRMP4 siRNA (CRMP4‐KD). (B) Bar graph showing that knockdown of CRMP4 significantly increased neurite length in HT22 cells. (C) Representative images of HT22 cells transfected with pEGFP‐N vector (Control) or pEGFP‐CRMP4 vector (CRMP4). (D) Bar graph showing that overexpression of CRMP4 (CRMP4) significantly decreased neurite length. Bars indicate mean ± SE of eight independent experiments (n = 8). Student's t‐tests were performed to compare the means, *P < 0.05. Scale bar: 50 μm.
Discussion
The present study revealed two morphological alterations in CRMP4‐KO mice in vivo. First, a deficiency in CRMP4 produced an alteration in the EPL and MCL thickness during the neonatal period. Secondly, MCs of CRMP4‐KO neonates had longer apical dendrites than WTs. In addition, our in vitro transfection studies showed that the expression of CRMP4 negatively affected the length of MAP2‐positive dendrites of the cultured MCs and HT22 cells.
Regarding the alteration of EPL thickness, this layer was thicker in CRMP4‐KO neonates than in WTs. Our results of the DiI‐tracing revealed longer apical dendrites in MCs of CRMP4‐KO neonates than WTs, suggesting that the thicker EPL in CRMP4‐deficient neonates was a result of the accelerated growth of MC apical dendrites, which are the main constituents of the EPL. However, other possibilities include that the thickness of the EPL may determine the length of the apical dendrites. That is, apical dendrites of MCs could grow until they reached the GL, resulting in longer apical dendrites of MCs in CRMP4 mutant than in WT mice. In adults, it is known that the main constituents of the EPL are MC apical and granule cell dendrites, and the latter create dendrodendritic circuits primarily on basal dendrites of MCs in the EPL (Yokoi et al. 1995; Isaacson & Strowbridge, 1998), although some tufted cells also exist in the EPL (Nagayama et al. 2014). However, there are only a few tufted cells (Mizuguchi et al. 2012) and dendrites of granule cells (Kishi, 1987) in the EPL at PD0, whereas abundant apical dendrites of MCs exist in the EPL from E17 (Blanchart et al. 2006). Therefore, it is understandable that the thickness of the EPL would be primarily determined by the length of the MC apical dendrites at PD0. As shown in Fig. 5, CRMP4 mRNA expression in WT mice was most abundant in the MCL at PD0 and decreased after that. On the other hand, our in vitro studies showed that the expression of the CRMP4 gene negatively regulates the elongation of MAP2‐positive dendrites of cultured MCs and HT22 cells (Figs 6 and 8). Taken together, these results suggest that the suppressive role of CRMP4 in the growth of MC apical dendrites was high in WT neonates and then gradually decreased during postnatal development. Combining the in vitro and in vivo data, we can speculate that CRMP4 gene expression could affect EPL thickness during development by regulating the growth of MC apical dendrites, although more experiments should be performed.
Another member of the CRMP family, CRMP2, was first identified as a mediator protein (Goshima et al. 1995) for the repulsive axon guidance molecule semaphorin 3A (Nakamura et al. 2000), which explains why CRMPs are regarded as axon regulator proteins. Furthermore, a series of studies have reported important roles for CRMP4 in the regulation of axonal growth (Cole et al. 2006; Alabed et al. 2007; Nagai et al. 2012; Khazaei et al. 2014; Tan et al. 2015). Cole et al. (2006) showed that overexpression of CRMP4 in hippocampal neurons increased axonal extension. In addition, Khazaei et al. (2014) showed that hippocampal neurons of CRMP4−/− mice exhibited a selective decrease in axon extension and reduced growth cone area, whereas overexpression of CRMP4 in these cells enhanced the formation and length of growth cone filopodia. All these studies support a promotive role for CRMP4 in axonal growth. In contrast, Alabed et al. (2007) identified CRMP4 as a convergent regulator of axonal outgrowth inhibition induced by myelin‐associated inhibitors and showed that GSK3‐beta regulates inhibition through CRMP4 (Alabed et al. 2010). In addition, Nagai et al. (2012) showed that CRMP4 mediates myelin‐associated glycoprotein (MAG)‐induced inhibition of axonal outgrowth. They also identified CRMP4 as a crucial factor that contributes to both axonal growth inhibition and scarring after spinal cord injury (Nagai et al. 2015). In contrast to these roles as a regulator of axonal growth, our DiI study showed a role for CRMP4 as a suppressor of the growth of MC apical dendrites. Because development of dendrites is influenced by axonal inputs (Callaway & Borrell, 2011), there is still the possibility that CRMP4 indirectly affects the growth of apical dendrites through the regulation of growth of axons that originate from other neurons and terminate on MC apical dendrites. However, the high expression levels of CRMP4 mRNA observed in MCs (Fig. 5B), as well as in our in vitro studies (Figs 6, 7, 8) support a direct suppressive effect of CRMP4 on dendrite elongation.
Niisato et al. (2012) reported a suppressive role for CRMP4 in the bifurcation of apical dendrites of CA1 pyramidal neurons, as well as the synergistic action of CRMP2 and CRMP4 in their branching points (Niisato et al. 2013). However, because the presence of long filopodia along MC apical dendrites in neonates makes the identification of the dendritic branch difficult as described by Matsutani & Yamamoto (2000), the present study focused on the length of dendrites.
Although the mechanisms underlying CRMP4 regulation of dendritic elongation remain to be determined, a relation between CRMP4 and components of dendritic cytoskeleton (e.g. microtubules and/or F‐actin, Georges et al. 2008) must be important. In fact, CRMP4 is known to bind directly to tubulin (Fukata et al. 2002) and tubulin‐β15 (Rembutsu et al. 2008), thereby promoting microtubule assembly (Khazaei et al. 2014). In addition, CRMP4 colocalizes with F‐actin (Rosslenbroich et al. 2005) and interacts with GAP‐43 (Kowara et al. 2007), regulating actin stabilization. Khazaei et al. (2014) revealed that CRMP4 promotes axonal growth through the regulation of both actin and microtubules, revealing a critical role in the regulation of cytoskeletal dynamics underlying growth cone development. Based on these and our results, CRMP4 promotes and suppresses axonal growth and dendritic elongation, respectively, by controlling cytoskeletal dynamics. The study of Cole et al. (2006) and our proteomics analysis suggest that posttranslational modification of CRMP4 regulates its function in neuronal plasticity in hyperalgesia (Fujisawa et al. 2008). Further studies, including modifications of CRMP4, will provide the molecular regulatory mechanism of CRMP4 in axonal and dendritic elongation.
Alterations in the layer thickness may reflect underlying pathological abnormalities produced by changes in cell death, survivability, migration, axonal projections, dendritic and spine arborization, and synaptic formation. The involvement of CRMP4 in neuronal apoptotic processes has been reported (Franken et al. 2003; Duplan et al. 2010). Calpain‐truncated CRMP‐3 and ‐4 act as proapoptotic effectors when cerebellar granule neurons undergo apoptosis (Liu et al. 2009). Nagai et al. (2015) recently showed that the number of apoptotic cells decreased in CRMP4‐KO mice after spinal cord injury, compared with WT (Nagai et al. 2015). In addition, we previously showed that a deficiency in CRMP4 affects the number of tyrosine hydroxylase‐immunoreactive (TH) neurons in the sexually dimorphic nucleus of the mouse preoptic area in mice (Iwakura et al. 2013). However, in the present study, few apoptotic cells were observed in the OB of either CRMP4‐KO or WT neonates. In addition, the number of cells in the MCL remained unchanged between these animals. Therefore, our results indicate that the thinner MCL observed in CRMP4‐KO mice was not a result of apoptosis. There is a possibility that the thinner MCL and increased MC density observed in CRMP4‐KO is just a secondary effect of EPL layer expansion. However, other possibilities such as impaired migration of MCs may be due to the alteration of MCL thickness in CRMP4‐KO mice. Some studies have reported the involvement of CRMPs in the neuronal migration during development. For example, Yamashita et al. (2006) demonstrated that CRMP1 mediates reelin signaling in cortical neuronal migration. Ip et al. (2011) showed that α2‐chimerin controls neuronal migration in the cerebral cortex via CRMP2. In addition, Morgan‐Smith et al. (2014) showed that GSK‐3 signaling, which is essential for radial migration of cortical neurons, is mediated by the phosphorylation of doublecortin and CRMP2. Investigation of the involvement of CRMP4 in neuronal migration is the focus of a planned future study aimed at tracing migrating neurons in CRMP4‐KO and WT mice.
We focused on the OB in the present study. However, our previous study showed changes in CRMP4 mRNA expression in the brain from the olfactory bulb to the cerebellum during development (Tsutiya & Ohtani‐Kaneko, 2012). Collectively, the present results and those of our previous study suggest that depending on the time and regional variation of CRMP4 gene expression, dendritic elongation could be differently regulated, and brain development could be influenced by CRMP4 gene expression. Moreover, numerous studies have suggested that CRMPs participate in the pathogenesis of neurodevelopmental disorders such as schizophrenia and ASD (Edgar et al. 2000; Johnston‐Wilson et al. 2000; Hong et al. 2005; Beasley et al. 2006; Bader et al. 2012; Braunschweig et al. 2013; Yamashita et al. 2013). Our recent study in CRMP4‐KO mice showed they exhibited altered olfactory ability, excitability, and expressions of GluR1 and GluR2 compared with WT pups, suggesting the physiological involvement of CRMP4 in neurodevelopmental disorders (Tsutiya et al. 2015). The present study revealed morphological alternations in CRMP4‐KO pups and shed new light on the role of CRMP4 during development.
Conclusions
Our morphological analysis of the OB in CRMP4‐KO mice revealed an alteration in the thickness of EPL and MCL during the neonatal period. In addition, we observed differences in length of MC apical dendrites between WT and CRMP4‐KO neonates. Furthermore, the length of MAP2‐positive dendrites of cultured MCs and HT22 cells was affected by CRMP4 gene expression. Therefore, this study revealed a new role for CRMP4 in the regulation of dendritic elongation.
Author contributions
A.T. and R.O‐K. designed the research, analyzed the data, and wrote the paper. Y.G. provided CRMP4‐KO mice and participated in the discussion of the work during the study. M.N. supported the study through animal care and work‐related discussions. A.T. mainly performed experiments. H.W. performed gene knockdown experiments. Y.N. supported the primary culture experiments.
Acknowledgements
We would like to thank Dr. Y. Yoshihara (Riken Brain Science Institute, Japan) and Dr. H. Okado (Tokyo Metropolitan Institute of Medical Science) for kindly providing the Tbx21 antibody and pEGFP‐N vector, respectively. This work was partly supported by JSPS KAKENHI grants (Nos 25430042 and 25‐7107 to R. Ohtani‐Kaneko and Atsuhiro Tsutiya, respectively) and the Grants‐in‐Aid for Scientific Research in a Priority Area from the Ministry of Education, Science, Sports and Culture, of Japan, to Yoshio Goshima (17082006). This study was also supported in part by a grant from the Program for the Strategic Research Foundation at Private Universities S1101017, organized by the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
References
- Alabed YZ, Pool M, Ong Tone S, et al. (2007) Identification of CRMP4 as a convergent regulator of axon outgrowth inhibition. J Neurosci 27, 1702–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabed YZ, Pool M, Ong Tone S, et al. (2010) GSK3 beta regulates myelin‐dependent axon outgrowth inhibition through CRMP4. J Neurosci 30, 5635–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura N, Menager C, Kawano Y, et al. (2005) Phosphorylation by Rho kinase regulates CRMP‐2 activity in growth cones. Mol Cell Biol 25, 9973–9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader V, Tomppo L, Trossbach SV, et al. (2012) Proteomic, genomic and translational approaches identify CRMP1 for a role in schizophrenia and its underlying traits. Hum Mol Genet 21, 4406–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista‐Brito R, Close J, Machold R, et al. (2008) The distinct temporal origins of olfactory bulb interneuron subtypes. J Neurosci 28, 3966–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley CL, Pennington K, Behan A, et al. (2006) Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: evidence for disease‐associated changes. Proteomics 6, 3414–3425. [DOI] [PubMed] [Google Scholar]
- Blanchart A, De Carlos JA, López‐Mascaraque L (2006) Time frame of mitral cell development in the mice olfactory bulb. J Comp Neurol 496, 529–543. [DOI] [PubMed] [Google Scholar]
- Braunschweig D, Krakowiak P, Duncanson P, et al. (2013) Autism‐specific maternal autoantibodies recognize critical proteins in developing brain. Transl Psychiatry 3, e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonviso N, Chaput MA, Scott JW (1991) Mitral cell‐to‐glomerulus connectivity: an HRP study of the orientation of mitral cell apical dendrites. J Comp Neurol 307, 57–64. [DOI] [PubMed] [Google Scholar]
- Callaway EM, Borrell V (2011) Developmental sculpting of dendritic morphology of layer 4 neurons in visual cortex: influence of retinal input. J Neurosci 31, 7456–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier E, Reibel S, Rogemond V, et al. (2003) Collapsin response mediator proteins (CRMPs): involvement in nervous system development and adult neurodegenerative disorders. Mol Neurobiol 28, 51–64. [DOI] [PubMed] [Google Scholar]
- Clepce M, Reich K, Gossler A, et al. (2013) Olfactory perception in schizophrenia: the rating range for hedonic judgements is increased during acute episodes. Psychiatry Res 208, 81–83. [DOI] [PubMed] [Google Scholar]
- Cole AR, Causeret F, Yadirgi G, et al. (2006) Distinct priming kinases contribute to differential regulation of collapsin response mediator proteins by glycogen synthase kinase‐3 in vivo. J Biol Chem 281, 16591–16598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EL, Thompson PM (2013) Typical and atypical brain development: a review of neuroimaging studies. Dialogues Clin Neurosci 15, 359–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplan L, Bernard N, Casseron W, et al. (2010) Collapsin response mediator protein 4a (CRMP4a) is upregulated in motoneurons of mutant SOD1 mice and can trigger motoneuron axonal degeneration and cell death. J Neurosci 30, 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Ginestet C, Feng Y, et al. (2013) Brain surface anatomy in adults with autism: the relationship between surface area, cortical thickness, and autistic symptoms. JAMA Psychiatry 70, 59–70. [DOI] [PubMed] [Google Scholar]
- Edgar PF, Douglas JE, Cooper GJ, et al. (2000) Comparative proteome analysis of the hippocampus implicates chromosome 6q in schizophrenia. Mol Psychiatry 5, 85–90. [DOI] [PubMed] [Google Scholar]
- Faedo A, Ficara F, Ghiani M, et al. (2002) Developmental expression of the T‐box transcription factor T‐bet/Tbx21 during mouse embryogenesis. Mech Dev 116, 157–160. [DOI] [PubMed] [Google Scholar]
- Franken S, Junghans U, Rosslenbroich V, et al. (2003) Collapsin response mediator proteins of neonatal rat brain interact with chondroitin sulfate. J Biol Chem 278, 3241–3250. [DOI] [PubMed] [Google Scholar]
- Fujisawa H, Ohtani‐Kaneko R, Naiki M, et al. (2008) Involvement of post‐translational modification of neuronal plasticity‐related proteins in hyperalgesia revealed by a proteomic analysis. Proteomics 8, 1706–1719. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, et al. (2002) CRMP‐2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol 4, 583–591. [DOI] [PubMed] [Google Scholar]
- Georges PC, Hadzimichalis NM, Sweet ES, et al. (2008) The yin‐yang of dendrite morphology: unity of actin and microtubules. Mol Neurobiol 35, 270–284. [DOI] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, et al. (1995) Collapsin‐induced growth cone collapse mediated by an intracellular protein related to UNC‐33. Nature 376, 509–514. [DOI] [PubMed] [Google Scholar]
- Hadjikhani N, Joseph RM, Snyder J, et al. (2005) Anatomical differences in the mirror neuron system and social cognition network in autism. Cereb Cortex 16, 1276–1282. [DOI] [PubMed] [Google Scholar]
- Hardan AY, Muddasani S, Vemulapalli M, et al. (2006) An MRI study of increased cortical thickness in autism. Am J Psychiatry 163, 1290–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong LE, Wonodi I, Avila MT, et al. (2005) Dihydropyrimidinase‐related protein 2 (DRP‐2) gene and association to deficit and nondeficit schizophrenia. Am J Med Genet B Neuropsychiatr Genet 136B, 8–11. [DOI] [PubMed] [Google Scholar]
- Hrdlicka M, Vodicka J, Havlovicova M, et al. (2011) Brief report: significant differences in perceived odor pleasantness found in children with ASD. J Autism Dev Disord 41, 524–527. [DOI] [PubMed] [Google Scholar]
- Imamura F, Nagao H, Naritsuka H, et al. (2006) A leucine‐rich repeat membrane protein, 5T4, is expressed by a subtype of granule cells with dendritic arbors in specific strata of the mouse olfactory bulb. J Comp Neurol 495, 754–768. [DOI] [PubMed] [Google Scholar]
- Imamura F, Ayoub AE, Rakic P, et al. (2011) Timing of neurogenesis is a determinant of olfactory circuitry. Nat Neurosci 14, 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JP, Shi L, Chen Y, et al. (2011) α2‐Chimaerin controls neuronal migration and functioning of the cerebral cortex through CRMP‐2. Nat Neurosci 15, 39–47. [DOI] [PubMed] [Google Scholar]
- Isaacson JS, Strowbridge BW (1998) Olfactory reciprocal synapses: dendritic signaling in the CNS. Neuron 20, 749–761. [DOI] [PubMed] [Google Scholar]
- Iwakura T, Sakoh M, Tsutiya A, et al. (2013) Collapsin response mediator protein 4 affects the number of tyrosine hydroxylase‐immunoreactive neurons in the sexually dimorphic nucleus in female mice. Dev Neurobiol 73, 502–517. [DOI] [PubMed] [Google Scholar]
- Johnston‐Wilson NL, Sims CD, Hofmann JP, et al. (2000) Disease‐specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. The Stanley Neuropathology Consortium. Mol Psychiatry 5, 142–149. [DOI] [PubMed] [Google Scholar]
- Khazaei MR, Girouard MP, Alchini R, et al. (2014) Collapsin response mediator protein 4 regulates growth cone dynamics through the actin and microtubule cytoskeleton. J Biol Chem 289, 30133–30143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi K (1987) Golgi studies on the development of granule cells of the rat olfactory bulb with reference to migration in the subependymal layer. J Comp Neurol 258, 112–124. [DOI] [PubMed] [Google Scholar]
- Kowara R, Me′nard M, Brown L, et al. (2007) Co‐localization and interaction of DPYSL3 and GAP43 in primary cortical neurons. Biochem Biophys Res Commun 363, 190–193. [DOI] [PubMed] [Google Scholar]
- Kubota M, Miyata J, Yoshida H, et al. (2011) Age‐related cortical thinning in schizophrenia. Schizophr Res 125, 21–29. [DOI] [PubMed] [Google Scholar]
- Kuperberg GR, Broome MR, McGuire PK, et al. (2003) Regionally localized thinning of the cerebral cortex in schizophrenia. Arch Gen Psychiatry 60, 878–888. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhou XW, Liu S, et al. (2009) Calpain‐truncated CRMP‐3 and ‐4 contribute to potassium deprivation‐induced apoptosis of cerebellar granule neurons. Proteomics 9, 3712–3728. [DOI] [PubMed] [Google Scholar]
- Mair RG, Gellman RL, Gesteland RC (1982) Postnatal proliferation and maturation of olfactory bulb neurons in the rat. Neuroscience 7, 3105–3116. [DOI] [PubMed] [Google Scholar]
- Malun D, Brunjes PC (1996) Development of olfactory glomeruli: temporal and spatial interactions between olfactory receptor axons and mitral cells in opossums and rats. J Comp Neurol 368, 1–16. [DOI] [PubMed] [Google Scholar]
- Matsutani S, Yamamoto N (2000) Differentiation of mitral cell dendrites in the developing main olfactory bulbs of normal and naris‐occluded rats. J Comp Neurol 418, 402–410. [PubMed] [Google Scholar]
- May T, Brewer WJ, Rinehart NJ, et al. (2011) Differential olfactory identification in children with autism and Asperger's disorder: a comparative and longitudinal study. J Autism Dev Disord 41, 837–847. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Naritsuka H, Mori K, et al. (2012) Tbr2 deficiency in mitral and tufted cells disrupts excitatory‐inhibitory balance of neural circuitry in the mouse olfactory bulb. J Neurosci 32, 8831–8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg PJ, Kamath V, Marchetto DM, et al. (2013) Meta‐analysis of olfactory function in schizophrenia, first‐degree family members, and youths at‐risk for psychosis. Schizophr Bull 40, 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan‐Smith M, Wu Y, Zhu X, et al. (2014) GSK‐3 signaling in developing cortical neurons is essential for radial migration and dendritic orientation. Elife 3, e02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaetova‐Ladinska EB, Arnold H, Jaros E, et al. (2004) Depletion of MAP2 expression and laminar cytoarchitectonic changes in dorsolateral prefrontal cortex in adult autistic individuals. Neuropathol Appl Neurobiol 30, 615–623. [DOI] [PubMed] [Google Scholar]
- Nagai J, Goshima Y, Ohshima T, et al. (2012) CRMP4 mediates MAG‐induced inhibition of axonal outgrowth and protection against Vincristine‐induced axonal degeneration. Neurosci Lett 519, 56–61. [DOI] [PubMed] [Google Scholar]
- Nagai J, Kitamura Y, Owada K, et al. (2015) Crmp4 deletion promotes recovery from spinal cord injury by neuroprotection and limited scar formation. Sci Rep 5, 8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama S, Homma R, Imamura F (2014) Neuronal organization of olfactory bulb circuits. Front Neural Circuits 8, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Kalb RG, Strittmatter GM (2000) Molecular basis of semaphorin‐mediated axon guidance. J Neurobiol 44, 219–229. [DOI] [PubMed] [Google Scholar]
- Niisato E, Nagai J, Yamashita N, et al. (2012) CRMP4 suppresses apical dendrite bifurcation of CA1 pyramidal neurons in the mouse hippocampus. Dev Neurobiol 72, 1447–1457. [DOI] [PubMed] [Google Scholar]
- Niisato E, Nagai J, Yamashita N, et al. (2013) Phosphorylation of CRMP2 is involved in proper bifurcation of the apical dendrite of hippocampal CA1 pyramidal neurons. Dev Neurobiol 73, 142–151. [DOI] [PubMed] [Google Scholar]
- Pilpel Y, Kolleker A, Berberich S, et al. (2009) Synaptic ionotropic glutamate receptors and plasticity are developmentally altered in the CA1 field of Fmr1 knockout mice. J Physiol 587, 787–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach TT, Massicotte G, Belin MF, et al. (2008) CRMP3 is required for hippocampal CA1 dendritic organization and plasticity. FASEB J 22, 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rembutsu M, Soutar MP, Van Aalten L, et al. (2008) Novel procedure to investigate the effect of phosphorylation on protein complex formation in vitro and in cells. Biochemistry 47, 2153–2161. [DOI] [PubMed] [Google Scholar]
- Rimol LM, Hartberg CB, Nesvåg R, et al. (2010) Cortical thickness and subcortical volumes in schizophrenia and bipolar disorder. Biol Psychiatry 68, 41–50. [DOI] [PubMed] [Google Scholar]
- Rosslenbroich V, Dai L, Baader SL, et al. (2005) Collapsin response mediator protein‐4 regulates F‐actin bundling. Exp Cell Res 310, 434–444. [DOI] [PubMed] [Google Scholar]
- Schmouth JF, Banks KG, Mathelier A, et al. (2012) Retina restored and brain abnormalities ameliorated by single‐copy knock‐in of human NR2E1 in null mice. Mol Cell Biol 32, 1296–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Cha C, Ye Y et al. (2015) CRMP4 and CRMP2 interact to coordinate cytoskeleton dynamics, regulating growth cone development and axon elongation. Neural Plast 2015, 947423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutiya A, Ohtani‐Kaneko R (2012) Postnatal alteration of collapsin response mediator protein 4 mRNA expression in the mouse brain. J Anat 221, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutiya A, Nishihara M, Goshima Y, et al. (2015) Mouse pups lacking collapsin response mediator protein 4 (CRMP4) manifest impaired olfactory function and hyperactivity in the olfactory bulb. Eur J Neurosci 42, 2335–2345. [DOI] [PubMed] [Google Scholar]
- Veyrac A, Giannetti N, Charrier E, et al. (2005) Expression of collapsin response mediator proteins 1, 2 and 5 is differentially regulated in newly generated and mature neurons of the adult olfactory system. Eur J Neurosci 21, 2635–2648. [DOI] [PubMed] [Google Scholar]
- Veyrac A, Reibel S, Sacquet J, et al. (2011) CRMP5 regulates generation and survival of newborn neurons in olfactory and hippocampal neurogenic areas of the adult mouse brain. PLoS ONE 6, e23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, Chaudhry R, Perera S, et al. (2013) Changes in cortical thickness in the frontal lobes in schizophrenia are a result of thinning of pyramidal cell layers. Eur Arch Psychiatry Clin Neurosci 263, 25–39. [DOI] [PubMed] [Google Scholar]
- Wilson CE, Palermo R, Burton AM, et al. (2011) Recognition of own‐ and other‐race faces in autism spectrum disorders. Q J Exp Psychol 64, 1939–1954. [DOI] [PubMed] [Google Scholar]
- Wu Y, Moriya‐Ito K, Iwakura T, et al. (2011) Sexually dimorphic effects of estrogen on spines in cultures of accessory olfactory bulb. Neurosci Lett 500, 77–81. [DOI] [PubMed] [Google Scholar]
- Yamashita N, Uchida Y, Ohshima T, et al. (2006) Collapsin response mediator protein 1 mediates reelin signaling in cortical neuronal migration. J Neurosci 26, 13357–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Morita A, Uchida Y, et al. (2007) Regulation of spine development by semaphorin3A through cyclin‐dependent kinase 5 phosphorylation of collapsin response mediator protein 1. J Neurosci 27, 12546–12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Mosinger B, Roy A, et al. (2011) CRMP5 (collapsin response mediator protein 5) regulates dendritic development and synaptic plasticity in the cerebellar Purkinje cells. J Neurosci 31, 1773–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Takahashi A, Takao K, et al. (2013) Mice lacking collapsin response mediator protein 1 manifest hyperactivity, impaired learning and memory, and impaired prepulse inhibition. Front Behav Neurosci 7, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi M, Mori K, Nakanishi S (1995) Refinement of odor molecule tuning by dendrodendritic synaptic inhibition in the olfactory bulb. Proc Natl Acad Sci U S A 92, 3371–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Abdullah JM (2013) The role of GluA1 in central nervous system disorders. Rev Neurosci 24, 499–505. [DOI] [PubMed] [Google Scholar]


